Abstract
The fundamental subunit of chromatin, the nucleosome, is not a static entity but can move along DNA via either thermal or enzyme-driven movements. Here we have monitored the movements of nucleosomes following deposition at well-defined locations on mouse mammary tumor virus promoter DNA. We found that the sites to which nucleosomes are deposited during chromatin assembly differ from those favored during thermal equilibration. Taking advantage of this, we were able to track the movement of nucleosomes over 156 bp and found that this proceeds via intermediate positions spaced between 46 and 62 bp. The remodeling enzyme ISWI was found to direct the movement of nucleosomes to sites related to those observed during thermal mobilization. In contrast, nucleosome mobilization driven by the SWI/SNF and RSC complexes were found to drive nucleosomes towards sites up to 51 bp beyond DNA ends, with little respect for the sites favored during thermal repositioning. The dynamic properties of nucleosomes we describe are likely to influence their role in gene regulation.
The bulk of eukaryotic genomes do not exist as free DNA, but are associated with histones to form nucleosomes. The structure of nucleosomes has been studied intensively since they were first identified in the 1970s (39). Now several high-resolution crystal structures provide detailed information describing how the four histone proteins are folded to form a histone octamer (11, 28, 46, 62, 66). DNA is bound to the surface of the histone octamer by over 140 atomic interactions. The most significant of these are hydrogen bonds between the phosphate backbone of the DNA and main chain atoms arranged in a helical path around the octamer surface. This large number of contacts contributes to the high resistance of nucleosomes to thermal and salt-induced dissociation. Given this stability, it is not surprising that the presence of nucleosomes on gene control elements can impede the function of regulatory proteins (59, 69). Indeed, alterations to chromatin structure are being found to play important roles in the regulation of many genes. Despite their resistance to dissociation, nucleosomes are not static entities but can move along DNA. As the nucleosomes of higher eukaryotes are separated by an average of 50 bp of DNA, the ability of nucleosomes to move along DNA provides a potent means by which access to regulatory elements might be altered without requiring the complete dissociation of the histone octamer.
Among the first evidence that nucleosomes can move along DNA was the observation that they can migrate off simian virus 40 minichromosomes onto DNA fragments in a slow reaction that is dependent on the physical association of the acceptor DNA (6). Observations consistent with salt-dependent nucleosome sliding were also made by Weischet et al. (65). It was not until the 1990s that this phenomenon was characterized more thoroughly by Bradbury and coworkers, who exploited the fact that nucleosomes located at different positions had altered mobilities during native polyacrylamide gel electrophoresis (48, 49). These studies revealed that it is a general property of nucleosomes assembled onto short DNA fragments that they can be shifted to different positions following thermal incubation. An alternative high-resolution approach to monitoring the location of nucleosomes along DNA was developed in the Richmond laboratory (18). Together with the studies of the Bradbury lab, this provided evidence that nucleosomes assembled onto short DNA fragments exhibited a tendency to migrate towards DNA ends during thermal repositioning (20, 49). As would be expected, these dynamic properties of nucleosomes have also been found to influence the ability of the transcriptional apparatus to gain access to chromatin (64).
Although nucleosomes assembled onto many DNA sequences exhibit some thermally driven mobility under physiologically relevant conditions, it has also been found that many ATP-dependent chromatin remodeling activities can accelerate this process. Studies of the Drosophila melanogaster ISWI-containing complexes NURF (27) and CHRAC (40) and the yeast SWI/SNF complex (33, 67) provided some of the first evidence that these complexes can alter the positions of nucleosomes along DNA. Subsequently, it has been found that many other enzymes, including RSC (45), Mi-2 (25), Xenopus laevis ISWI (24), and human Brg1 (3), can alter the positions of nucleosomes along DNA. Although the function of ATP-dependent remodeling enzymes is not fully understood, it appears that they play important roles in the regulation of gene expression (17, 30, 31, 42, 61), DNA replication (8), chromatin assembly (32), and recombination and DNA repair (1, 2, 34).
Despite the fundamental importance that nucleosome mobility is likely to play in many of these processes, little is understood about the mechanisms by which nucleosomes move along DNA. Here we have monitored the movement of nucleosomes away from the sites at which they are deposited during chromatin assembly. We find that nucleosomes assembled onto sequences from the mouse mammary tumor virus (MMTV) long terminal repeat (LTR) exhibit an unexpected preference for movements in the range of 40 to 62 bp. This preference for larger movements is expected to influence the contribution of nucleosomes to regulatory pathways. We use this knowledge of the behavior of nucleosomes during thermal movement to gain new insights into the function of ATP-dependent remodeling activities. These reveal that whereas ISWI relocates nucleosomes to positions similar to those observed during thermal redistribution, the SWI/SNF and RSC complexes are capable of moving the nucleosome dyad to a site 22 bp from the DNA ends. This is expected to result in the loss of up to 51 bp of DNA from one side of the nucleosome, providing a possible explanation for why SWI/SNF-related complexes disrupt nucleosome integrity to a greater extent than ISWI-related proteins.
MATERIALS AND METHODS
Nucleosome assembly.
All nucleosomes are described by the length of flanking DNA extensions upstream and downstream of the 147-bp nucleosome core particle encompassing the specified dyad position. For the initial assembled nucleosome positions, the nucleosome core position is denoted A and B for NucA and NucB located with the dyad at +70 and −127, respectively, relative to the transcription start site +1 (12).
DNA for nucleosome assembly was generated by PCR using a template sequence derived directly from a native MMTV isolate (20) and appropriate PCR primers for the specified fragments. 5′ Cy5 dye labels (MWG; ThermoHybaid) were incorporated during oligonucleotide synthesis where necessary. Products of standard PCRs (Bioline) were precipitated and then purified by ion-exchange chromatography on a 1-ml Source HQ column (Pharmacia) developed in a gradient from 0 to 800 mM NaCl in 10 mM TrisCl (pH 7.5)-0.1 mM EDTA. Radioactively labeled DNA was prepared by 5′ end labeling using polynucleotide kinase (NEB) and [γ-32P]ATP (ICN), with the nonlabeled end protected from reaction by a 5′ Cy5 label. Nucleosomes were assembled as described previously (20, 47).
Nucleosome redistribution reactions.
All reactions contained 2 pmol of nucleosomes in a final volume of 10 μl. For thermal shifting, final reaction conditions were 50 mM TrisCl (pH 7.5)-150 mM NaCl, except for the experiment represented in Fig. 1A and B, which used 50 mM TrisCl (pH 7.5). ATP-dependent remodeling was carried out under final reaction conditions of 50 mM TrisCl (pH 7.5)-50 mM KCl-1 mM MgCl2-1 mMmg-ATP for the times specified and was stopped by chilling on ice and the addition of 125 ng of HindIII-digested λ DNA (Promega). Native gels and site-directed mapping were performed as described previously (18, 19). Results plotted in Fig. 1C were obtained with nucleosomes assembled using wild-type X. laevis recombinant histone octamers and visualized by fluorescent gel imaging (Fuji). Experiments shown in all other figures were carried out with the same histones, except for H4 with the point mutation S47C, onto which the cysteaminyl EDTA mapping reagent was attached (18). Native gel electrophoresis controls revealed no differences in the positions to which wild-type and reagent coupled octamer were moved during thermal redistribution (data not shown).
FIG. 1.
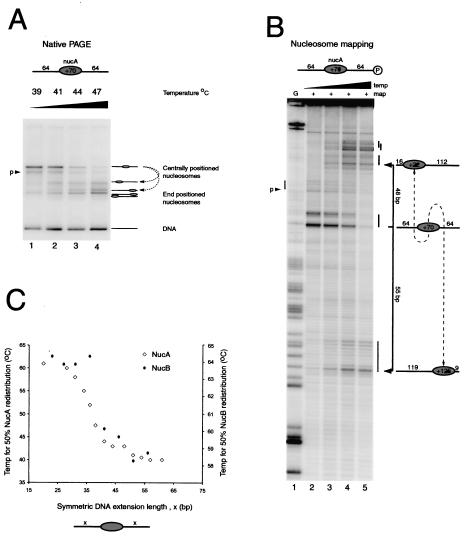
Increased linker DNA length reduces the temperature required for thermally driven nucleosome redistribution. (A) Native gel electrophoresis (PAGE) of nucleosomes assembled onto the 64A64 DNA fragment following incubation at 39, 41, 44, and 47°C (lanes 1 to 4, respectively). Most of the nucleosomes start as a discrete slowly migrating species, with a small proportion as a second species labeled “p.” Following thermal incubation, faster migrating species are generated. (B) Site-directed mapping of the same samples as for panel A (lanes 2 to 5, respectively). Nucleosomes generate a characteristic pattern of two cleavage sites separated by 7 bp and allow determination of nucleosome positions at base pair resolution using the G ladder in lane 1, as diagrammed to the right (nucleosomes are not to scale). A small amount of mapping is also observed at a second site labeled “p.” After thermal incubation, mapping products accumulate within 16 bp of either end of the DNA fragment. (C) Plot of the temperature required to shift 50% of nucleosomes from their initial central location on DNA fragments to the symmetric extensions of flanking DNA. Temperatures required for shifting on the MMTV NucA (open diamonds; left axis) and MMTV NucB (filled diamonds; right axis) positioning sequences are shown.
Purification of remodeling activities.
ISWI was purified as described by Corona et al. (9), SWI/SNF was purified as described by Neely et al. (51), and RSC was purified as described by Saha et al. (56). The concentration of ISWI was determined based upon its predicted extinction coefficient (A276 for 1 M ISWI = 119,425). Concentrations of the SWI/SNF complex and RSC were determined by comparing the intensities with which subunits were stained to those of standards.
RESULTS
Increased linker DNA length reduces the temperature required to shift nucleosomes assembled at MMTV NucA and NucB.
In order to investigate how nucleosomes move along DNA, we have taken advantage of the ability of MMTV LTR DNA to direct the assembly of nucleosomes to specific positions on DNA (20). This makes it possible to design DNA fragments on which nucleosomes will be assembled predominantly at a central location. Initially, nucleosomes were assembled onto a 275-bp DNA fragment in which the MMTV NucA positioning sequence is flanked on both sides by 64 bp of additional DNA. We have adopted a nomenclature by which we refer to this nucleosome as 64A64. Native gel electrophoresis of 64A64 nucleosomes reveals that they form one predominant species (Fig. 1A, lane 1).
In order to confirm that the nucleosomes adopt a central location on the DNA fragment, we used site-directed nucleosome mapping (18). Briefly, this involves the tethering of cysteaminyl EDTA to a recombinant histone octamer via a thiol group introduced at H4 cysteine 47. The chelation of Fe2+ ions by this reagent provides a means of generating localized hydroxyl radicals that are capable of cleaving DNA, but only within a range of approximately 1.5 nm (15). The site at which the reagent is attached on histone H4 comes closest to contacting the DNA 2 bp from either side of the nucleosome dyad, meaning that the sites of cleavage can be used to determine nucleosome positions at base pair resolution. In the case of the 64A64 fragments, the major sites of cleavage show that nucleosomes were predominantly located at the expected site, with the nucleosome dyad at position +70 relative to the MMTV transcriptional start site (Fig. 1B, lane 2).
Thermal incubation has previously been reported to have the general effect of shifting nucleosomes from central locations towards the ends of DNA fragments (20, 49, 57). We next investigated whether this was also the case for nucleosomes assembled on the 64A64 fragment. Figure 1A shows that following incubation at successively higher temperatures, the mobility of the nucleosome during electrophoresis increased. Site-directed mapping was used to confirm that this alteration to nucleosome mobility reflected the movement of nucleosomes from the center of the DNA fragment to positions located within 16 bp of either end (Fig. 1B). By performing thermal shifting over a fine range of temperatures we could calculate the temperature required to shift half of the nucleosomes from their original positions. We also noted that even under conditions of incomplete shifting, both native gel electrophoresis and site-directed mapping indicate that nucleosomes were found to be located either at the center of the DNA fragment or near either DNA end, but not in between.
In order to investigate this more thoroughly, we determined the effect of altering the length of DNA flanking the nucleosome on the temperature required for 50% shifting. A series of 13 fragments were generated in which the DNA flanking the central nucleosome was varied from 20 to 64 bp. Remarkably, it was found that significantly higher temperatures were required to move nucleosomes assembled onto the short DNA fragments than those on larger fragments (Fig. 1C). Extensions of 25 bp or less required temperatures of about 60°C, whereas extensions of over 42 bp could be shifted at temperatures below 45°C. This indicates that nucleosomes assembled at the MMTV NucA sequence can more readily be moved distances of 42 bp or more than shorter distances.
In order to determine whether this behavior is unique to MMTV NucA, we used the MMTV NucB positioning sequence (55) to carry out a similar investigation. As observed previously (20), we found that significantly higher temperatures are required to shift these nucleosomes. Despite the differences in the absolute temperature required, shifting from NucB sequences displayed a length dependence that is very similar to that observed for NucA (Fig. 1C).
Manipulation of DNA fragment length can impose an orientation to nucleosome movement.
The approach described in the previous section involves monitoring the movement of nucleosomes from their initial locations to positions close to the DNA ends. Although the propensity of nucleosomes to move to positions closer to the ends of short DNA fragments appears to be a general phenomenon, the basis for this preference is not fully understood and is probably not relevant in vivo, where few nucleosomes are positioned adjacent to termini. Thus, it would be preferable to study the movement of nucleosomes between locations that are not close to DNA ends. Our observations made using symmetric DNA fragments indicate that nucleosomes assembled at NucA display a preference for movements of >40 bp. This suggests that by assembling nucleosomes onto DNA fragments such that they are positioned asymmetrically, it might be possible to bias thermally driven movement towards the longer end.
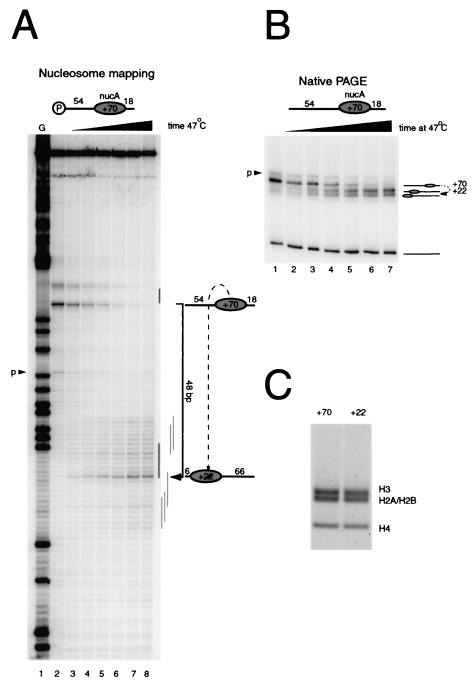
Figure 2A shows how this approach has been used to characterize the movement of nucleosomes assembled onto the DNA fragment 54A18. Prior to thermal incubation, site-directed mapping revealed a single major location (+70) with 18 bp of DNA on the downstream side of the nucleosome and 54 bp on the upstream side (Fig. 2A, lane 2). Consistent with this, the nucleosome migrated predominantly as a discrete species during native gel electrophoresis (Fig. 2B, lane 1). Following incubation for increasing lengths of time at 47°C, a loss of the site-directed mapping signal at the original location was observed, together with a concomitant increase in cleavage at new sites centered 48 bp along the 54-bp extension at +22 relative to the MMTV transcriptional start site (Fig. 2A, lanes 4 to 8). Notably, no nucleosomes were detected moving in the opposite direction to the short end of the DNA fragment. This suggests that NucA can be shifted more readily when a long DNA arm is available, consistent with the observations made using symmetric nucleosomes.
FIG. 2.
Oriented nucleosome redistribution. By creating DNA fragments with long linker DNA on one side and short linker DNA on the other, it is possible to direct thermal nucleosome redistribution onto the longer DNA arm. (A) Site-directed mapping at base pair resolution (lane 2) shows that nucleosomes are initially deposited as expected at +70 relative to the MMTV transcriptional start site, with a minor population of nucleosomes also detectable at position +53, denoted “p.” Following incubation at 47°C for 4, 8, 16, 32, 64, and 120 min (lanes 3 to 8), a loss of mapping is observed at the +70 location concurrently with an accumulation of new cleavage sites around the new major location at +22. No intermediate mapping sites are observable. (B) Native gel electrophoresis (PAGE) of the same samples is consistent with the expected migration of nucleosomes to the positions mapped in panel A. (C) A thermal shifting reaction as for panel B was performed on a preparative scale. The unshifted (+70) and major shifted (+22) species were excised. Protein was extracted from the gel slices, which were then resolved by SDS gel electrophoresis. No change in histone content was detected.
Although the major new location at +22 is 48 bp from the original location, at the longer time points it is clear that some nucleosomes also move to positions at and up to 4 bp beyond the end of the DNA fragment. A small proportion of the nucleosomes also move to more internal sites up to 18 bp from the end. These observations are consistent with native gel analysis of nucleosome shifting on this DNA fragment (Fig. 2B). Nucleosomes at the initial location disappear during the course of thermal incubation, and a major new species accumulates with a faster mobility, as expected for a nucleosome shifted to a site 6 bp from one end of the DNA fragment. A proportion of still faster migrating species also accumulates during the time course, consistent with the accumulation of some nucleosomes at the end of the DNA fragment, as observed by site-directed mapping. In order to confirm that this faster migrating species contained all four histones, the shifted and unshifted bands were excised from a gel and the histones were examined by SDS electrophoresis (Fig. 2C).
Stepwise movement of nucleosomes over 156 bp.
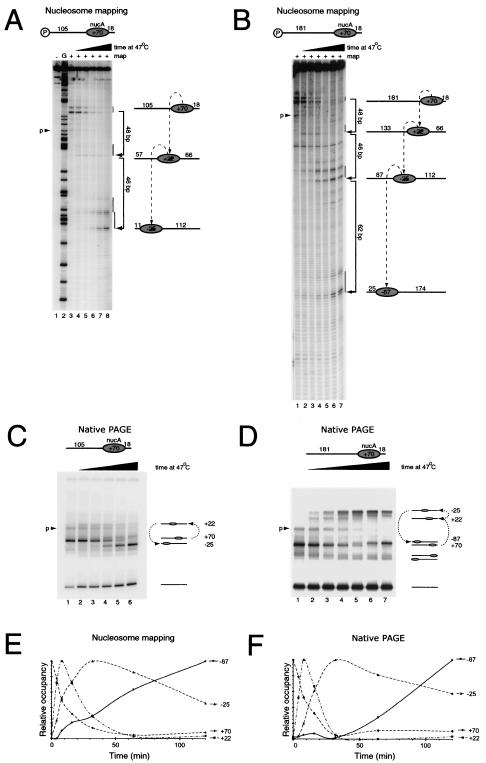
We next sought to investigate the effect of increasing the length of the 54-bp extension. Site-directed mapping reveals that nucleosomes assembled on the 105A18 fragment were initially positioned at the expected location (+70) and that following thermal incubation the proportion of nucleosomes at this location decreased (Fig. 3A, lanes 3 to 8). The major site at which nucleosomes accumulate is located 94 bp along the DNA fragment at position −25. However, careful inspection of the mapping gel reveals that a small amount of signal first accumulates and then decreases at +22 bp, 48 bp from the starting location. This is exactly the same position as the major destination for nucleosomes moving along the 54A18 fragment. Thus, the location at +22 has the properties expected of a short-lived intermediate during the transit of nucleosomes between the initial location at +70 and the destination at −25.
FIG. 3.
Sequential movement of nucleosomes over 154 bp. (A) Site-directed mapping on the fragment 105A18 indicates that nucleosomes are initially assembled as before, with the dyad at +70 (lane 3). Lanes 4 to 8 show that during incubation at 47°C for 4, 8, 16, 32, and 64 min, mapping is lost at +70 and transiently accumulated at +22 prior to accumulation during the last time points at −25 relative to the MMTV transcriptional start site. Lane 1, control in which the reagent-attached nucleosomal DNA was not subjected to site-directed mapping; lane 2, G ladder. (B) Tracking of the redistribution of nucleosomes assembled onto the fragment 181A18 during incubation at 47°C for 4, 8, 16, 32, 64, and 120 min (lanes 2 to 7). Nucleosomes initially accumulate at +22, subsequently at −25, and finally at −85. (C and D) Native gel electrophoresis (PAGE) of the same samples as for panels A and B, respectively. In all panels, minor amounts of nucleosomes assembled at +53 are indicated by “p.” (E and F) Quantitation of the amount of material at each position in panels B and D. These graphs illustrate how the relative amounts of nucleosomes at each position change over time.
As in the case of the 54A18 fragment, native gel analysis of the nucleosome on 105A18 during thermal shifting correlates with what we observed by site-directed mapping. The nucleosome migrates initially predominantly as a single discrete species, consistent with the movement of a nucleosome at the initial location (Fig. 3C, lane 1, +70). Following thermal incubation, a small quantity of a slowly migrating species first accumulates and subsequently decays (Fig. 3C, lanes 2 to 5, +22). This is likely to represent the more centrally located intermediate at +22 generated as a result of the first 48-bp step observed by site-directed mapping (Fig. 3A, lanes 4 to 7, +22). At later time points, the major product to accumulate has a faster mobility than the initial nucleosome (Fig. 3C, lane 6). This is consistent with a nucleosome located at −25, as observed by site-directed mapping (Fig. 3A, lane 8).
Nucleosome movement on the fragment 181A18 also showed a loss of cleavage at the initial position followed by an accumulation of cleavage first at +22, 48 bp from the initial location, as observed with the 54A18 and 105A18 fragments (Fig. 3B, lanes 1 to 4). Next, cleavage was observed to accumulate at a site 46 bp further along the DNA at −25, the same site nucleosomes moved to on the 105A18 fragment (Fig. 3B, lanes 4 to 6). Finally, at the later time points cleavage of DNA occurred at sites 62 bp further upstream at −87, which is 156 bp from the initial location (Fig. 3B, lanes 6 and 7). As with the shorter fragments, there is no evidence for movement of these nucleosomes along the shorter arm of DNA. Native gel electrophoresis gave the same results for the accumulation and decay of species, with mobilities consistent with those identified by site-directed mapping (Fig. 3D). Quantitation of the different nucleosomal species over the time course showed that both site-directed mapping and native gel electrophoresis provide evidence for a similar temporal progression (Fig. 3E and F). No further alterations to nucleosome positioning were observed during extended incubations for up to 480 min, suggesting that nucleosomes reached equilibrium during the course of these reactions (data not shown). In combination, these data suggest that nucleosomes assembled at the +70 NucA location move sequentially along the DNA during incubation at 47°C in a sequence of movements of 48, 46, and 62 bp.
DNA sequence directs the location to which nucleosomes move.
There are two major explanations for how this stepwise behavior could arise. If the mechanism for nucleosome redistribution involves large stepwise movements rather than an incremental progression along DNA, then this could result in stepwise movements similar to those we observed. The transit of a loop around the octamer surface similar to that observed during transit of RNA polymerases around nucleosomes could cause this (60; http://www.dundee.ac.uk/biocentre/toh/mech3a.html). A second explanation for the stepwise behavior is that DNA sequences that represent successively more favorable sites for positioning nucleosomes during thermal redistribution are located at intervals of 48, 46, and 62 bp along the DNA fragment.
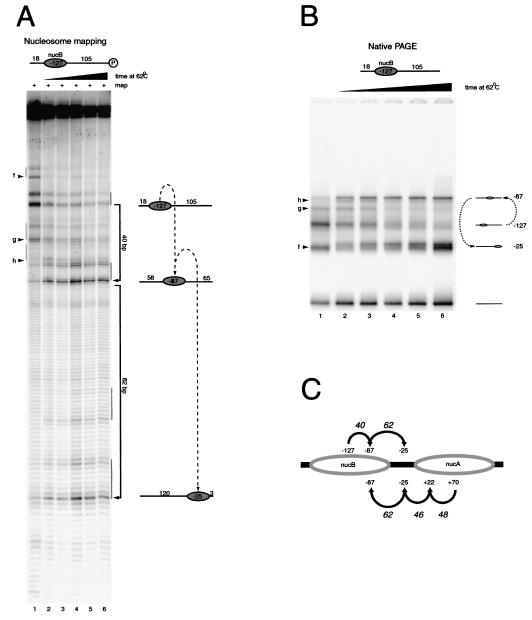
In order to distinguish between these possibilities, we monitored the movement of a nucleosome over the same DNA sequence, but starting from a different location. Nucleosomes were assembled at the NucB positioning sequence 197 bp upstream of NucA, with 105 bp of DNA on the downstream side that overlaps the DNA fragments used for Fig. 3. Following thermal incubation, site-directed mapping and gel shift assays indicated that this nucleosome moves in steps of 40 and 62 bp (Fig. 4A and B). This involves NucB moving to the −87 and −25 locations that were also occupied during redistribution of NucA (Fig. 3). The locations to which each nucleosome moves are summarized in Fig. 4C. The fact that nucleosomes from different starting locations move through the same positions suggests that the stepwise behavior we observe results from a sequential hierarchy of more favorable locations. Any mechanism for nucleosome redistribution that enables nucleosomes to scan along DNA fragments would favor these stable locations. The reason we do not detect nucleosomes at sites between the more favored locations could be that their stabilities are so low that occupancy is undetectable by site-directed mapping or native gel shift assays. However, it is also possible that a combination of stepwise movements and a preference for the final location could explain the redistribution pattern observed.
FIG. 4.
Redistribution of nucleosomes from the MMTV NucB positioning sequence. (A) Site-directed mapping of nucleosomes assembled on the DNA fragment 18B105 results in the generation of a major signal at −127 (lane 1). Minor positions are also observed at the positions indicated by “f,” “g,” and “h.” Following thermal incubation at 62°C for 3.75, 7.5, 15, 30, and 60 min (lanes 2 to 6), new nucleosome locations are observed at −87 and later at −25. (B) Native gel electrophoresis (PAGE) results in the generation of a series of bands consistent with those observed during site-directed mapping. (C) Schematic summarizing the nucleosome movements detected in the experiments for Fig. 3 and this figure. Nucleosomes were tracked moving over the same sequences from different directions. Despite their different origins, both NucA and NucB move through the −25 and −87 positions. This suggests that DNA sequence plays an important role in determining the sites to which nucleosomes are relocated.
Nucleosome movement driven by the ISWI protein.
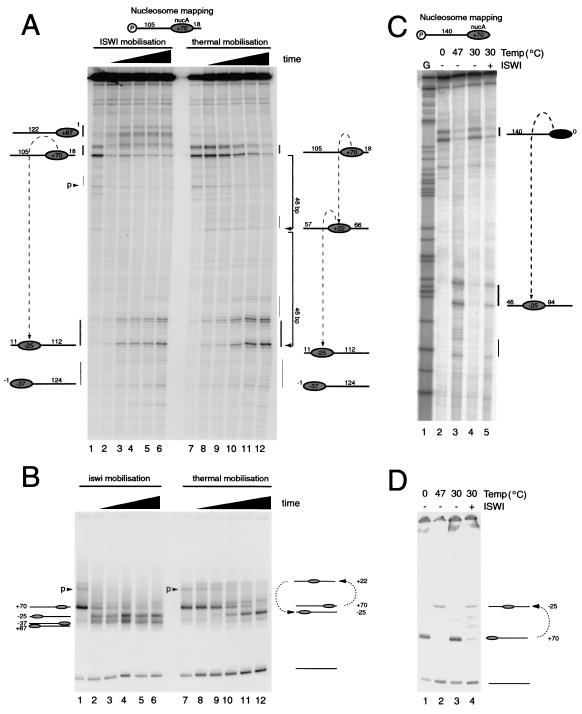
Having monitored the movements of NucA during thermal incubation, we next investigated how this nucleosome was repositioned by the remodeling protein ISWI. Figure 5 shows a direct comparison between the movements of nucleosomes driven by ISWI and temperature following assembly onto the fragment 105A18. In both cases a loss of nucleosomes at the initial position is observed. It is also notable that in both cases the major new location to which nucleosomes migrate is position −25. However, there are also differences in the behavior of nucleosomes following ISWI-driven mobilization. Firstly, following mobilization driven by ISWI, a proportion of nucleosomes migrate to the short downstream DNA arm at +87. It is also notable that whereas during thermally driven nucleosome movement an intermediate species is seen at +22, this is not detectable during ISWI-driven nucleosome movement.
FIG. 5.
Nucleosome redistribution driven by ISWI protein. (A) Redistribution of nucleosomes assembled onto the DNA fragment 105A18 directed by ISWI (lanes 2 to 6) is compared to thermal redistribution (lanes 8 to 12). Nucleosomes were incubated with 0.4 pmol of ISWI for 4, 8, 16, 32, and 64 min (lanes 2 to 6). For comparison, nucleosomes were incubated at 47°C for the same times (lanes 8 to 12). (B) Native gel shift assay of the same samples as in panel A. (C) Nucleosome redistribution by thermal incubation and ISWI on the fragment 140A0 using site-directed mapping. Lanes 1 and 4 show that nucleosomes are initially located at the terminal position +70 on this DNA fragment, but following thermal incubation at 47°C (lane 2) or treatment with 0.9 pmol of ISWI (lane 4) for 60 min, nucleosomes are relocated predominantly to the −25 location. (D) Native gel electrophoresis of the samples in panel C.
Our observation that nucleosome movement driven by ISWI results in a bias in favor of more terminal locations is consistent with previous reports that ISWI moves nucleosomes from central positions toward DNA ends (7). However, the accumulation of nucleosomes at positions related to those observed during thermal equilibration raises the possibility that ISWI may not always move nucleosomes to positions closer to DNA ends. In order to test this further, we investigated whether ISWI is capable of moving terminally located nucleosomes assembled onto the fragment 140A0. Figure 5C illustrates that ISWI is capable of relocating nucleosomes, albeit with reduced efficiency, from the initial terminal location to the position −25, which is 46 bp from the nearest DNA end. Thus, the presence of thermodynamically favored locations plays a dominant role in determining the positions to which nucleosomes are moved by ISWI.
Nucleosome movement driven by the SWI/SNF and RSC complexes.
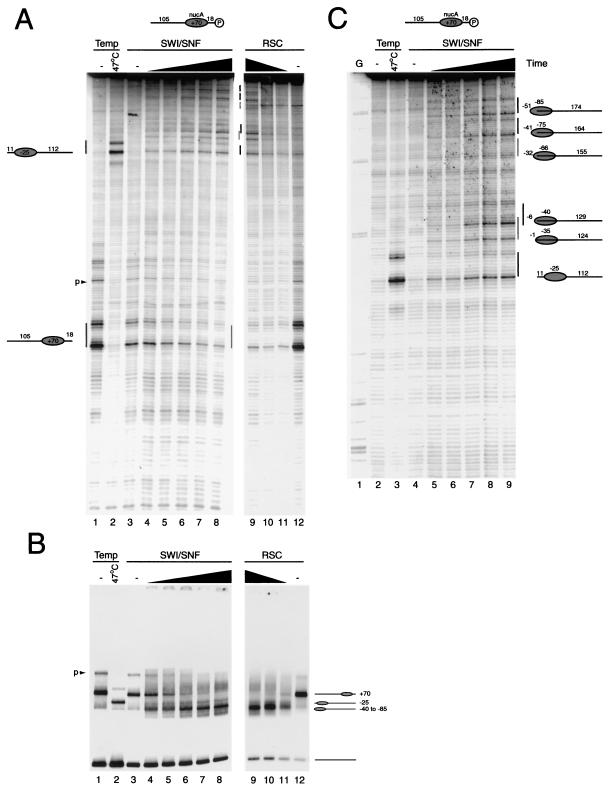
The yeast SWI/SNF and RSC complexes are also capable of altering the positions of nucleosomes along DNA. However, these complexes have also been reported to be capable of generating disrupted nucleosomes which cannot be explained by the relocation of nucleosomes to different sites alone (4, 43, 50, 58). We found that the SWI/SNF and RSC complexes functioned very similarly to each other but relocated nucleosomes to positions that differed from those observed during thermal mobilization or ISWI-driven nucleosome redistribution. This is illustrated using the 105A18 fragment labeled at the downstream end in Fig. 6. This shows that only a small proportion of nucleosomes accumulate at the −25 location favored during thermal and ISWI redistribution. The majority of the new cleavage sites suggest that nucleosomes have moved to positions beyond the end of the DNA fragment. To resolve these more clearly, the same samples were subjected to extended electrophoresis (Fig. 6B). This shows that cleavage by the mapping reagent accumulates at sites as little as 22 bp from the end of the DNA fragment (Fig. 6B). Labeling of this DNA fragment on the other strand revealed that some nucleosomes are also repositioned off the downstream end of the fragment (data not shown). Consistent with this, native gel electrophoresis revealed the accumulation of species with high mobility. Although the native gels indicate that a fairly discrete fast migrating species is generated, site-directed mapping occurs at several sites. However, it should be noted that movement of nucleosomes beyond the end of a DNA fragment may not increase electrophoretic mobility beyond that of an end-positioned nucleosome. Thus, the product of remodeling reactions observed on the native gel may consist of a mixture of nucleosomes positioned at and beyond the end of the DNA fragment.
FIG. 6.
Nucleosome redistribution driven by the yeast SWI/SNF and RSC complexes. (A) The fate of nucleosomes assembled onto the fragment 105A18 radiolabeled at the downstream end was monitored by site-directed mapping following incubation at 47°C for 1 h (lane 2) or treatment with 0.25 pmol of SWI/SNF complex and ATP for 4, 8, 16, 32, and 64 min (lanes 4 to 8). Nucleosomes were incubated with 0.12 pmol of RSC complex for 16, 8, and 4 min (lanes 9 to 11). (B) Native gel electrophoresis of the same samples as in panel A. (C) Extended electrophoresis was performed on the reactions from panel B to enable mapping locations towards the end of the fragment to be identified at base pair resolution.
DISCUSSION
We have monitored the motion of nucleosomes undergoing thermally driven movement following assembly onto MMTV promoter DNA. We find that nucleosomes can quantitatively vacate sites onto which they have been deposited during assembly. The ability of the MMTV promoter to direct assembly of nucleosomes at sites that are not favored during thermal equilibration provides a powerful system to track the progress of nucleosomes as they move along DNA. We have been able to monitor the movement of nucleosomes over 156 bp. During the course of their progress over this distance, we have detected the presence of nucleosomes at intermediate locations. Thus, the nucleosomes appear to move along the DNA as a result of three consecutive movements of 48, 46, and 62 bp.
Consequences of stepwise nucleosome redistribution.
Local chromatin structure, and more specifically the precise positioning of nucleosomes over promoters, can play a critical role in determining how genes respond to signaling pathways (41). We have observed a clear preference for nucleosomes to be repositioned in increments of 40 to 60 bp on naturally occurring promoter DNA. A consequence of nucleosomes moving in this way will be that the accessibility of DNA is altered over a greater range than would occur if nucleosomes moved in single-base increments. Thus, the preference we have observed for nucleosomes to move in a stepwise fashion could act to amplify the effects of signaling pathways at promoters. It remains to be established how generally this behavior applies to other regions of the genome. However, it does appear that activation of the beta interferon promoter involves the movement of a nucleosome over 36 bp, and chromatin transitions dependent on the action of the yeast ISWI complexes are of this order at the PHO3, MET17, and REC104 promoters but smaller at the DRS2 promoter (22, 38). Movements of this size in vivo might influence the movement of neighboring nucleosomes, causing transitions to chromatin structure to be propagated over multiple nucleosomes. This appears to be the case at PHO8, MET17, and PHO3 (23, 38). It is also likely that DNA binding proteins will influence the behavior of nucleosomes in at least some cases (35, 53).
Our previous study of nucleosome redistribution on MMTV DNA sequences used shorter DNA fragments which precluded the unexpectedly large preferred movements observed here (20). Some other previous studies of nucleosome positioning use fragment lengths which may also restrict mobility, but movements of this order have been observed on DNA fragments derived from the beta interferon (42), mouse ribosomal DNA (40), and Drosophila Hsp70 (27) genes. Although movements in the range of 40 to 60 bp appear common, we would anticipate that a strong rotational phasing sequence could cause the appearance of intermediates spaced at approximately 10-bp intervals.
Nucleosome positions favored during chromatin assembly and thermal equilibration.
One of the key observations we make is that nucleosomes quantitatively vacate the sites at which they are deposited during chromatin assembly. Thus, the sites preferred by nucleosomes during thermal equilibration are not the same as those favored during chromatin assembly. This supports a previous argument that the sites selected by nucleosomes during chromatin assembly are not necessarily the same as those favored by intact nucleosomes (13). A possible explanation for this is that the sites selected by the tetramer during chromatin assembly differ from those favored by the intact octamer once assembly is complete. In many previous studies of thermally driven nucleosome movement, a significant proportion of the movement resulted from the movement of nucleosomes from central positions towards DNA ends. In these cases it was possible to argue that the energetic penalty associated with the DNA crossover at the entry and exit sites of the nucleosome could override sequence-based determinants of nucleosome positioning and bias movement towards terminal locations (20, 57). However, through the selection of appropriate lengths of DNA fragment, here we have been able to study nucleosome redistribution on fragments for which the starting and final locations are not at DNA ends (Fig. 3C and D; Fig. 5C and D).
ISWI-driven nucleosome mobilization.
Our characterization of the way nucleosomes behave during thermal mobilization on MMTV NucA sequences provides a framework against which we can compare their movement by ATP-dependent chromatin remodeling activities. In the case of the ISWI protein, we found that the thermodynamically favored positioning sequences were the primary determinants of where nucleosomes were relocated. Although we selected a quantity of ISWI that enabled nucleosome redistribution to be monitored over a convenient time course, reactions performed in the presence of a molar excess of ISWI resulted in more rapid redistribution to the same locations (data not shown). Thus, the nucleosome-positioning properties of the underlying DNA sequence appear to be the primary determinants of where nucleosomes move during ISWI-driven mobilization. These observations are reminiscent of a previous characterization of nucleosome shifting by the ISWI-containing remodeling activity of NURF (35). This study showed that NURF relocated nucleosomes to positions that were closely related to those occupied during thermal redistribution. Thus, ISWI within a native remodeling complex displays properties that are similar to those we describe here. However, not all ISWI-containing complexes behave in this way. ACF, CHRAC, and ISW2 have all been found to relocate nucleosomes to more central locations on nucleosomal DNA (14, 35, 36). Thus, the way in which ISWI functions in nucleosome mobilization reactions can be altered by the components it associates with.
Superimposed on the characteristics that are shared with thermal mobilization, ISWI has a greater tendency to drive nucleosomes towards the ends of DNA fragments (Fig. 5), consistent with the previously reported movement of nucleosomes from central to terminal locations in the presence of ISWI (14). An explanation for this may be that the action of ISWI makes nucleosomes move in a more processive fashion than when they are allowed to equilibrate thermally. The ISWI protein may achieve this through translocation along DNA (21, 68).
The ability of ISWI to move nucleosomes both towards and away from DNA ends has important implications for how ISWI may engage with nucleosomes during its functioning. The ability of ISWI to move nucleosomes right up to the edge of a DNA fragment and especially the ability of ISWI to effect this type of movement more efficiently than occurs by thermal shifting (Fig. 5A and B) suggest that ISWI does not require contacts with DNA on the destination side of the nucleosome. Our observation that ISWI can also remove nucleosomes from DNA ends suggests that contacts are not required on the departure side in order for ISWI to push nucleosomes along DNA. This is most readily explained if ISWI functions predominantly through DNA contacts within nucleosomes.
Nucleosome mobilization by SWI/SNF and RSC complexes.
The SWI/SNF and RSC complexes were found to drive histone octamers to positions at and beyond DNA ends, with little regard for the sites favored during thermal and ISWI-driven nucleosome movement. The detection of site-directed mapping cleavage at sites with characteristic 7-bp spacing as little as 24 bp from the end of the DNA fragment indicates that at least the H3/H4 tetramer must be relocated to these positions (Fig. 6). The simplest explanation for this is that nucleosomes are moved to positions beyond the ends of the DNA fragments such that up to 51 bp of DNA is lost from one side of the nucleosome. Although this involves the loss of a remarkable number of histone-DNA contacts, the majority of the strongest interactions between the H3/H4 tetramer and DNA would be retained. Consistent with this, the mobility of these nucleosomes is similar to that of end-positioned nucleosomes on this DNA fragment. However, it remains possible that additional alterations to nucleosome structure are also involved. While this report was in preparation, Kassabov et al. also elegantly demonstrated that SWI/SNF is capable of removing 50 bp from the edge of nucleosomes. Cross-linking to some of the sites used by Kassabov et al. suggests that DNA from the exposed end of the fragment can loop back to occupy some of the exposed DNA binding sites on the other side of the octamer (37). Although we are unable to determine whether this also occurs in the reactions described here, it is possible that the curvature and flexibility of DNA do not permit this on all DNA fragments. Fan et al. have also recently reported the movement of nucleosomes to locations up to and beyond DNA ends (16). While these observations relate to mononucleosomal fragments, we have recently observed that nucleosomes can be forced into overlapping positions on multinucleosome templates (M. Bruno; A. Flaus, and T. Owen-Hughes, submitted for publication).
The redistribution of nucleosomes to such extreme sites beyond the ends of DNA fragments may provide an explanation for previously observed alterations to chromatin structure that result from the action of SWI/SNF-related complexes. For example, the movement of nucleosomes this far beyond DNA ends would be expected to alter accessibility to nucleases and transcription factors (10, 50). Movement of a nucleosome past one end of a DNA fragment will expose sites that were previously buried significantly within the nucleosome. Sites that were initially located close to the dyad would be expected to become significantly more accessible as they become located closer to the edge of the nucleosome (54). As nucleosomes can be moved beyond either end, accessibility would be altered throughout the length of nucleosome core particles and might provide an opportunity for the binding of multiple transcription factors (52). This ability to expose sites that were initially located within nucleosomes could be confused with the generation of loops on the surface of nucleosomes. While this is possible, we have found that following remodeling of the 105A18 nucleosome by RSC and SWI/SNF, it is possible to account for the majority of the original mapping signals at the new locations (data not shown). This suggests that following remodeling few, if any, of the nucleosomal fragments are distorted such that DNA is moved outside the 1.5-nm range of hydroxyl radicals at their attachment site close to the nucleosome dyad.
The movement of nucleosomes up to 50 bp beyond the ends of DNA fragments will also cause nucleosomes assembled on short fragments to accumulate both DNA that is not bound to histones and unfilled DNA binding sites on the surface of the histone octamer. The association of the unbound DNA from one nucleosome with the DNA binding sites available on a different nucleosome could result in the formation of dinucleosome-like particles (43, 58). However, we did not detect the formation of significant quantities of dinucleosome-like particles in the remodeling reactions described here. This could be because we used lower quantities of remodeling complexes in our assays than have been used previously. The ability of SWI/SNF to move nucleosomes to locations well beyond DNA ends must involve the expenditure of sufficient energy to unwind significant amounts of DNA from nucleosomes. This might also be of relevance to the apparent loss of DNA content in SWI/SNF-remodeled nucleosomes (5) and the stable alterations to the linking number of plasmid DNA (26, 45). Movement of nucleosomes further beyond the end of DNA fragments could lead to the destabilization of the histone octamer, which could result in the displacement of some or all histones in trans (44, 52, 67).
In contrast to the assortment of disrupted structures that can be generated by SWI/SNF-related complexes, many ISWI complexes, with the exception of the NURF complex (63), do not appear to disrupt the integrity of nucleosomes other than to alter their positioning (36, 40). Why should the positions to which nucleosomes are moved by ISWI and SWI/SNF be so different? Both complexes are driven by Snf2 family proteins sharing the same helicase-like motifs and both have been found to be capable of generating superhelical torsion in DNA fragments (29) that may occur as a result of translocation around the DNA helix (56, 68). Possible explanations include differences in the amount of energy, processivity, or orientation the ISWI and Snf2p motors exert on nucleosomal DNA. It is also possible that the dependence of ISWI activity upon the H4 tail epitope provides a constraint on the extent to which nucleosomes can be redistributed.
Acknowledgments
We thank Chris Stockdale, Helder Ferreira, and Iestyn Whitehouse for preparation of material; Anjanabha Saha and Brad Cairns for providing strain BCY211; Kristen Neely and Jerry Workman for strain YJW426; and David Corona and Peter Becker for the ISWI expression construct. We also thank all members of the lab and the Division of Gene Regulation for helpful discussions.
This work was funded by a Wellcome Trust Senior Fellowship to T.O.H. A.F. was supported by HFSP and EMBO fellowships.
REFERENCES
- 1.Alexeev, A., A. Mazin, and S. C. Kowalczykowski. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10:182-186. [DOI] [PubMed] [Google Scholar]
- 2.Alexiadis, V., and J. T. Kadonaga. 2002. Strand pairing by Rad54 and Rad51 is enhanced by chromatin. Genes Dev. 16:2767-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoyagi, S., and J. J. Hayes. 2002. hSWI/SNF-catalyzed nucleosome sliding does not occur solely via a twist-diffusion mechanism. Mol. Cell. Biol. 22:7484-7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aoyagi, S., G. Narlikar, C. Zheng, S. Sif, R. E. Kingston, and J. J. Hayes. 2002. Nucleosome remodeling by the human SWI/SNF complex requires transient global disruption of histone-DNA interactions. Mol. Cell. Biol. 22:3653-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bazett-Jones, D. P., J. Cote, C. C. Landel, C. L. Peterson, and J. L. Workman. 1999. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol. Cell. Biol. 19:1470-1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beard, P. 1978. Mobility of histones on the chromatin of simian virus 40. Cell 15:955-967. [DOI] [PubMed] [Google Scholar]
- 7.Brehm, A., G. Langst, J. Kehle, C. R. Clapier, A. Imhof, A. Eberharter, J. Muller, and P. B. Becker. 2000. dMi-2 and ISWI chromatin remodelling factors have distinct nucleosome binding and mobilization properties. EMBO J. 19:4332-4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, N., R. A. Poot, I. Kukimoto, C. Garcia-Jimenez, G. Dellaire, and P. D. Varga-Weisz. 2002. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat. Genet. 32:627-632. [DOI] [PubMed] [Google Scholar]
- 9.Corona, D. F., G. Langst, C. R. Clapier, E. J. Bonte, S. Ferrari, J. W. Tamkun, and P. B. Becker. 1999. ISWI is an ATP-dependent nucleosome remodeling factor. Mol. Cell 3:239-245. [DOI] [PubMed] [Google Scholar]
- 10.Côté, J., J. Quinn, J. L. Workman, and C. L. Peterson. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265:53-60. [DOI] [PubMed] [Google Scholar]
- 11.Davey, C. A., D. F. Sargent, K. Luger, A. W. Maeder, and T. J. Richmond. 2002. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 A resolution. J. Mol. Biol. 319:1097-1113. [DOI] [PubMed] [Google Scholar]
- 12.Donehower, L. A., B. Fleurdelys, and G. L. Hager. 1983. Further evidence for the protein coding potential of the mouse mammary tumor virus long terminal repeat: nucleotide sequence of an endogenous proviral long terminal repeat. J. Virol. 45:941-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drew, H. R. 1991. Can one measure the free energy of binding of the histone octamer to different DNA sequences by salt-dependent reconstitution? J. Mol. Biol. 219:391-392. [DOI] [PubMed] [Google Scholar]
- 14.Eberharter, A., S. Ferrari, G. Langst, T. Straub, A. Imhof, P. Varga-Weisz, M. Wilm, and P. B. Becker. 2001. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. EMBO J. 20:3781-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebright, Y. W., Y. Chen, P. S. Pendergrast, and R. H. Ebright. 1992. Incorporation of an EDTA-metal complex at a rationally selected site within a protein: application to EDTA-iron DNA affinity cleaving with catabolite gene activator protein (CAP) and Cro. Biochemistry 31:10664-10670. [DOI] [PubMed] [Google Scholar]
- 16.Fan, H. Y., X. He, R. E. Kingston, and G. J. Narlikar. 2003. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell 11:1311-1322. [DOI] [PubMed] [Google Scholar]
- 17.Fazzio, T. G., C. Kooperberg, J. P. Goldmark, C. Neal, R. Basom, J. Delrow, and T. Tsukiyama. 2001. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol. Cell. Biol. 21:6450-6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flaus, A., K. Luger, S. Tan, and T. J. Richmond. 1996. Mapping nucleosome position at single base-pair resolution by using site-directed hydroxyl radicals. Proc. Natl. Acad. Sci. USA 93:1370-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flaus, A., and T. J. Richmond. 1999. Base-pair resolution mapping of nucleosomes in vitro. Methods Mol. Biol. 119:45-60. [DOI] [PubMed] [Google Scholar]
- 20.Flaus, A., and T. J. Richmond. 1998. Positioning and stability of nucleosomes on MMTV 3′LTR sequences. J. Mol. Biol. 275:427-441. [DOI] [PubMed] [Google Scholar]
- 21.Fyodorov, D. V., and J. T. Kadonaga. 2002. Dynamics of ATP-dependent chromatin assembly by ACF. Nature 418:896-900. [DOI] [PubMed] [Google Scholar]
- 22.Goldmark, J. P., T. G. Fazzio, P. W. Estep, G. M. Church, and T. Tsukiyama. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103:423-433. [DOI] [PubMed] [Google Scholar]
- 23.Gregory, P. D., A. Schmid, M. Zavari, M. Munsterkotter, and W. Horz. 1999. Chromatin remodelling at the PHO8 promoter requires SWI-SNF and SAGA at a step subsequent to activator binding. EMBO J. 18:6407-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guschin, D., T. M. Geiman, N. Kikyo, D. J. Tremethick, A. P. Wolffe, and P. A. Wade. 2000. Multiple ISWI ATPase complexes from Xenopus laevis: functional conservation of an ACF/CHRAC homolog. J. Biol. Chem. 275:35248-35255. [DOI] [PubMed] [Google Scholar]
- 25.Guschin, D., P. A. Wade, N. Kikyo, and A. P. Wolffe. 2000. ATP-dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry 39:5238-5245. [DOI] [PubMed] [Google Scholar]
- 26.Guyon, J. R., G. J. Narlikar, S. Sif, and R. E. Kingston. 1999. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol. Cell. Biol. 19:2088-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamiche, A., R. Sandaltzopoulos, D. A. Gdula, and C. Wu. 1999. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell 97:833-842. [DOI] [PubMed] [Google Scholar]
- 28.Harp, J. M., B. L. Hanson, D. E. Timm, and G. J. Bunick. 2000. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D Biol. Crystallogr. 56:1513-1534. [DOI] [PubMed] [Google Scholar]
- 29.Havas, K., A. Flaus, M. Phelan, R. Kingston, P. A. Wade, D. M. Lilley, and T. Owen-Hughes. 2000. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103:1133-1142. [DOI] [PubMed] [Google Scholar]
- 30.Hirschhorn, J. N., S. A. Brown, C. D. Clark, and F. Winston. 1992. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 6:2288-2298. [DOI] [PubMed] [Google Scholar]
- 31.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 32.Ito, T., M. E. Levenstein, D. V. Fyodorov, A. K. Kutach, R. Kobayashi, and J. T. Kadonaga. 1999. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 13:1529-1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaskelioff, M., I. M. Gavin, C. L. Peterson, and C. Logie. 2000. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell. Biol. 20:3058-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaskelioff, M., S. Van Komen, J. E. Krebs, P. Sung, and C. L. Peterson. 2003. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278:9212-9218. [DOI] [PubMed] [Google Scholar]
- 35.Kang, J. G., A. Hamiche, and C. Wu. 2002. GAL4 directs nucleosome sliding induced by NURF. EMBO J. 21:1406-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kassabov, S. R., N. M. Henry, M. Zofall, T. Tsukiyama, and B. Bartholomew. 2002. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell. Biol. 22:7524-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kassabov, S. R., B. Zhang, J. Persinger, and B. Bartholomew. 2003. SWI/SNF unwraps, slides and rewraps the nucleosome. Mol. Cell 11:391-403. [DOI] [PubMed] [Google Scholar]
- 38.Kent, N. A., N. Karabetsou, P. K. Politis, and J. Mellor. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 15:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kornberg, R. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184:868-871. [DOI] [PubMed] [Google Scholar]
- 40.Langst, G., E. J. Bonte, D. F. Corona, and P. B. Becker. 1999. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97:843-852. [DOI] [PubMed] [Google Scholar]
- 41.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110:261-271. [DOI] [PubMed] [Google Scholar]
- 42.Lomvardas, S., and D. Thanos. 2001. Nucleosome sliding via TBP DNA binding in vivo. Cell 106:685-696. [DOI] [PubMed] [Google Scholar]
- 43.Lorch, Y., B. R. Cairns, M. Zhang, and R. D. Kornberg. 1998. Activated RSC-nucleosome complex a persistently altered form of the nucleosome. Cell 94:29-34. [DOI] [PubMed] [Google Scholar]
- 44.Lorch, Y., M. Zhang, and R. D. Kornberg. 1999. Histone octamer transfer by a chromatin-remodelling complex. Cell 96:389-392. [DOI] [PubMed] [Google Scholar]
- 45.Lorch, Y., M. Zhang, and R. D. Kornberg. 2001. RSC unravels the nucleosome. Mol. Cell 7:89-95. [DOI] [PubMed] [Google Scholar]
- 46.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 47.Luger, K., T. J. Rechsteiner, A. J. Flaus, M. M. Waye, and T. J. Richmond. 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 272:301-311. [DOI] [PubMed] [Google Scholar]
- 48.Meersseman, G., S. Pennings, and E. M. Bradbury. 1991. Chromatosome positioning on assembled long chromatin. Linker histones affect nucleosome placement on 5S rDNA. J. Mol. Biol. 220:89-100. [DOI] [PubMed] [Google Scholar]
- 49.Meersseman, G., S. Pennings, and E. M. Bradbury. 1992. Mobile nucleosomes—a general behavior. EMBO J. 11:2951-2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narlikar, G. J., M. L. Phelan, and R. E. Kingston. 2001. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell 8:1219-1230. [DOI] [PubMed] [Google Scholar]
- 51.Neely, K. E., A. H. Hassan, C. E. Brown, L. Howe, and J. L. Workman. 2002. Transcription activator interactions with multiple SWI/SNF subunits. Mol. Cell. Biol. 22:1615-1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Owen-Hughes, T., R. T. Utley, J. Cote, C. L. Peterson, and J. L. Workman. 1996. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science 273:513-516. [DOI] [PubMed] [Google Scholar]
- 53.Pazin, M. J., P. Bhargava, E. P. Geiduschek, and J. T. Kadonaga. 1997. Nucleosome mobility and the maintenance of nucleosome positioning. Science 276:809-812. [DOI] [PubMed] [Google Scholar]
- 54.Polach, K. J., and J. Widom. 1995. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J. Mol. Biol. 254:130-149. [DOI] [PubMed] [Google Scholar]
- 55.Richard-Foy, H., and G. L. Hager. 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 6:2321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saha, A., J. Wittmeyer, and B. R. Cairns. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 16:2120-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakaue, T., K. Yoshikawa, S. H. Yoshimura, and K. Takeyasu. 2001. Histone core slips along DNA and prefers positioning at the chain end. Phys. Rev. Lett. 87:078105. [DOI] [PubMed] [Google Scholar]
- 58.Schnitzler, G., S. Sif, and R. E. Kingston. 1998. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell 94:17-27. [DOI] [PubMed] [Google Scholar]
- 59.Simpson, R. T. 1990. Nucleosome positioning can affect the function of a cis-acting DNA element in vitro. Nature 343:387-389. [DOI] [PubMed] [Google Scholar]
- 60.Studitsky, V. M., D. J. Clark, and G. Felsenfeld. 1994. A histone octamer can step around a transcribing polymerase without leaving the template. Cell 76:371-382. [DOI] [PubMed] [Google Scholar]
- 61.Sudarsanam, P., V. R. Iyer, P. O. Brown, and F. Winston. 2000. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:3364-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suto, R. K., M. J. Clarkson, D. J. Tremethick, and K. Luger. 2000. Crystal structure of a nucleosome core particle containing the variant histone H2A. Z. Nat. Struct. Biol. 7:1121-1124. [DOI] [PubMed] [Google Scholar]
- 63.Tsukiyama, T., and C. Wu. 1995. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83:1011-1020. [DOI] [PubMed] [Google Scholar]
- 64.Ura, K., J. J. Hayes, and A. P. Wolffe. 1995. A positive role for nucleosome mobility in the transcriptional activity of chromatin templates: restriction by linker histones. EMBO J. 14:3752-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weischet, W. O., J. R. Allen, G. Riedel, and K. E. Van Holde. 1979. The effects of salt concentration and H-1 depletion on the digestion of calf thymus chromatin by micrococcal nuclease. Nucleic Acids Res. 6:1843-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White, C. L., R. K. Suto, and K. Luger. 2001. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 20:5207-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400:784-787. [DOI] [PubMed] [Google Scholar]
- 68.Whitehouse, I., C. Stockdale, A. Flaus, M. D. Szczelkun, and T. Owen-Hughes. 2003. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol. Cell. Biol. 23:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Workman, J. L., and R. G. Roeder. 1987. Binding of transcription factor TFIID to the major late promoter during in vitro nucleosome assembly potentiates subsequent initiation by RNA polymerase II. Cell 51:613-622. [DOI] [PubMed] [Google Scholar]