Abstract
Aryl hydrocarbon receptor (AHR) is a transcription factor whose activity is regulated by environmental agents, including several carcinogenic agonists. We measured recruitment of AHR and associated proteins to the human cytochrome P4501A1 gene promoter in vivo. Upon treatment with the agonist β-naphthoflavone, AHR is rapidly associated with the promoter and recruits the three members of the p160 family of coactivators as well as the p300 histone acetyltransferase, leading to recruitment of RNA polymerase II (Pol II) and induction of gene transcription. AHR, coactivators, and Pol II cycle on and off the promoter, with a period of ∼60 min. In contrast, the chemopreventative AHR ligand 3,3′-diindolylmethane promotes AHR nuclear translocation and p160 coactivator recruitment but, remarkably, fails to recruit Pol II or cause histone acetylation. This novel mechanism of receptor antagonism may account for the antitumor properties of chemopreventative compounds targeting the AHR.
The aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor belonging to the basic helix-loop-helix/Per-ARNT-Sim family of proteins (12). The AHR mediates the toxic effects of several chemical carcinogens, including polycyclic and halogenated aromatic hydrocarbons. These are but examples of the diverse ligands for the AHR, which include dietary compounds, natural and synthetic flavonoids, natural products, and pharmaceuticals (6). Prior to ligand binding, AHR exists in the cytoplasm in a complex with heat shock protein 90 (24), the cochaperone p23 (14), and the immunophilin homolog XAP2 (3). Following ligand binding, AHR moves to the nucleus, dissociates from the chaperone complex, and forms a heterodimer with the basic helix-loop-helix/Per-ARNT-Sim protein ARNT. This heterodimer binds to xenobiotic response elements (XREs) in the promoter and enhancer regions of target genes to regulate their transcription. Induction of cytochrome P4501A1 (CYP1A1) expression has been studied extensively as a model of AHR action (35).
Although structurally unrelated, AHR activity shares several features with members of the nuclear receptor superfamily. These transcription factors recruit a host of cofactor proteins to gene promoters in order to regulate transcription. Several nuclear receptor coactivators also interact with the AHR, including ERAP140 (22), RIP140 (20), p300, CBP (16), BRG-1 (34), and the three members of the p160 family of coactivators: NCoA1 (SRC-1), NCoA2 (GRIP-1 and TIF-2), and NCoA3 (AIB-1, p/CIP, and ACTR) (1). AHR interacts with these factors via its C-terminal transactivation domain (19), and ARNT may also be involved in recruiting cofactors to the promoter. The cofactors are involved in recruiting of additional proteins, ATP-dependent chromatin remodeling, and acetylation of promoter histones. The net effect of these activities is to relax chromatin, reposition nucleosomes, and facilitate recruitment of RNA polymerase II (Pol II).
AHR and its agonists have been implicated in the initiation and progression of cancers in multiple organs (25). However, data from cultured cells and animal models indicate that AHR ligands can inhibit formation and proliferation of breast tumors (10, 17). This antitumorigenic activity has led to proposals that AHR ligands could be used in treatment of breast cancer (11, 28). Of these ligands, 3,3′-diindolylmethane (DIM) has drawn significant interest, because DIM and related compounds are naturally occurring chemopreventative agents found in cruciferous vegetables (2). DIM inhibits proliferation of estrogen-responsive breast cancer cells via the AHR but is a weak partial agonist for CYP1A1 induction that antagonizes the effect of full AHR agonists (4, 5). Due to these properties, DIM has been labeled a selective AHR modulator (27), analogous to selective estrogen receptor modulators used in breast cancer treatment. The mechanism for selectivity of response to DIM is unknown.
We have used chromatin immunoprecipitation (ChIP) to monitor recruitment of AHR, coactivators, and Pol II to, and acetylation of histones on, the CYP1A1 promoter in vivo. The results show that an AHR agonist causes cycles of receptor and cofactor recruitment leading to gene transcription. In contrast, the selective AHR modulator DIM recruited a subset of cofactors but failed to cause histone acetylation or effective polymerase recruitment. These results suggest a mechanism for the chemopreventative activity of DIM and related compounds.
MATERIALS AND METHODS
Antibodies, cell culture, and treatment.
Antibodies used for ChIP include the following: for AHR, SA-210 (Biomol, Plymouth Meeting, Pa.); for NCoA1, SRC1Ab1 (Neomarkers, Fremont, Calif.) and S-19 (Santa Cruz Biotechnology, Santa Cruz, Calif.); for NCoA2, GRIP1Ab1 (Neomarkers) and C-20 (Santa Cruz); for NCoA3, rabbit polyclonal antibody (this laboratory) and N-17 (Santa Cruz); for p300, RW-128 (D. Livingston, Dana-Farber) and C-20 (Santa Cruz); for CBP, AC-26 (7); for Pol II, 8WG16 (Covance, Richmond, Calif.); and for acetyl-histone H4, 06-866 (Upstate, Lake Placid, N.Y.). MCF-7 breast carcinoma cells (American Type Culture Collection, Rockville, Md.) were cultured in Dulbecco's modified essential medium with 10% fetal bovine serum, l-glutamine, and 0.01 mg of insulin/ml. Two days prior to treatment, cells were split 1:3. Compounds were dissolved in fresh 37°C medium from 1,000× stocks in dimethyl sulfoxide (DMSO), and the medium was added to cell monolayers. DIM (98% pure) was from Biomol. All other chemicals, including β-naphthoflavone (BNF) and α-naphthoflavone (ANF), were from Sigma (St. Louis, Mo.).
ChIP.
MCF-7 cells in 150-mm-diameter dishes were treated with compounds for the times indicated in the figures, rinsed with 37°C phosphate-buffered saline (PBS), and cross-linked with 1% formaldehyde in PBS at 37°C for 10 min. Cells were then rinsed twice with ice-cold PBS, scraped into 1 ml of ice-cold PBS with protease inhibitors (Roche, Mannheim, Germany), and pelleted. Pellets were rinsed with 1 ml of ice-cold PBS with protease inhibitors, centrifuged again, and lysed in 300 μl of lysis buffer (1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 50 mM Tris [pH 8.1], protease inhibitors) for 10 min on ice. This solution was sonicated three times for 15 s each with a Dismembrator 300 (Fisher). The sonication had been shown to yield DNA fragments averaging ∼1 kb in length (31). The soluble chromatin was then centrifuged at 16,000 × g for 10 min at 4°C, and 100-μl aliquots were diluted to 1 ml in dilution buffer (1% Triton X-100, 2 mM EDTA, 20 mM Tris, 150 mM NaCl, protease inhibitors). An aliquot of soluble chromatin was also set aside as the input fraction. Diluted chromatin was precleared with protein-A/G agarose (45 μl of a 50% slurry; Santa Cruz), 2 μg of sheared salmon sperm DNA, and 5 μl of preimmune serum at 4°C with agitation for 2 h. Protein-A/G agarose beads were pelleted, the supernatant was transferred to a new tube, and 1 μg of antibody was added for immunoprecipitation overnight at 4°C with agitation. Protein-A/G agarose and 2 μl of salmon sperm DNA were added for 1 h, and the pellets were washed sequentially for 10 min at 4°C with buffer I (0.1% SDS, 2 mM EDTA, 20 mM Tris, 150 mM NaCl), buffer II (0.1% SDS, 2 mM EDTA, 20 mM Tris, 500 mM NaCl), buffer III (1% LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris), and Tris-EDTA buffer. The resulting pellets were resuspended in 100 μl of elution buffer (1% SDS, 0.1 M sodium bicarbonate, 0.2 M NaCl) and heated at 65°C for 6 h or overnight (input samples were included). DNA was purified using a PCR purification kit (Qiagen) and eluted in 50 μl. DNA content was quantified using the PicoGreen double-stranded-DNA kit (Molecular Probes, Eugene, Oreg.).
PCR and real-time PCR.
ChIP DNA (5 μl) was amplified by PCR with primers 5′CACCCGCCACCCTTCGACAGTTCT3′ and 5′CTCCCGGGGTGGCTAGTGCTTTGA3′ (amplifying the region from 784 to 1,156 bp upstream of the CYP1A1 transcription start site) by using the following cycles: 95°C for 3 min; 40 cycles of 95°C for 45 s, 58°C for 45 s, and 70°C for 1 min; and 70°C for 5 min. Nonspecific primers, amplifying a region from 3,152 to 3,528 bp 5′ of the start site, were used as a negative control (5′AGACGCTCCTCACTTTCCAGACTG3′ and 5′CGCCGCCACGCCTGACTG3′). For real-time PCR, SYBR Green master mix (Applied Biosystems, Bedford, Mass.) was used to amplify a smaller sequence (5′ACGCAGACCTAGACCCTTTGC3′ and 5′CGGGTGCGCGATTGAA3′) in an ABI 7700 sequence detection system. CYP1A1 promoter content was normalized to DNA content for each sample and then normalized to time zero (no ligand) or DMSO-treated cells. Between-replicate standard errors within real-time PCR experiments were always less than 5% of the mean.
Real-time RT-PCR.
Total RNA was isolated from MCF-7 cells in 100-mm-diameter dishes by using an RNeasy kit (Qiagen), with on-column DNase treatment to remove contaminating genomic DNA. Real-time reverse transcription PCR (RT-PCR) was performed on 500 ng of RNA by using SYBR Green master mix and MultiScribe reverse transcriptase (Applied Biosystems) according to the manufacturer's protocol. For CYP1A1 mRNA, the primers used were 5′CGGCCCCGGCTCTCT3′ and 5′GTGTCGGAAGGTCTCCCAGGAT3′, and for hnRNA, the primers were 5′TTGTGATCCCAGGCTCCAAGA3′ and 5′GGAGGCACCAAAATGTTCCTTT3′. Expression was normalized to the time zero sample. Amplification of specific targets was verified by gel electrophoresis and dissociation curves. Controls without reverse transcriptase confirmed the absence of DNA.
RESULTS
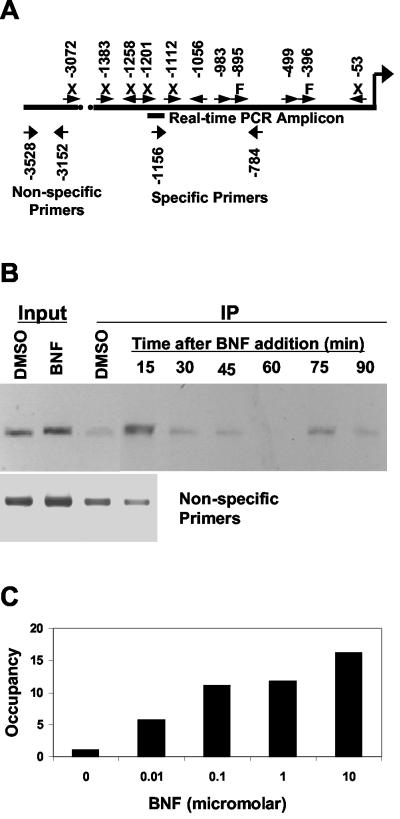
The human CYP1A1 gene is regulated by AHR through XREs in its promoter (18). We designed primers that flanked active response elements (Fig. 1A). ChIP was used to measure occupancy of this region by the AHR in MCF-7 cells following agonist (BNF) treatment. Occupancy of the promoter by AHR increased sharply after 15 min of BNF treatment, declined through 60 min, and then peaked again at 75 min before declining at 90 min (Fig. 1B). This cycling, with a period of ∼1 h, is similar to results for the estrogen receptor in the same cells (31). The timing of the first peak in occupancy (for both AHR and cofactors) (see below) varies slightly between experiments and depends on the method of treatment (data not shown). BNF selectively increased occupancy of the region of the promoter containing active XREs compared to that of a region further upstream and to treatment with a solvent (Fig. 1B).
FIG. 1.
ChIP of AHR at the CYP1A1 promoter. (A) Locations of XREs, PCR primers, and the real-time PCR amplicon on the CYP1A1 promoter, relative to the transcription start site. Data on activity of individual XREs (orientation shown by arrows) are from reference 18. X, not occupied in vivo (as determined by DNase footprinting) and not active in reporter assays; F, occupied but not active; no label, occupied and active. (B) MCF-7 cells were treated with BNF (1 μM) or DMSO (0.1%). ChIP was performed as described in Materials and Methods. DNA from input or immunoprecipitation (IP) fractions of DMSO- or BNF-treated cells was amplified using primers for regions with (specific) (top panel) or without (nonspecific) XREs. (C) Cells were treated for 75 min with the indicated concentrations of BNF. Following ChIP, recovery of CYP1A1 promoter was measured by real-time PCR and normalized to a solvent control.
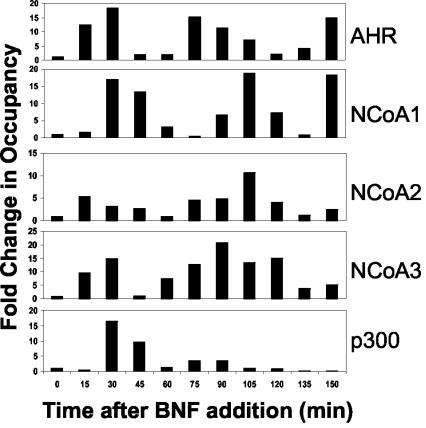
In order to quantify the changes in promoter occupancy by the AHR and other factors, quantitative real-time PCR was used to measure recovery of the CYP1A1 promoter by ChIP. Using this method, we found that promoter occupancy by AHR depended on the BNF dose (Fig. 1C). The cycles in promoter occupancy could also be measured quantitatively, showing that binding of XREs by AHR increased almost 20-fold following BNF treatment, declined to near-baseline levels, and then increased again in regular cycles (Fig. 2, top panel).
FIG. 2.
Time course of promoter occupancy by AHR and associated cofactors. ChIP was performed as described in the legend to Fig. 1 with antibodies against the indicated proteins, promoter content was assessed by real-time PCR, and the results were normalized to time zero (no ligand). Results shown are representative of those from at least three independent experiments with two different antibodies for each of the cofactors.
Next, the timing of recruitment of cofactors to the promoter was monitored. Each of the three members of the p160 family of coactivators is recruited to the CYP1A1 promoter in a cyclical fashion, with periods similar to that of the AHR (Fig. 2). NCoA2 and NCoA3 appear to follow the pattern of AHR recruitment, while NCoA1 lags by 15 min. The acetyltransferase p300 associates with the CYP1A1 promoter in a transient fashion, and very little is recruited following the first cycle of occupancy.
Coincident with recruitment of the AHR and cofactors to the CYP1A1 promoter, Pol II is recruited (Fig. 3). Like that of AHR and the cofactors, association of Pol II with the promoter is cyclic. Transcription of CYP1A1 mRNA, measured by RT-PCR of unprocessed hnRNA (9), follows Pol II recruitment and peaks when Pol II is absent from the promoter. Accumulation of processed mRNA over time reflects the rate of transcription, as shown by the comparison between the calculated integration of hnRNA and the measured level of mRNA (Fig. 3, bottom panel).
FIG. 3.
Time course of RNA polymerase recruitment and transcription at the CYP1A1 promoter. Treatment and ChIP were as described in the legend to Fig. 2. Total RNA was isolated, DNase treated, and amplified with primers recognizing hnRNA (one primer in an exon and one in an adjacent intron) or processed mRNA (primers in adjacent exons). Expression was normalized to time zero. The line in the bottom panel represents the level of integrated hnRNA (rescaled for comparison) for each time point, indicating that total mRNA corresponds to the accumulated new transcripts. Results shown are representative of those from at least three independent experiments.
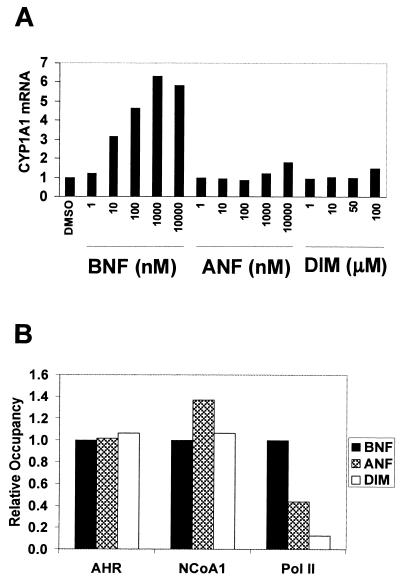
Several antagonists of the AHR block translocation to the nucleus in vivo and XRE binding in vitro (13). In contrast, some AHR partial agonists have been shown by mobility shift assays to induce binding of the receptor to XREs in vitro. We wished to determine whether the ligands ANF and DIM cause recruitment of AHR to XREs in vivo. In contrast to BNF, ANF caused little, and DIM no, induction of CYP1A1 expression after 2.5 h of treatment (Fig. 4A). Binding to XREs in vivo and subsequent recruitment of cofactors were measured by ChIP (Fig. 4B). Both ANF and DIM were as efficient as BNF at recruiting AHR and NCoA1 to the CYP1A1 promoter. However, the antagonists differed from BNF in their ability to recruit Pol II: AHR occupied by ANF recruited only 40% as much Pol II, and DIM recruited only 15%. The timing of recruitment of these factors was similar for ANF and BNF (data not shown); recruitment by DIM lagged slightly (see below).
FIG. 4.
CYP1A1 induction and factor recruitment following treatment with BNF, ANF, or DIM. (A) Cells were treated with the indicated compounds for 2.5 h, and CYP1A1 mRNA was measured as described in the legend to Fig. 3. (B) Cells were treated with BNF, ANF (10 μM), or DIM (100 μM) for 30 min (BNF and ANF) or 60 min (DIM). Maximal promoter occupancy occurred at these times. Recruitment of each factor was assessed by ChIP and normalized to BNF for comparison. Results shown are representative of those from at least three independent experiments.
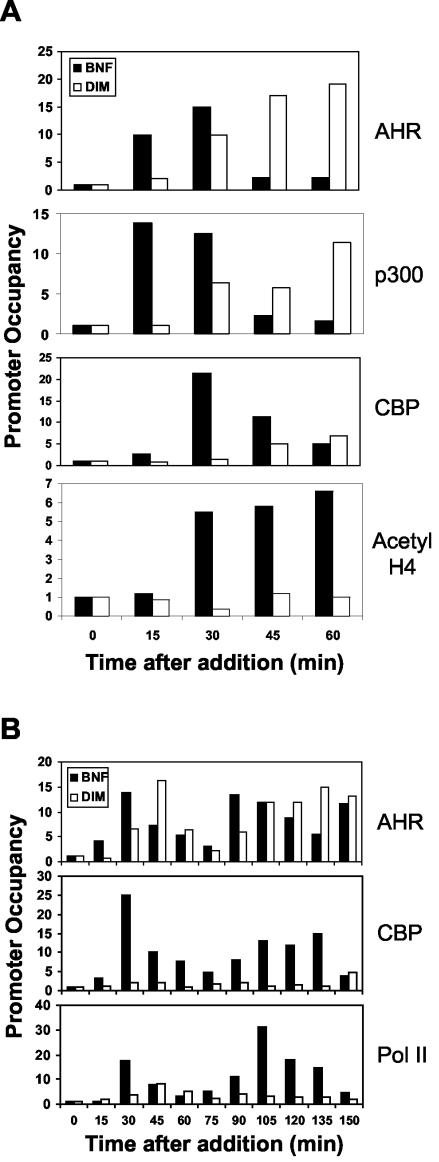
In order to determine the mechanism of differential Pol II recruitment by antagonists, we chose to use DIM to examine other intermediary steps in the process of transcription initiation. Again, AHR was recruited to similar extents by DIM and BNF, although it was recruited 15 to 30 min later by DIM (Fig. 5A). Measurement of histone acetyltransferase (HAT) recruitment showed that while BNF and DIM treatments recruited p300 to similar extents, DIM was much less effective at recruiting CBP. Significantly, promoter H4 histones were not acetylated in response to DIM treatment. At later times after treatment (Fig. 5B), AHR was recruited to similar extents and in a cyclic fashion in response to both BNF and DIM, but CBP and Pol II recruitment was severely impaired in cells treated with DIM.
FIG.5.
Factor recruitment and histone acetylation in response to BNF or DIM. ChIP was performed with antibodies against the indicated proteins as described in the legend to Fig. 2. (A) Recruitment of receptor and HATs and acetylation of histone H4 over the course of 60 min. (B) Confirmation of similar levels of AHR recruitment, and a relative lack of CBP and Pol II recruitment by DIM, over 2.5 h. Results shown are representative of those from at least three independent experiments.
DISCUSSION
This study demonstrates the recruitment of AHR and associated cofactors to the CYP1A1 promoter in vivo. Ordered and cyclical association of AHR and coactivators led to histone acetylation, Pol II recruitment, and gene transcription. Furthermore, unlike results with other AHR antagonists or the active antagonism seen with steroid hormone receptors, DIM caused association of the AHR with the promoter and recruitment of coactivators. DIM antagonism of CYP1A1 induction appears to result from poor recruitment of critical HATs. This novel mechanism of antagonism may form the basis for the chemopreventative effects of DIM and related compounds.
AHR and cofactor recruitment seen here agrees with findings of previous reports. Rapid association of AHR with the XRE coincides with results of a study showing that green fluorescent protein-tagged AHR is mostly nuclear 15 min after treatment with an agonist and is forming nuclear foci by 30 min (8). Each of the three p160 coactivators interacts with the mouse CYP1A1 promoter in vivo (1), although ChIP was performed at a single time point after treatment, so it is not clear whether mouse AHR and cofactors also show cycles of association. Concomitant recruitment of p160 coactivators and p300 has been seen with the estrogen, androgen, and thyroid hormone receptors (31-33). Transient, single-cycle association of p300 with the promoter is also a common factor among these receptors.
The results presented in Fig. 2 appear to show that NCoA1, NCoA2, and NCoA3 are each present on the promoter simultaneously. However, since ChIP examines a population of cells, with only ∼20% of the total promoters recovered following immunoprecipitation (Hestermann and Brown, unpublished results), these results do not demonstrate whether multiple members of the p160 family are recruited to the same promoter. Furthermore, with many XREs present, it is possible that individual AHR (or ARNT) proteins recruit separate p160s to different response elements on the same promoter. With DNA sheared to an average of 1 kb, ChIP lacks sufficient resolution to determine occupancy of individual, closely spaced response elements.
Ligand-induced transcription factor antagonists act through a variety of mechanisms. Ligands for both the estrogen (30, 31) and androgen (32) receptors cause recruitment of corepressors and histone deacetylases, rather than that of coactivators and HATs, to promoters, leading to active repression of transcription. The glucocorticoid receptor suppresses NF-κB-mediated responses by inhibiting phosphorylation of the C-terminal domain, rather than recruitment, of Pol II (23). Several flavone antagonists of the AHR block translocation to the nucleus in vivo and XRE binding in vitro (13). In contrast, DIM was able to recruit AHR and some cofactors to the promoter but failed to bring about histone acetylation or Pol II recruitment. Taken together, the results indicate that these transcription factors are susceptible to interference at multiple steps in the transcriptional regulation pathway, presenting several opportunities for therapeutic intervention.
Our work suggests a novel mechanism of AHR antagonism. Dramatic differences between the transcriptional outcomes of exposure to AHR agonists and exposure to antagonists occur at a step after ligand binding, nuclear translocation, and p160 coactivator recruitment but before recruitment of HATs, histone acetylation, and recruitment of Pol II. Since local histone acetylation may be required for Pol II recruitment, it is most likely that failure to recruit critical HATs is the key difference between agonistic and antagonistic effects. Our data suggest that ligand-dependent changes in AHR conformation, either alone or with p160 coactivators, affect the stability of HAT, specifically CBP, binding. Failure to acetylate histone H4 reflects the role of CBP in specifically acetylating lysines 8 and 12 of H4 (21). A lack of histone H4 acetylation has also been correlated with inhibition of CYP1A1 induction by NF-κB activation (15).
Ligands like DIM that allow DNA binding and coactivator recruitment but inhibit CYP1A1 transcription likely interfere with carcinogenesis through parallel pathways. First, they inhibit tumor initiation by blocking CYP1A1 induction (and enzymatic activity [5]) and thus preventing metabolism by CYP1A1 of procarcinogens into genotoxic forms. Second, proliferation of estrogen-dependent tumors is also inhibited because AHR interferes with estrogen receptor-mediated transcription and cell proliferation through competition for promoter binding sites (29) and/or common cofactors (26). Mechanism-based design of compounds that specifically block HAT recruitment by AHR in a gene- and/or target cell-specific manner may allow new cancer chemopreventative agents targeting AHR, analogous to the selective estrogen receptor modulators that have proven to be of great clinical value in the treatment of breast cancer.
Acknowledgments
This work was supported by grants from the National Cancer Institute, the Department of Defense Breast Cancer Research Program, and the Claudia Adams Barr Program in Cancer Research (M.B.) and by a National Research Service Award from the National Institutes of Health (E.V.H.).
We thank David Livingston for providing antibodies and Mitch Lazar and Mark Hahn for suggestions on the manuscript.
REFERENCES
- 1.Beischlag, T. V., S. Wang, D. W. Rose, J. Torchia, S. Reisz-Porszasz, K. Muhammad, W. E. Nelson, M. R. Probst, M. G. Rosenfeld, and O. Hankinson. 2002. Recruitment of the NCoA/SRC-1/p160 family of transcriptional coactivators by the aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator complex. Mol. Cell. Biol. 22:4319-4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Broadbent, T. A., and H. S. Broadbent. 1998. The chemistry and pharmacology of indole-3-carbinol (indole-3-methanol) and 3-(methoxymethyl)indole. [Part II.] Curr. Med. Chem. 5:469-491. [PubMed] [Google Scholar]
- 3.Carver, L. A., and C. A. Bradfield. 1997. Ligand-dependent interaction of the aryl hydrocarbon receptor with a novel immunophilin homolog in vivo. J. Biol. Chem. 272:11452-11456. [DOI] [PubMed] [Google Scholar]
- 4.Chen, I., A. McDougal, F. Wang, and S. Safe. 1998. Aryl hydrocarbon receptor-mediated antiestrogenic and antitumorigenic activity of diindolylmethane. Carcinogenesis 19:1631-1639. [DOI] [PubMed] [Google Scholar]
- 5.Chen, I., S. Safe, and L. Bjeldanes. 1996. Indole-3-carbinol and diindolylmethane as aryl hydrocarbon (Ah) receptor agonists and antagonists in T47D human breast cancer cells. Biochem. Pharmacol. 51:1069-1076. [DOI] [PubMed] [Google Scholar]
- 6.Denison, M. S., and S. R. Nagy. 2003. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu. Rev. Pharmacol. Toxicol. 43:309-334. [DOI] [PubMed] [Google Scholar]
- 7.Eckner, R., J. W. Ludlow, N. L. Lill, E. Oldread, Z. Arany, N. Modjtahedi, J. A. DeCaprio, D. M. Livingston, and J. A. Morgan. 1996. Association of p300 and CBP with simian virus 40 large T antigen. Mol. Cell. Biol. 16:3454-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbi, C., T. Misteli, and G. L. Hager. 2002. Recruitment of dioxin receptor to active transcription sites. Mol. Biol. Cell 13:2001-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elferink, C. J., and J. J. Reiners. 1996. Quantitative RT-PCR on CYP1A1 heterogeneous nuclear RNA: a surrogate for the in vitro transcription run-on assay. BioTechniques 20:470-477. [DOI] [PubMed] [Google Scholar]
- 10.Gierthy, J. F., J. A. Bennett, L. M. Bradley, and D. S. Cutler. 1993. Correlation of in vitro and in vivo growth suppression of MCF-7 human breast cancer by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Cancer Res. 53:3149-3153. [PubMed] [Google Scholar]
- 11.Greenlee, W. E., L. J. Hushka, and D. R. Hushka. 2001. Molecular basis of dioxin actions: evidence supporting chemoprotection. Toxicol. Pathol. 29:6-7. [DOI] [PubMed] [Google Scholar]
- 12.Hankinson, O. 1995. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 35:307-340. [DOI] [PubMed] [Google Scholar]
- 13.Henry, E. C., A. S. Kende, G. Rucci, M. J. Totleben, J. J. Willey, S. D. Dertinger, R. S. Pollenz, J. P. Jones, and T. A. Gasiewicz. 1999. Flavone antagonists bind competitively with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) to the aryl hydrocarbon receptor but inhibit nuclear uptake and transformation. Mol. Pharmacol. 55:716-725. [PubMed] [Google Scholar]
- 14.Kazlauskas, A., S. Sundstrom, L. Poellinger, and I. Pongratz. 2001. The hsp90 chaperone complex regulates intracellular localization of the dioxin receptor. Mol. Cell. Biol. 21:2594-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ke, S., A. B. Rabson, J. F. Germino, M. A. Gallo, and Y. Tian. 2001. Mechanism of suppression of cytochrome P-450 1A1 expression by tumor necrosis factor-alpha and lipopolysaccharide. J. Biol. Chem. 276:39638-39644. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi, A., K. Numayama-Tsuruta, K. Sogawa, and Y. Fujii-Kuriyama. 1997. CBP/p300 functions as a possible transcriptional coactivator of Ah receptor nuclear translocator (Arnt). J. Biochem. (Tokyo) 122:703-710. [DOI] [PubMed] [Google Scholar]
- 17.Kociba, R. J., D. G. Keyes, J. E. Beyer, R. M. Carreon, C. E. Wade, D. A. Dittenber, R. P. Kalnins, L. E. Frauson, C. N. Park, S. D. Barnard, R. A. Hummel, and C. G. Humiston. 1978. Results of a two-year chronic toxicity and oncogenicity study of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in rats. Toxicol. Appl. Pharmacol. 46:279-303. [DOI] [PubMed] [Google Scholar]
- 18.Kress, S., J. Reichert, and M. Schwarz. 1998. Functional analysis of the human cytochrome P4501A1 (CYP1A1) gene enhancer. Eur. J. Biochem. 258:803-812. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, M. B., and G. H. Perdew. 1999. Nuclear receptor coactivator SRC-1 interacts with the Q-rich subdomain of the AhR and modulates its transactivation potential. Gene Expr. 8:273-286. [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar, M. B., R. W. Tarpey, and G. H. Perdew. 1999. Differential recruitment of coactivator RIP140 by Ah and estrogen receptors. Absence of a role for LXXLL motifs. J. Biol. Chem. 274:22155-22164. [DOI] [PubMed] [Google Scholar]
- 21.Ludlam, W. H., M. H. Taylor, K. G. Tanner, J. M. Deny, R. H. Goodman, and S. M. Smolik. 2002. The acetyltransferase activity of CBP is required for wingless activation and H4 acetylation in Drosophila melanogaster. Mol. Cell. Biol. 22:3832-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen, T. A., D. Hoivik, J. E. Lee, and S. Safe. 1999. Interactions of nuclear receptor coactivator/corepressor proteins with the aryl hydrocarbon receptor complex. Arch. Biochem. Biophys. 367:250-257. [DOI] [PubMed] [Google Scholar]
- 23.Nissen, R. M., and K. R. Yamamoto. 2000. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 14:2314-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perdew, G. H. 1988. Association of the Ah receptor with the 90-kDa heat shock protein. J. Biol. Chem. 263:13802-13805. [PubMed] [Google Scholar]
- 25.Poland, A., and J. C. Knutson. 1982. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annu. Rev. Pharmacol. Toxicol. 22:517-554. [DOI] [PubMed] [Google Scholar]
- 26.Reen, R. K., A. Cadwallader, and G. H. Perdew. 2002. The subdomains of the transactivation domain of the aryl hydrocarbon receptor (AhR) inhibit AhR and estrogen receptor transcriptional activity. Arch. Biochem. Biophys. 408:93-102. [DOI] [PubMed] [Google Scholar]
- 27.Safe, S., and A. McDougal. 2002. Mechanism of action and development of selective aryl hydrocarbon receptor modulators for treatment of hormone-dependent cancers. Int. J. Oncol. 20:1123-1128. [PubMed] [Google Scholar]
- 28.Safe, S., F. Wang, W. Porter, R. Duan, and A. McDougal. 1998. Ah receptor agonists as endocrine disruptors: antiestrogenic activity and mechanisms. Toxicol. Lett. 102-103:343-347. [DOI] [PubMed] [Google Scholar]
- 29.Safe, S., M. Wormke, and I. Samudio. 2000. Mechanisms of inhibitory aryl hydrocarbon receptor-estrogen receptor crosstalk in human breast cancer cells. J. Mammary Gland Biol. Neoplasia 5:295-306. [DOI] [PubMed] [Google Scholar]
- 30.Shang, Y., and M. Brown. 2002. Molecular determinants for the tissue specificity of SERMs. Science 295:2465-2468. [DOI] [PubMed] [Google Scholar]
- 31.Shang, Y., X. Hu, J. DiRenzo, M. A. Lazar, and M. Brown. 2000. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843-852. [DOI] [PubMed] [Google Scholar]
- 32.Shang, Y., M. Myers, and M. Brown. 2002. Formation of the androgen receptor transcription complex. Mol. Cell 9:601-610. [DOI] [PubMed] [Google Scholar]
- 33.Sharma, D., and J. D. Fondell. 2002. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. USA 99:7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang, S., and O. Hankinson. 2002. Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J. Biol. Chem. 277:11821-11827. [DOI] [PubMed] [Google Scholar]
- 35.Whitlock, J. P. 1999. Induction of cytochrome P4501A1. Annu. Rev. Pharmacol. Toxicol. 39:103-125. [DOI] [PubMed] [Google Scholar]