Abstract
The receptor for insulin-like growth factor I (IGF-IR) controls normal and pathological growth of cells. DNA repair pathways represent an unexplored target through which the IGF-IR signaling system might support pathological growth leading to cellular transformation. However, this study demonstrates that IGF-I stimulation supports homologous recombination-directed DNA repair (HRR). This effect involves an interaction between Rad51 and the major IGF-IR signaling molecule, insulin receptor substrate 1 (IRS-1). The binding occurs within the cytoplasm, engages the N-terminal domain of IRS-1, and is attenuated by IGF-I-mediated IRS-1 tyrosine phosphorylation. In the absence of IGF-I stimulation, or if mutated IGF-IR fails to phosphorylate IRS-1, localization of Rad51 to the sites of damaged DNA is diminished. These results point to a direct role of IRS-1 in HRR and suggest a novel role for the IGF-IR/IRS-1 axis in supporting the stability of the genome.
The receptor for insulin-like growth factor I (IGF-IR) is a multifunctional, membrane-associated tyrosine kinase. The ligand-activated receptor sends strong antiapoptotic and mitogenic signals supporting both normal and pathological growth of the cell (4, 24, 35). During transformation, an increase in the cell's propensity to accumulate mutations may develop as a result of the deregulation of normal mechanisms controlling faithful and unfaithful DNA repair. Scattered reports indicate that the IGF-IR may have functions affecting DNA repair. These include enhanced radioresistance proportional to the IGF-IR protein level in both mouse embryo fibroblasts and breast tumor cells (51), enhanced DNA repair via the IGF-I-activated p38 signaling pathway in response to UV-mediated DNA damage (16), and delayed UVB-induced apoptosis via IGF-I-mediated activation of Akt, resulting in enhanced repair of DNA cyclobutane thymidine dimers in keratinocytes (11). In contrast, one report notes a delay in DNA repair of potentially lethal radiation damage observed following IGF-I and insulin treatments of A549 cells (19). Although these few reports suggest the role of IGF-I in drug resistance and possibly in DNA repair, it remains unclear whether the receptor affects DNA repair directly, or whether its strong antiapoptotic properties (9, 42, 53) simply increase the resistance to genotoxic agents.
Here we investigate the effect of the IGF-IR signaling system on DNA repair of double-strand breaks (DSBs). DSBs, which are usually lethal, arise from ionizing radiation, from a variety of genotoxic chemicals including cisplatin, and also when replication forks encounter single-strand DNA breaks (17, 20). Early cellular events in the detection of DSBs include activation of the protein kinases ATM (ataxia telangiectasia mutated), ATR, and DNA protein kinase (DNA-PK), which in turn phosphorylate histone H2AX (γ-H2AX) within megabase pair regions surrounding DSBs, which “attracts” DNA repair machinery (6, 28). To support cell survival after DSBs, at least one of the two major DNA repair mechanisms has to be activated: (i) homologous recombination-directed DNA repair (HRR), which is usually error free, or (ii) nonhomologous end joining (NHEJ), which is error prone. In proliferating cells HRR seems to predominate, while quiescent cells favor NHEJ (17). The choice between DNA repair mechanisms is controlled, at least partially, by the availability of a DNA template. Proliferating cells utilize the newly synthesized template supplied by the process of DNA replication. Cells arrested in G1 face the more difficult task of searching for a template on the homologous chromosome. Alternatively, quiescent cells may simply link the ends of a DSB without any template by using the end binding Ku70/Ku80 complex and DNA-PK, followed by DNA ligation with XRCC4-ligase 4 (26, 32). This rapid repair of DNA by NHEJ, however, may cost the cell a gain or loss of base pairs (17).
Rad51 is a structural and functional eukaryotic homologue of the bacterial RecA recombinase, which is considered the key enzyme for HRR (5, 48, 55). Following detection of DSBs, the breast cancer susceptibility gene (BRCA2) is suspected to mediate translocation of Rad51 to the damaged DNA (10). In parallel, the ATM-activated 5′-3′ endonuclease complex (Rad50-MRE11-NBS1) exposes both 3′ ends of DNA at the DSB (31, 59). The ends are initially protected by the single-stranded DNA (ssDNA) binding protein replication protein A, which is subsequently replaced by Rad51 in a process that involves initial binding of Rad52 into the ssDNA-RPA complex (7, 13). As a result, newly formed Rad51 nucleoprotein filaments are directly involved in homology searching and strand invasion, which in eukaryotic cells require additional cooperation (ATPase activity) from Rad54 (43, 48, 49).
Our results indicate that the signal from activated IGF-IR enhances HRR by a mechanism that controls the translocation of Rad51 to the sites of damaged DNA (nuclear foci). This effect involves a direct interaction between Rad51 and the major IGF-IR signaling molecule, insulin receptor substrate 1 (IRS-1). The binding is direct, occurs within the perinuclear region of the cell, involves the N-terminal portion of IRS-1, and is negatively regulated by IGF-I-mediated IRS-1 tyrosine phosphorylation. Importantly, cells with low levels of the IGF-IR, or cells expressing an IGF-IR mutant that fails to phosphorylate IRS-1, retain Rad51 within the perinuclear compartment and show significantly less DNA repair by HRR. These results suggest a novel function for the IGF-IR/IRS-1 pathway that involves regulation of the intracellular trafficking of Rad51 to the site of damaged DNA—a crucial step in the process of DNA repair by homologous recombination.
MATERIALS AND METHODS
Cell lines.
The cell lines used are derived from 3T3-like fibroblasts isolated from mouse embryos with a targeted disruption of the IGF-IR gene (R− cells) (2). Following stable transfection with the human IGF-IR cDNA under control of the rat (−2350 to +1640) IGF-IR promoter, several clones expressing different levels of the IGF-IR, ranging from 3,000 to 30,000 IGF-IR molecules/cell, were selected and characterized (37, 41). R12 expresses 3,000, R508 expresses 15,000, and R600 expresses 30,000 molecules of IGF-IR/cell, as evaluated by Scatchard analysis (37). In addition, R−GR96 cells express an IGF-IR double mutant (Y950F + Δ1245) in the absence of the wild-type (wt) IGF-IR. The R−GR96 cells fail to phosphorylate IRS-1 following IGF-I stimulation. We have also generated a new cell line (R600/GR171) by transducing R600 fibroblasts with the pGR171 retroviral vector containing the PH/PTB domain of IRS-1 (39). All R− derivatives, with the exception of R−GR96 and R600/PH/PTB, were selected in G418 (1 mg/ml). R600/GR171 cells were selected in puromycin (2 μg/ml), and R−GR96 cells were routinely cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and hygromycin (250 μg/ml).
HRR.
Plasmid pDR-GFP (generously provided by M. Jasin, Sloan-Kettering Cancer Center, New York, N.Y.) (33) was stably introduced, by using a calcium phosphate reagent (Promega, Madison, Wis.), into R12, R508, and R600 fibroblasts. Stable clones were selected in puromycin (2 μg/ml) and characterized by PCR and Southern blotting to estimate DR-GFP copy numbers (see below). pDRGFP contains a nonactive green fluorescent protein (GFP) gene (SceGFP) as a recombination reporter and a fragment of the GFP gene as a donor for homologous repair. The SceGFP cassette has an inactivating insertion, which consists of two stop codons and a restriction site for the rare-cutting endonuclease I-SceI. When I-SceI is expressed in DR-GFP-expressing clones, it inflicts DSBs within the SceGFP fragment, providing a signal for homologous recombination and the reconstruction of functional GFP. To analyze the effectiveness of HRR, cells were transiently transfected with 3 μg of pCβA-Sce and 1 μg of pDsRed1-Mito (Clontech, Palo Alto, Calif.) by utilizing the Fugene 6 reagent (Roche, Indianapolis, Ind.). pCβA-Sce contains I-Sce I cDNA to generate DSBs in SceGFP cDNA (33), and pDsRed1-Mito contains red fluorescent protein with a mitochondrial localization signal to control for the efficiency of transfection. DNA repair by HRR was evaluated by counting cells with both green nuclear fluorescence and red mitochondrial fluorescence versus all positively transfected cells (red and green cells versus only red cells) at 72 h after transfection. Cells showing green nuclear fluorescence only were sporadically detected; however, these cells were not considered in the calculations.
PCR and Southern blotting.
PCR and Southern blotting were utilized to screen for DR-GFP and to determine DR-GFP copy numbers in selected clones of R−-derived cell lines. Genomic DNA was extracted with a Puregene DNA isolation kit (Gentra Systems, Minneapolis, Minn.) according to the manufacturer's recommendations. Southern blotting was carried out according to methodologies described previously (25). The GFP probe (800 bp) was obtained from pDR-GFP by SalI/HindIII digestion. To amplify GFP cDNA from cell lines stably transfected with the DR-GFP expression vector, 1 μg of genomic DNA and GFP-specific primers (right primer, 5′-CCTTCGGGCATGGCGGACTTG-3′; left primer, 5′-AGGGCGTTCGGCTTCTGG-3′) were utilized in a typical PCR (36).
NHEJ.
The cell-free NHEJ assay was used (21) with some modifications. An aliquot of 107 mouse embryonic fibroblasts was washed three times with ice-cold phosphate-buffered saline (PBS) and lysed in hypotonic buffer A (10 mM HEPES, 1.5 mM MgCl2, 10 mM KCl [pH 7.5], 2 μg of aprotinin/ml, 2 μg of leupeptin/ml, 0.5 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol, 25 mM NaF, and 0.2 mM NaVO3) for 10 min on ice. Following centrifugation at 6,000 × g for 3 min, nuclear pellets were resuspended in buffer B (20 mM HEPES, 25% glycerol, 500 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA [pH 7.5], and protease inhibitors as in buffer A), and the samples were repeatedly frozen in liquid nitrogen. Following centrifugation (at 30,000 × g for 30 min), supernatants were dialyzed overnight against buffer C (25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 10% glycerol, and proteinase inhibitors as in buffer A), and aliquots were stored at −70°C. NHEJ reactions were performed under the following conditions: 10 μg of nuclear lysate, 1 mM ATP, 0.25 mM deoxynucleoside triphosphates, 25 mM Tris acetate (pH 7.5), 100 mM potassium acetate, 10 mM magnesium acetate, and 1 mM dithiothreitol. After 5 min of preincubation at 37°C, the reaction mixture was supplemented with the substrate [200 ng of XhoI-XbaI-linearized pBluescript KS(+)]. The reaction mixture was incubated for 1 h at 37°C to ligate the plasmid and was treated with proteinase K (1 μg per reaction at 65°C for 30 min) to digest DNA-bound proteins. Products of NHEJ reactions were resolved in a 0.5% agarose gel containing 0.5 μg of ethidium bromide/ml. For each experiment, the sensitivity of the assay was additionally evaluated by running control samples in which increasing amounts of the substrate (ranging from 0 to 500 ng), and increasing amounts of the nuclear extract (ranging from 0 to 20 μg) were evaluated.
Western blotting and immunoprecipitation.
For IRS-1 immunoprecipitation and Western blotting, we used a polyclonal rabbit anti IRS-1 antibody that recognizes the C-terminal portion of IRS-1 (Upstate Biotechnology Inc. [UBI], Lake Placid, N.Y.). To detect the PH/PTB domain of IRS-1, we employed a rabbit antibody that recognizes an epitope at the amino terminus of IRS-1 (A-19; Santa Cruz Biotechnology, Santa Cruz, Calif.). To determine the tyrosine phosphorylation of IRS-1, quiescent cells (48 h in serum-free medium [SFM]) were stimulated with IGF-I (50 ng/ml) or were left without stimulation, and total proteins were extracted 15 min later. IRS-1 was immunoprecipitated from 500 μg of protein extract with a rabbit anti-IRS-1 antibody (Transduction Laboratories, Lexington, Ky.) and agarose-conjugated protein A (Calbiochem, San Diego, Calif.). Corresponding blots were first developed with an anti-phosphotyrosine antibody (PY20; Transduction Laboratories) and then reprobed with an anti-IRS-1 antibody against the N-terminal portion of IRS-1 (A-19; Santa Cruz). To evaluate levels of selected DNA repair proteins, monolayer cultures were lysed on ice with 400 μl of lysis buffer (50 mM HEPES [pH 7.5], 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 10% glycerol, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 0.2 mM sodium orthovanadate, and 10 μg of aprotinin/ml). To improve recovery of DNA-bound proteins, DNase was added to the lysis buffer. Aliquots (50 μg) were separated on a 4 to 15% gradient sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel (Bio-Rad) and transferred to nitrocellulose membranes. The resulting blots were probed with the following rabbit polyclonal antibodies: anti-Rad51 (Ab-1; Oncogene), anti-Rad52 (H-300; Santa Cruz), anti-Rad54 (H-152; Santa Cruz), anti-Ku70 (Serotec, Oxford, United Kingdom), and anti-Ku80 (Serotec). An anti Grb-2 antibody (Transduction Laboratories) was used as a control to monitor equal loading conditions (12).
Glutathione S-transferase (GST) pulldown assay.
Previously described protocols for in vitro translation and pulldown reactions were followed (22). Five overlapping IRS-1 truncation mutants (54) and in vitro-translated, [35S]methionine-labeled, full-length RAD51 were used. Fusion proteins were generated on the basis of the pGEX-5x-1 vector expressed in isopropyl-β-d-thiogalactopyranoside (IPTG)-induced bacterial cultures and were purified on glutathione-agarose beads. RAD51 cDNA in the pcDNA3 expression vector was in vitro translated by utilizing the Promega protocol based on wheat germ extract and T7 RNA polymerase.
Immunofluorescence.
All cells were cultured on poly-d-lysine-coated Lab-Tek chamber slides (Nalge Nunc International). Before immunostaining, cells were differentially treated: (i) quiescent cells were obtained by a 48-h incubation in SFM, (ii) exponentially growing cells were cultured in the presence of 10% FBS, and (iii) IGF-I (50 ng/ml) was applied either to cells cultured in 10% FBS or to cells in SFM. Finally, cisplatin (1 μg/ml) treatment was applied to exponentially growing or quiescent cultures in the presence or absence of IGF-I stimulation, at different time points ranging from 6 to 24 h. For immunostaining, cells were fixed and permeabilized with a buffer containing 0.02% Triton X-100 and 4% formaldehyde in PBS. Fixed cells were washed three times in PBS and blocked in 1% bovine serum albumin (BSA) for 30 min at 37°C. RAD51 was detected by a mouse anti-RAD51 monoclonal antibody (UBI) followed by a fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse secondary antibody (Molecular Probes, Eugene, Oreg.). IRS-1 was detected with a rabbit anti-IRS-1 antibody (UBI), which recognizes the C-terminal portion of the IRS-1 protein, and a rhodamine-conjugated goat anti-rabbit secondary antibody (Molecular Probes). Finally, phospho-histone H2AX was detected by a mouse monoclonal antibody which recognizes phosphorylated serine within the amino acid (aa) 134-to-142 fragment of human histone H2A.X (UBI) and a rhodamine-conjugated goat anti-mouse secondary antibody (Molecular Probes). Negative controls were performed in the presence of irrelevant, anti-bromodeoxyuridine antibody instead of the primary antibodies. In all cases DNA was counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). Specific staining was visualized with an inverted Olympus 1X70 fluorescence microscope equipped with a Cook Sensicom ER camera (Olympus America, Inc., Melville, N.Y.). In some cases, a series of three-dimensional images of each individual picture were deconvoluted to one two-dimensional image and resolved by adjusting the signal cutoff to near-maximal intensity to increase resolution. Final images were prepared with Adobe Photoshop to demonstrate subcellular localization and colocalization of detected proteins.
Cell cycle analysis.
Aliquots of cells (106/ml) were fixed in 70% ethanol for 30 min at 4°C, cells were centrifuged at 390 × g for 5 min, and the resulting pellets were resuspended in 1 ml of freshly prepared propidium iodide-RNase A solution for 30 min at 37°C. Cell cycle distribution was analyzed by FACScalibur (Becton Dickinson, Franklin Lakes, N.J.) using the CellQuest program (44).
Clonogenic assay.
Cells were plated at 5 × 102 on 12-well culture plates (Costar) in DMEM supplemented with 10% FBS with or without IGF-I (50 ng/ml). Following attachment, cells were treated with cisplatin at 0, 0.2, 0.5, and 1.0 μg/ml. Colonogenic potential was evaluated at 10 days after treatment by counting stained colonies (0.25% Coomassie brilliant blue in methanol).
RESULTS
IGF-I protects against cisplatin-induced cytotoxicity.
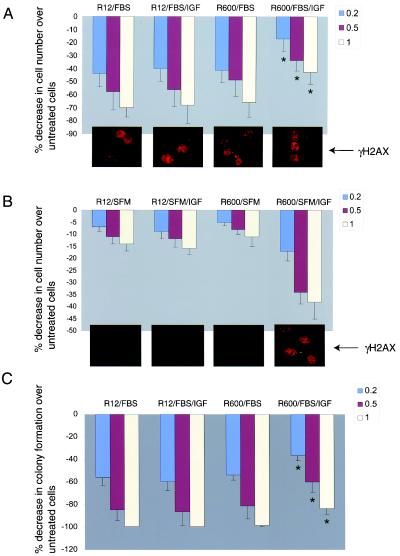
Our initial experiments indicate that exponentially growing fibroblasts are more resistant to the genotoxic agents cisplatin and mitomycin when the culture medium that already contains 10% FBS is additionally supplemented with IGF-I (50 ng/ml). We further analyzed this observation by use of mouse embryonic fibroblasts, in which the magnitude of the response to IGF-I depends on the number of IGF-IR molecules (37, 41). Originally developed in R. Baserga's laboratory, R12 and R600 fibroblasts express 3,000 and 30,000 IGF-IR molecules/cell, respectively (37, 41). Independently of the different levels of IGF-IR, these two cell lines show a very similar sensitivity to cisplatin when cultured in 10% FBS without IGF-I supplementation (Fig. 1A). Following addition of IGF-I, exponentially growing R12 cells, which are essentially not responsive to IGF-I (37), were still sensitive to cisplatin. R600 cells, which are IGF-I responsive, became significantly more resistant (Fig. 1A). Starting from the lowest concentration examined, 0.2 μg/ml, cisplatin caused a dramatic 44% ± 12% decrease in the survival of R12 fibroblasts at 48 h after treatment, and the survival rate did not change in the presence of IGF-I (Fig. 1A; compare R12/FBS and R12/FBS/IGF). In contrast, after IGF-I stimulation, R600 fibroblasts showed significant 2.4-, 1.4-, and 1.5-fold decreases in cell loss at 0.2, 0.5, and 1.0 μg of cisplatin/ml, respectively.
FIG. 1.
Effects of IGF-I on cell survival. (A and B) Exponentially growing and quiescent cultures of R12 fibroblasts, which express 3,000 IGF-IR molecules/cell, and R600 fibroblasts, which express 30,000 IGF-IR molecules/cell, were treated with cisplatin at 0.2 (blue bars) 0.5 (red bars), and 1.0 (yellow bars) μg/ml. SFM consisting of DMEM with 0.1% BSA and 2 mM glutamine was additionally supplemented either with 10% FBS or IGF-I (50 ng/ml), or both. Cell losses were evaluated at 48 h following cisplatin treatment. Data are expressed as percentages of decrease in the number of cells in comparison to the number of cells in untreated cultures and were collected from three separate experiments in which two cultures were used for each experimental point (n = 6). (Insets) γ-H2AX immunolabeling in R12 and R600 fibroblasts at 6 h following cisplatin treatment (1 μg/ml). (C) Clonogenic growth evaluated under the culture conditions described for panel A. Cells were plated at 5 × 102/35-mm culture dish, allowed to attach, and subsequently treated with the indicated concentrations of cisplatin. The percentages of decrease in colony formation at 0.2, 0.5, and 1.0 μg of cisplatin/ml were calculated from the number of colonies formed in the absence of cisplatin. Data are averages from three experiments performed in duplicate (n = 6). Asterisks indicate values that are statistically significant (P < 0.05).
It is noteworthy that quiescent R12 or R600 cells, cultured in SFM, are much more resistant to cisplatin than cells growing in the presence of serum. At the concentration of 1 μg/ml, which eliminated more than 70% of R12 and R600 cells growing in FBS (Fig. 1A), only about 15% cell loss was observed in SFM (Fig. 1B). Addition of IGF-I to quiescent cultures of R600 cells (Fig. 1B, R600/SFM/IGF) triggered an effect opposite that observed in serum-stimulated cultures (Fig. 1; compare panels A and B). In SFM, IGF-I triggered massive loss of R600 fibroblasts (R600/SFM/IGF) and had practically no effect on R12 fibroblasts (R12/SFM/IGF). Note, however, that R12 cells do not replicate DNA following IGF-I stimulation (37). DNA replication is required for cisplatin to generate lethal DSBs; therefore, it is not surprising that quiescent cultures survive the treatment. On the other hand, IGF-I-mediated cell proliferation and subsequent generation of DSBs, when replication forks encounter cisplatin-mediated primary DNA lesions, may cause cell death independently of the fact that IGF-I may also improve DNA repair. Therefore, the conditions under which IGF-I effects on DNA repair could actually be seen require similar rates of DNA replication and similar rates of initial DNA damage.
The effectiveness of cisplatin-mediated formation of DSBs was measured indirectly by immunocytochemical detection of phosphorylated γ-H2AX (6, 40). The insets below each histogram in Fig. 1A and B show representative images indicating the extent of DNA damage observed in R12 and R600 cells under different culture conditions. Proliferating R12 and R600 fibroblasts showed comparable levels of γ-H2AX immunolabeling at 6 h after cisplatin treatment, suggesting that the IGF-I-mediated resistance to cisplatin observed in R600 fibroblasts (Fig. 1A) does not depend on different sensitivities of R12 and R600 cells to cisplatin treatment. In SFM, quiescent R12 and R600 cells showed very low levels of γH2AX, and the addition of IGF-I to serum-free cultures activated γ-H2AX in R600 fibroblasts but not in R12 fibroblasts.
IGF-I-mediated improvement in short-term survival in the presence of cisplatin was additionally evaluated by the clonogenic assay. In the absence of IGF-I, proliferating cultures of R600 (R600/FBS) and R12 (R12/FBS) fibroblasts showed quite similar clonogenic survival after cisplatin treatment (Fig. 1C). Supplementation of the medium with IGF-I (FBS/IGF) significantly improved the clonogenic growth of R600 cells but did not affect this parameter in R12 cells (Fig. 1C; compare R12/FBS/IGF and R600/FBS/IGF).
In addition, we measured the clonogenic survival of two new cell lines (R−GR96 and R600/PH/PTB) which express mutated variants of IGF-IR and IRS-1, respectively. The sensitivities to cisplatin of these two new cell lines were similar to that observed in R12 fibroblasts (data not shown), indicating that interference with the IGF-IR signaling system either at the level of IRS-1 tyrosine phosphorylation (GR96 mutant) or by a dominant-negative action of the PH/PTB domain (GR171 mutant) (see Materials and Methods) attenuated IGF-I-mediated protection against cisplatin in this experimental setting.
Effects of IGF-I on DNA repair of DSBs.
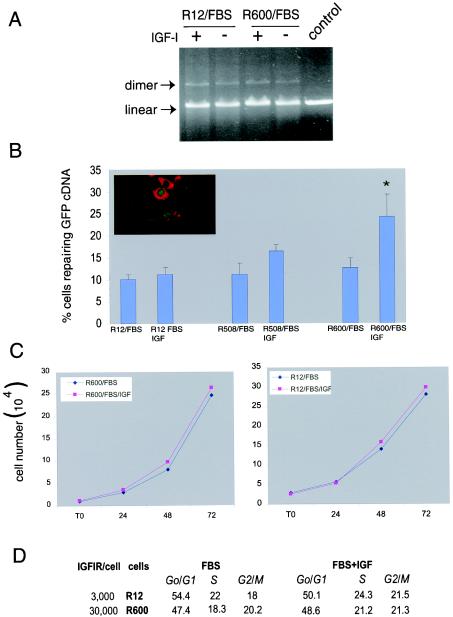
Because DSBs are the most lethal of DNA lesions caused by cisplatin, we asked whether molecular mechanisms involved in the repair of DSBs contribute to IGF-I-mediated drug resistance. NHEJ is considered the most effective way to quickly ligate DSBs, supporting cell survival. We have examined the potential effects of IGF-I on NHEJ by utilizing a cell-free NHEJ assay (21). The results in Fig. 2A show almost identical levels of in vitro ligation of linear pBluscript KS(+) measured in the presence of nuclear extracts from exponentially growing R600 and R12 fibroblasts, in the presence or absence of IGF-I stimulation. These in vitro results indicate that IGF-I-mediated resistance to cisplatin does not rely on an additional activation of NHEJ.
FIG. 2.
Effects of IGF-I on DNA repair. (A) Evaluation of NHEJ by a cell-free system. Exponentially growing R600 and R12 fibroblasts in the presence of 10% FBS were stimulated (+) or not stimulated (−) with IGF-I (50 ng/ml). Corresponding nuclear extracts were utilized to ligate linear pBluscript KS(+) (see Materials and Methods). The DNA bands are visualized on a 0.5% agarose gel containing ethidium bromide. The control reaction represents linear plasmid DNA not treated with nuclear extracts. Arrows indicate positions of the linear plasmid and the dimer. (B) R12, R508, and R600 fibroblasts (37) were stably transfected with the pDR-GFP plasmid and analyzed for reconstitution of functional GFP. Cells cultured in 10% FBS with or without IGF-I (50 ng/ml) were transiently transfected with two expression vectors; the first containing SceI cDNA, to generate DSBs in GFP cDNA, and the second containing red fluorescent protein with a mitochondrial localization signal to control for the efficiency of transfection. DNA repair by HRR was evaluated by counting cells with both green nuclear fluorescence and red mitochondrial fluorescence versus all positively transfected cells (cells with red fluorescence only) (see inset). The histogram shows the percentage of cells capable of repairing GFP cDNA. The results were collected from three separate experiments, in duplicate, in which 500 positively transfected cells were counted in at least 10 randomly selected microscopic fields (n = 6). The asterisk indicates a value that is statistically significant (P < 0.05). (C) Growth curves of R12 and R600 fibroblasts. Cells were plated at 2 × 104/35-mm culture dish either in 10% FBS alone or 10% FBS plus IGF-I (50 ng/ml). Cells were counted at time zero (T0) (after attachment) and at 24, 48, and 72 h after plating. Results are averages from two experiments performed in duplicate (n = 4). (D) The two cell lines described in the legend to panel C were labeled with propidium iodide and analyzed for cell cycle distribution by flow cytometry. Note that very similar patterns of cell cycle distribution were detected in the presence (FBS + IGF) and absence (FBS) of IGF-I stimulation.
The assay to evaluate DSB repair by HRR is based on reconstruction of the wt GFP from two nonfunctional heteroallelic fragments of GFP cDNA delivered into cells by the pDRGFP expression vector (34). The DR-GFP construct was introduced into R12 and R600 fibroblasts. Integration of the construct was evaluated by Southern blotting and PCR (data not shown), and stable clones with a single copy were analyzed for GFP recombination. Proliferating cells carrying the integrated DR-GFP construct were transiently transfected with two additional expression vectors; the first containing the rare-cutting endonuclease I-SceI, to generate DSBs in GFP cDNA, and the second containing red fluorescent protein with a mitochondrial localization signal, to measure the efficiency of transfection (see Materials and Methods and Fig. 2B inset). In the absence of exogenous IGF-I, similar rates of GFP recombination were observed in proliferating cultures of R12 and R600 cells (Fig. 2B). Differences started to emerge when SceI-induced DNA breaks were repaired in the presence of exogenous IGF-I (10% FBS supplemented with 50 ng of IGF-I/ml). A significant twofold increase (P < 0.03) in the restoration of GFP function was observed in R600 cells. Similarly, another cell line, R508, which expresses intermediate levels of the IGF-IR, 15, 000 IGF-IR molecules/cell (37), showed an average of 30% improvement in GFP repair after IGF-I stimulation. Finally, R12 fibroblasts, which are not responsive to IGF-I, did not improve the rate of GFP repair above the level routinely detected in the absence of IGF-I (Fig. 2B). Altogether, these results indicate the presence of an IGF-IR-inducible component in HRR.
Although the results demonstrate a significant increase in HRR in IGF-I-responsive cell lines, this effect could be still indirect, depending on the IGF-I-mediated acceleration of cell proliferation. To control this parameter, and to ensure a similar rate of cell cycle progression, all experiments were carried out in the presence of 10% FBS with or without IGF-I. Cell cycle distribution measured by flow cytometry (Fig. 2D) and growth curves (Fig. 2C) indicate that supplementation of the medium, which already contained 10% FBS, with IGF-I (50 ng/ml) affected the rate of cell proliferation only minimally. This indicates that under conditions in which the rate of cell proliferation (Fig. 2C and D) and the extent of DNA damage (Fig. 1A and B insets) are comparable, IGF-I treatment enhances HRR in a manner that is proportional to the level of IGF-IR.
Effects of IGF-I on the expression and subcellular localization of Rad51 proteins.
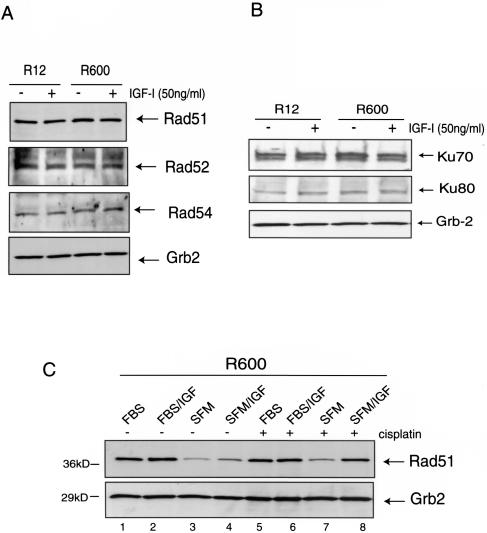
Total protein levels of Rad51 and its major partners in HRR, Rad52 and Rad54, were not affected either by the level of IGF-IR expression or by IGF-I stimulation in proliferating cultures (Fig. 3A). Similarly, other DNA repair proteins such as KU70 and KU80, for NHEJ (Fig. 3B), and MSH2, MSH6, and PMS1, for mismatch repair (data not shown), also were not affected, suggesting that IGF-I-mediated effects on DNA repair are not regulated on the level of gene expression or protein stability. Since Rad51 expression is cell cycle dependent (15), we evaluated Rad51 protein levels in quiescent (SFM) and proliferating cultures of R600 fibroblasts stimulated either with IGF-I (Fig. 3C, lanes 4 and 8) or with 10% FBS (lanes 1 and 5), in the presence or absence of cisplatin. The Western blot depicted in Fig. 3C confirms cell cycle-dependent regulation of Rad51 expression in our system and indicates that cell proliferation per se, rather than IGF-I-specific stimulation, controls Rad51 protein levels.
FIG. 3.
Effects of IGF-I on the expression of DNA repair proteins. (A and B) Protein extracts were isolated from quiescent (48 h in SFM), IGF-I-stimulated (50 ng/ml), or serum-stimulated (10% FBS) R12 and R600 fibroblasts. Total proteins (50 μg/ml) were separated by polyacrylamide gel electrophoresis, and the resulting nitrocellulose filters were probed with antibodies against proteins involved in HRR (Rad51, Rad52, and Rad54) (A) or NHEJ (KU70 and KU80) (B). (C) Rad51 protein levels in R600 fibroblasts cultured in either 10% FBS (FBS), 10% FBS +IGF-I (FBS/IGF), SFM, or SFM supplemented with IGF-I (SFM/IGF), in the presence (+) or absence (−) of cisplatin (1 μg/ml). An anti-Grb2 antibody was used to monitor equal loading conditions.
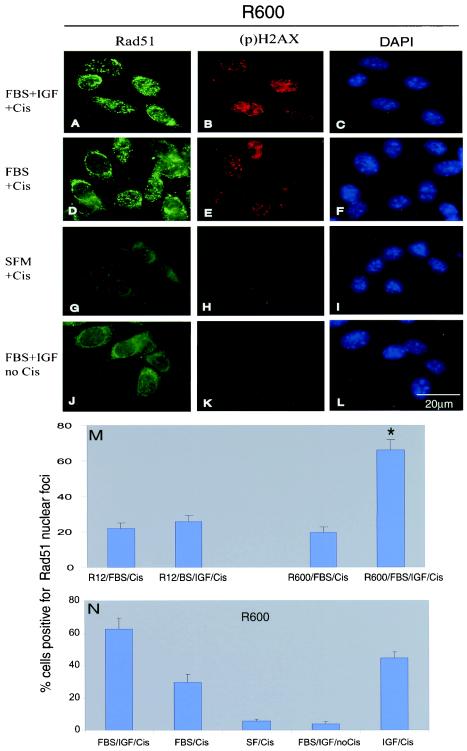
By taking advantage of the fact that Rad51 forms functional complexes at DNA lesions (nuclear foci), immunofluorescent labeling techniques have been developed to visualize these structures (34). We next asked whether IGF-I affects the subcellular distribution of Rad51 following cisplatin-induced DNA damage. Large numbers of Rad51 nuclear foci were observed following IGF-I stimulation (Fig. 4A) and, to a lesser extent, after serum stimulation (Fig. 4D). Importantly, we did not observe any difference in the initial levels of cisplatin-induced DNA damage (evaluated by formation of γ-H2AX foci) between proliferating R600 fibroblasts cultured in the presence or absence of IGF-I (Fig. 4B and E). Despite that, the effectiveness of Rad51 nuclear focus formation was significantly higher in R600 fibroblasts stimulated with IGF-I (Fig. 4M). This IGF-I-mediated improvement in Rad51 nuclear focus formation was not observed in R12 cells (Fig. 4M). As expected, the level of Rad51 nuclear focus formation was very low in the absence of cisplatin, or when cisplatin treatment was applied to R600 fibroblasts cultured in SFM (Fig. 4; compare panels G and J, and quantification in panel N).
FIG. 4.
Effects of IGF-I on subcellular localization of Rad51. (A through L) R600 fibroblasts expressing 30,000 IGF-IR molecules/cell (37) were utilized to evaluate the formation of Rad51 nuclear foci (A, D, G, and J) and the extent of DNA damage by monitoring of the phosphorylated form of histone H2AX (B, E, H, and K) under the indicated culture conditions. (C, F, I, and L) Corresponding images in which cells were labeled with DAPI to indicate the exact positions of the nuclei. (M and N) Quantification of the cisplatin-mediated recruitment of Rad51 into nuclear foci evaluated in R600 and R12 fibroblasts. Cells characterized by more than 10 foci per nucleus were considered positive. Results are averages from three experiments with three repetitions (n = 9) and standard deviations. Asterisk indicates a value that is statistically significant (P < 0.05).
Quantitatively, R600 fibroblasts show almost a threefold (P < 0.001) increase in the number of cells positive for Rad51 nuclear foci in the presence of IGF-I. In contrast, R12 fibroblasts did not improve the recruitment of Rad51 into damaged DNA after IGF-I stimulation (Fig. 4M). Interestingly, the pattern of IGF-I-mediated increase in HRR (Fig. 2B) is very similar to the pattern of IGF-I-mediated enhancement in the formation of Rad51 nuclear foci (Fig. 4M). This may suggest a functional connection between these two processes.
In addition, multiple control conditions (Fig. 4N) indicate that (i) cisplatin treatment applied to proliferating cultures of R600 fibroblasts (IGF/Cis, FBS/Cis, and FBS/IGF/Cis) induces the formation of Rad51 nuclear foci, (ii) treatment of R600 cells with IGF-I enhances the process of Rad51 nuclear focus formation (compare IGF/Cis and IGF/FBS/Cis with FBS/Cis), (iii) about 5% of cells are capable of forming Rad51 nuclear foci in the absence of cisplatin treatment (IGF/noCis), which likely reflects the repair of DSBs formed spontaneously during DNA replication in rapidly proliferating cultures, and (iv) cisplatin treatment applied to quiescent cells (SFM/Cis) resulted in a very low level of Rad51 and γ-H2AX nuclear focus formation (Fig. 4G, H, and N), confirming that cisplatin requires DNA replication to generate DSBs and to enable subsequent recruitment of Rad51.
Rad51 interacts with IRS-1.
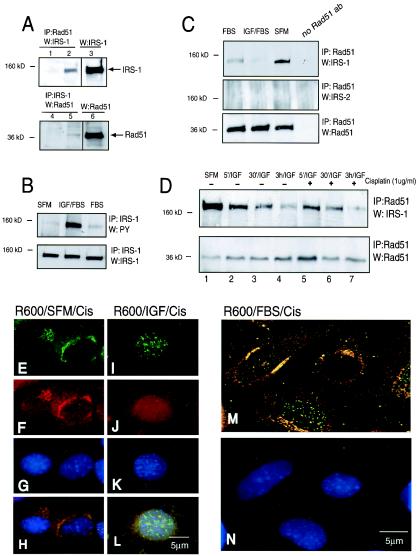
We have attempted to determine how the signal from activated IGF-IR facilitates the recruitment of Rad51 into DSBs. First we asked whether any of the known IGF-IR signaling molecules interacts with Rad51. The screening included IGF-IR itself, as well as SHC, 14-3-3, IRS-1, phosphatidylinositol 3-kinase, and C-terminal src kinase-signaling proteins known to directly bind activated IGF-IR. The molecule that formed a detectable immunocomplex with Rad51 was IRS-1 (Fig. 4). IRS-1 is the major signaling molecule for both the insulin and IGF-I receptors (58). In its role as a docking protein, IRS-1 transduces both metabolic and growth-promoting signals from activated insulin and IGF-I receptors (27). However, recent studies indicate that IRS-1 can be translocated to the nucleus (22, 50). The results depicted in Fig. 5A show by reciprocal immunoprecipitation and Western blotting that IRS-1 and Rad51 interact with each other. The IRS-1-Rad51 complex is clearly detectable, however the bands are significantly weaker after immunoprecipitation than corresponding Western blots, which were run without immunoprecipitation (Fig. 5A; compare lane 2 with lane 3 and lane 5 with lane 6). This may suggest that the interaction is not very efficient. In this experiment cell lysates were collected from R600 cells growing in 10% FBS—the condition under which IRS-1 is weakly phosphorylated on tyrosine residues (Fig. 5B). We then asked whether the IRS-1-Rad51 interaction depends on IRS-1 tyrosine phosphorylation. Surprisingly, the strongest interaction between IRS-1 and Rad51 was detected in SFM (Fig. 5C), the condition under which IRS-1 is not phosphorylated (Fig. 5B). Cells stimulated with IGF-I or, to a lesser extent, cells stimulated with 10% FBS showed an apparent decrease in the interaction between Rad51 and IRS-1 (Fig. 5C). Partial attenuation of the binding between IRS-1 and Rad51 was observed as early as 5 min following IGF-I stimulation, and decreased steadily over a period of 3 h (Fig. 5D, lanes 1 to 4). A similar pattern of Rad51 downregulation was observed in the presence of cisplatin (Fig. 5D, lanes 5 to 7). Under control conditions, the IRS-1-Rad51 complex was not detected when the anti-Rad51 antibody was replaced with an irrelevant antibody (Fig. 5A, lanes 1 and 4) or when an anti-IRS-1 primary antibody was replaced with an anti-IRS-2 antibody (Fig. 5C, center), additionally confirming the formation of a specific complex between IRS-1 and Rad51 in the absence of IGF-I stimulation.
FIG. 5.
Phosphorylation-dependent interaction between Rad51 and IRS-1. (A) Immunoprecipitation (IP) and Western blotting (W) targeting Rad51 and IRS-1 were carried out initially in serum-stimulated cultures of R600 fibroblasts. Portions (500 μg) of total protein lysates were IP with an anti-Rad51 antibody, and the corresponding blot was developed with an anti-IRS-1 antibody (lane 2). The conditions were reversed: protein lysates were IP with an anti-IRS-1 antibody, and the blot was developed with an anti-Rad51 antibody (lane 5). Lanes 3 and 6, IRS-1 and Rad51 protein levels in 50 μg of corresponding protein lysates. Lanes 1 and 4, negative controls in which IP reactions were performed in the presence of irrelevant antibodies instead of the Rad51 or IRS-1 primary antibody. (B) Protein lysates were collected from quiescent R600 fibroblasts cultured in SFM and stimulated with IGF-I or serum (FBS). Cell lysates were IP with an anti-IRS-1 antibody, and the corresponding blots were developed with antiphosphotyrosine (PY) and anti-IRS-1 antibodies. (C) R600 fibroblasts were treated as in the experiment for which results are shown in panel B. Cell lysates were IP with an anti-Rad51 antibody, and the corresponding blots were developed with anti-IRS-1, anti-IRS-2, and anti-Rad51 antibodies. Note an apparent downregulation of the binding between IRS-1 and Rad51 following IGF-I and serum stimulation.(D) IRS-1-Rad51 interaction evaluated in quiescent R600 cells (SFM) and at 5 min (5′/IGF), 30 min (30′/IGF), and 3 h (3 h/IGF) following IGF-I (50 ng/ml) stimulation, in the presence (+) or absence (−) of cisplatin (1 μg/ml). (E through N) Subcellular localization of IRS-1 and Rad51. Quiescent (serum starved for 48 h) (E through H) and IGF-I-stimulated (I through L) cultures of R600 fibroblasts were treated with cisplatin for 6 h. The cells were triple labeled with anti-IRS (red; rhodamine-conjugated secondary antibody) and anti-Rad51 (green; FITC-conjugated secondary antibody) antibodies and with DAPI (blue nuclear staining). (E and I) Rad51 immunolabeling; (F and J) IRS-1 immunolabeling; (G and K) DAPI nuclear staining; (H and L) superimposition of images depicted in panels E, F, and G with images from panels I, J, and K, respectively. (M and N) Digital sectioning of the fluorescent image by deconvolution software indicates that colocalization of IRS-1 and Rad51 (yellow fluorescence) is detected mostly within perinuclear regions (M). By this approach, nuclear foci of Rad51 are predominantly green. A corresponding image labeled with DAPI is shown (N). Original magnification for all images, ×100.
To further characterize the interaction between IRS-1 and Rad51, we utilized double immunocytochemical labeling with anti-Rad51 and anti-IRS-1 antibodies (Fig. 5E to N). In these experiments, subcellular localizations of the complex were examined in quiescent (SFM) or IGF-I stimulated R600 fibroblasts. These conditions were selected because the strongest IRS-1-Rad51 binding occurs in SFM, and IGF-I treatment alone or in the presence of serum strongly impairs the binding (Fig. 5C and D). Immunolabeling with an anti-Rad51 antibody revealed an abundance of Rad51 nuclear foci when cisplatin treatment was applied to IGF-I-stimulated cultures (Fig. 5I). In contrast, perinuclear Rad51 aggregates were observed when cisplatin was added to quiescent cultures (Fig. 5E). In the same microscopic fields, an anti-IRS-1 antibody, visualized as red fluorescence, showed diffuse cytoplasmic and partial nuclear labeling in the presence of IGF-I (Fig. 5F and J, respectively). In addition, intensive perinuclear labeling was detected in the absence of IGF-I stimulation (Fig. 5F). Superimposition of the images from Fig. 5E (Rad51), F (IRS-1), and G (DAPI nuclear staining) indicates possible perinuclear IRS-1-Rad51 colocalization by yellow fluorescence (Fig. 5H). The same analysis applied to IGF-I-stimulated cells (Fig. 5L) showed predominantly green fluorescence within the nuclear foci, with a few yellow aggregates detected mostly in the cytoplasm. Some colocalization of IRS-1 and Rad51 could also be detected within the nuclear compartment, but only in those cells which were characterized by an abundance of Rad51 nuclear foci (data not shown). Considering the thickness of the cell, these occasional yellow nuclear signals could well represent interference between green and red fluorescence from different layers within the cell.
To address this issue, we have utilized deconvolution software, which permits collection of fluorescent digital images from sequential 0.5-μm-thick digital sections through a single cell. The culture condition under which we could best observe both perinuclear Rad51 and Rad51 nuclear foci was cisplatin treatment of cells growing in the presence of 10% FBS without IGF-I. Figure 5M shows a representative image from such a culture double immunolabeled with anti-Rad51 (green) and anti-IRS-1 (red) antibodies. Two cells within the image are negative for Rad51 nuclear foci, and these two cells show an apparent perinuclear colocalization (yellow) of IRS-1 and Rad51. The third cell, which demonstrates Rad51 nuclear foci (green), does not show any yellow fluorescence within the nucleus, although some Rad51-IRS-1 complexes are still detectable around the nucleus. This may suggest that the interaction between Rad51 and IRS-1 occurs predominantly in the cytoplasm, under culture conditions which favor the hypophosphorylated form of IRS-1. Treatment with IGF-I applied either to exponentially growing (10% FBS) or to quiescent (SFM) cells decreases perinuclear Rad51 levels and significantly enhances the formation of Rad51 nuclear foci in the presence of cisplatin (Fig. 4, compare panels A and D; Fig. 5, compare panels E, I, and M).
Interference with the Rad51-IRS-1 complex.
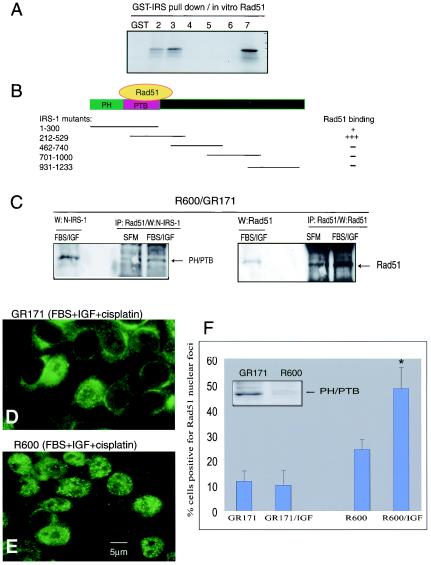
A series of overlapping truncation mutants of IRS-1 were employed in a GST pulldown assay to characterize the region responsible for the interaction with Rad51. The strongest Rad51 binding was mapped within the region between IRS-1 aa 212 to 529 (Fig. 6A). This region of IRS-1 contains the PTB domain and a short fragment of the C-terminal tail (27, 57). The other mutant capable of pulling down Rad51 is composed of the first 300 aa, which contains both the PH and PTB domains of the IRS-1. The overlapping portion between the two binding mutants contains almost the entire PTB domain, indicating that this is the most probable binding site for Rad51 (Fig. 6B). We have verified the binding between Rad51 and the PTB domain in newly generated R600 cells which were stably transfected with the PH/PTB fragment of IRS-1 (R600/GR171). As indicated in Fig. 6C, an anti-Rad51 antibody was capable of precipitating both Rad51 and the PH/PTB domain from the R600/GR171 cells. Since the PH/PTB fragment binds Rad51 both in SFM and following IGF-I stimulation, we have asked whether PH/PTB could act in a dominant-negative manner and attenuate the translocation of Rad51 into nuclear foci following IGF-I stimulation. In this scenario the PH/PTB fragment is expected to compete against the release of Rad51 from the IRS-1 perinuclear complexes after IGF-I stimulation. Figure 6D shows that in the presence of cisplatin, IGF-I treatment did not enhance the formation of Rad51 nuclear foci in R600/GR171 fibroblasts. As expected, parental R600 fibroblasts were much more efficient in directing Rad51 into cisplatin-damaged DNA (Fig. 6E). Quantitatively, only 10% of R600/GR171 fibroblasts were positive for Rad51 nuclear foci following cisplatin treatment, and this low incidence of Rad51 binding to DSBs did not change following IGF-I stimulation (Fig. 6F). In parallel experiments R600 fibroblasts responded to IGF-I stimulation by increasing the number of cells with Rad51 nuclear foci from 24 to 49% (P < 0.01).
FIG. 6.
Characterization of the IRS-1-Rad51 binding. (A) Pulldown reaction between GST-IRS fusion proteins (five overlapping IRS-1 truncation mutants [lanes 2 to 6]) and in vitro-translated, [35S]methionine-labeled, full-length RAD51. Following pulldown, two IRS-1 truncation mutants, GST-IRS-1(1-300) and GST-IRS-1(212-529), show interaction with Rad51 (lanes 2 and 3). All other mutants (lanes 4 to 6) and GST alone (lane 1) are completely negative. Lane 7, aliquot of the in vitro-translated Rad51used in pulldown reactions (positive control). (B) Diagram of five overlapping IRS-1 mutants, with their sizes and the potential Rad51 binding region. (C) Interaction between the PH/PTB domain and Rad51 confirmed by immunoprecipitation (IP) and Western blotting (W) in R600 fibroblasts stably transfected with PH/PTB cDNA in the retroviral expression vector R600/GR171. The antibody to detect the PH/PTB fragment of IRS-1 was raised against the N-terminal portion of IRS-1 (Santa Cruz). (D and E) Immunolabeling with an anti-Rad51 antibody applied to IGF-I-stimulated R600/GR171cells (D) and to parental R600 fibroblasts (E). The cells were treated with cisplatin (1 μg/ml) for 6 h. (F) Quantification of the data from panels D and E. Cells characterized by more than 10 foci per nucleus were considered positive. Results are averages from three experiments performed in duplicate (n = 6) with standard deviations. Asterisk indicates a value that is statistically significant (P < 0.05). (Inset) Western blot processed with an anti-N-terminal IRS-1 antibody to detect PH/PTB protein levels in parental R600 fibroblasts and in R600 cells stably transfected with PH/PTB cDNA under the control of the cytomegalovirus promoter (GR171).
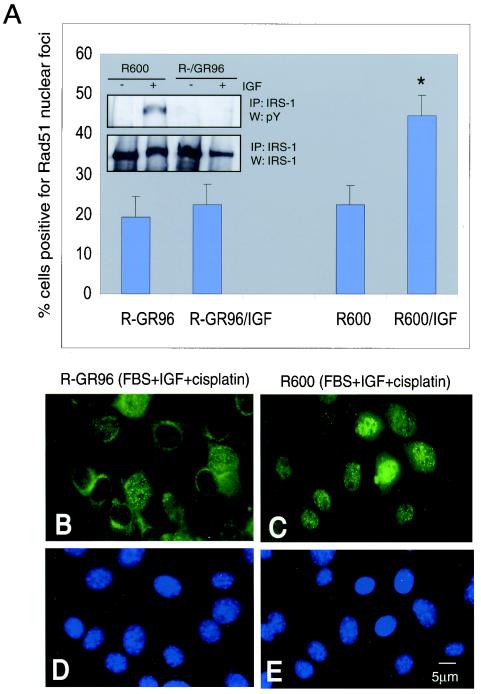
To further investigate the interaction between hypophosphorylated IRS-1 and Rad51, we utilized R− fibroblasts (IGF-IR gene knockout) stably transfected with a specific double mutant of IGF-IR (F950Y + Δ1250). An important feature of this mutant is its inability to phosphorylate IRS-1 on tyrosine residues following IGF-I stimulation (Fig. 7A inset). As shown in Fig. 7A and B, IGF-I treatment in the presence of cisplatin did not support the formation of Rad51 nuclear foci in R− fibroblasts stably expressing the F950Y + Δ1250 mutant (R−/GR96 cells), and nearly 80% of R−/GR96 cells demonstrated perinuclear Rad51 (Fig. 7B). Again, the same treatment applied in the presence of wt IGF-IR efficiently translocated Rad51 to the damaged DNA of R600 fibroblasts (Fig. 7; compare panels B and C).
FIG. 7.
Interference with Rad51-IRS-1 binding. To investigate the interaction between hypophosphorylated IRS-1 and Rad51, we utilized a specific double mutant of IGF-IR (F950Y + Δ1250), designated GR96. (A) Exponentially growing cells that express wt IGF-IR (R600 fibroblasts) or the F950Y + Δ1250 mutant of IGF-IR (R−GR96) were treated with cisplatin (1 μg/ml) for 6 h in the presence (R−GR96/IGF and R600/IGF) or absence (R−GR96 and R600) of IGF-I (50 ng/ml). Following immunofluorescent labeling with an anti-Rad51 antibody, the formation of Rad51 nuclear foci was evaluated under a fluorescent microscope. Cells characterized by more than 10 foci per nucleus were considered positive. Results are averages from three experiments performed in duplicate (n = 6). Asterisk indicates a value that is statistically significant (P < 0.05). (Inset) Immunoprecipitation (IP) and Western blotting (W) in which IRS-1 was immunoprecipitated from 500 μg of total proteins isolated from quiescent (−) or IGF-I-stimulated (+) R600 or R−GR96 fibroblasts. Corresponding blots were probed with an antiphosphotyrosine (PY) antibody and with an anti IRS-1 antibody. (B and C) Immunofluorescent labeling with an anti-Rad51 antibody. In comparison to cells which express wt IGF-IR (R600), R−GR96 cells, which do not phosphorylate IRS-1 (panel A inset), show predominantly cytoplasmic localization of Rad51. (D and E) DAPI nuclear staining in corresponding images. Original magnification, ×40.
The collected results indicate that two independent molecular manipulations which attenuate the binding between Rad51 and IRS-1, overexpression of the PH/PTB domain and hypophosphorylation of IRS-1 on tyrosine residues, inhibit the translocation of Rad51 to the site of damaged DNA. These results support the contribution of the IGF-IR/IRS-1 signaling system in controlling the recruitment of Rad51 into the damaged DNA and may explain the increased efficiency of DNA repair by homologous recombination following IGF-I stimulation.
DISCUSSION
Through the experiments described in this report, we have attempted to determine whether and how the signal from activated IGF-IR affects DNA repair via homologous recombination. The relevance of this question is derived from previous work establishing that IGF-IR sends a strong mitogenic signal (3), has overwhelming anti-apoptotic properties (30, 52), and contributes to the development of drug resistance (51). By utilizing an experimental setting specifically designed to study IGF-IR-mediated cellular responses, we have noticed that proliferating cells expressing high levels of the IGF-IR are better protected, in the presence of IGF-I, against the genotoxic action of cisplatin. This initial observation directed our attention toward two possible mechanisms: the well-established antiapoptotic properties of the IGF-IR and an unexplored possibility that the IGF-IR could facilitate DNA repair of lethal DSBs. Our results show that (i) IGF-I treatment enhances HRR in a manner that is proportional to the level of IGF-IR expression; (ii) this IGF-I-mediated response seems to be controlled at the level of Rad51 translocation to the nucleus; (iii) it involves a direct interaction between Rad51 and the major IGF-IR signaling molecule, IRS-1; and (iv) the binding is direct, as shown by a GST pulldown assay, is localized within the perinuclear region of the cell, involves the N-terminal portion of IRS-1, and is negatively regulated by IGF-I-mediated IRS-1 tyrosine phosphorylation. Finally, (v) in the absence of IGF-I stimulation, or if IGF-I treatment is applied to cells expressing an IGF-IR mutant that fails to phosphorylate IRS-1, Rad51 is partially retained in the perinuclear cytoplasm and its translocation to nuclear foci is impaired.
A highly unstable karyotype is one of the hallmarks of the transformed phenotype. It involves both changes in chromosome number and an enhanced rate of intrachromosomal rearrangements and translocations (46, 56). Cells which are prone to develop these genomic aberrations may occasionally acquire a growth advantage, which in consequence may lead to the selection of the transformed phenotype. Chromosomal rearrangements are a consequence of the loss of fidelity in repairing DNA DSBs. These lethal DNA lesions are repaired in surviving cells by two primary pathways: (i) NHEJ, considered unfaithful, which can result in the gain or loss of nucleotide sequences, and (ii) the predominantly faithful HRR (17). However, this general classification, which divides DNA repair processes into error-proof HRR and error-prone NHEJ, is not always accurate. Recent studies in a mouse model indicate that the absence of either pathway can lead to severe genomic instability (14). This could be partially explained by the fact that NHEJ, although considered unfaithful, also participates in capping chromosome ends, protecting them from end-to-end fusions (1, 8). The prominent role of HRR in maintaining chromosome stability can be also challenged. As has been demonstrated recently, enhanced drug resistance in BCR-ABL-transformed cells is caused by a combination of antiapoptotic signals and enhanced HRR. In this case, HRR introduces a high rate of mutations by a mechanism that may involve recruitment of unfaithful DNA polymerases to the site of DNA repair (45). Although it is still difficult to draw the line between faithful and unfaithful DNA repair mechanisms and their contributions to genomic instability, inherited genetic defects in HRR pathways are clearly associated with an unstable genome and a high predisposition to cancer (47).
Beyond that, very little is known about the cellular signaling mechanism(s) that could modulate the effectiveness of DNA repair. One recently reported example of such signaling regulation involves BCR-ABL-mediated enhancement of Rad51 expression. The possible contribution of this aberrant pathway to the development of drug resistance in leukemias and lymphomas suggests a functional link between nonreceptor tyrosine kinases and HRR (45). The question as to whether surface receptors with tyrosine kinase activity are also able to control HRR remains to be answered. The only connection that has been established so far includes ATM kinase and the IGF-IR (29). ATM is known to play a critical role in triggering multiple cellular responses following DNA damage, including the initial steps of HRR (31). In addition, ATM kinase controls IGF-IR promoter activity (29). This might partially explain low levels of the IGF-IR protein in cell lines from AT-defective individuals (29) and the association with growth abnormalities, insulin resistance, and neurodegeneration in ataxia telangiectasia patients (23).
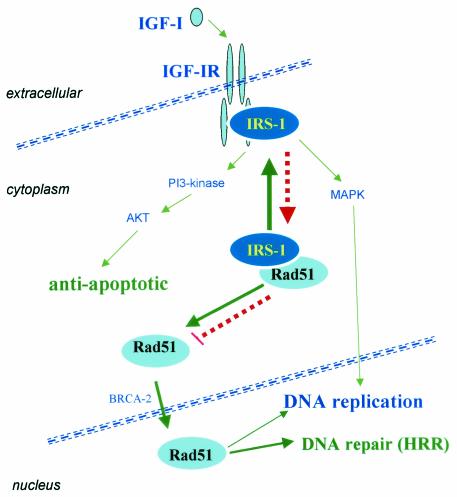
Previous experiments on the effects of the IGF-IR on cell growth and survival together with our present findings allow us to postulate a sequence of molecular events (depicted in Fig. 8) by which IGF-IR triggers multiple signals that lead to synchronized activation of (i) cell proliferation, (ii) protection from apoptosis, and (iii) DNA repair by homologous recombination. IRS-1 seems to play a central role in this model. In the absence of IGF-I stimulation (red arrows), hypophosphorylated IRS-1 stays within the perinuclear region, with Rad51 restricting its availability and potential recruitment to DSBs. Following ligand binding (green arrows), activated IGF-IR recruits and phosphorylates IRS-1 on multiple tyrosines. This dramatically decreases the affinity of IRS-1 for Rad51 and engages phospho-IRS-1 in multiple signaling events supporting IGF-I-mediated cell proliferation and cell survival. If at this time DSBs are formed, either naturally or by treatment with genotoxic agents, the cell will be ready to repair them faithfully by HRR. Therefore, IGF-IR-mediated coordination of growth responses with DNA repair may be critical to ensure the stability of the genome during normal development. Alternatively, during transformation, cells often develop alternative pathways to support proliferation (different growth factors, cytokines, their receptors, cellular and viral oncoproteins), likely leaving IRS-1 hypophosphorylated. In this stage, IRS-1 shows high affinity to Rad51 in the cytoplasm. This may result in insufficient recruitment of Rad51 to the sites of DSBs and a low level of DNA repair by homologous recombination. If proliferating cells are exposed to genotoxic agents, the excess of DSBs may trigger apoptosis, or if unfaithful repair mechanisms predominate, additional mutations could accumulate in surviving cells—possibly contributing to the selection of a malignant phenotype. In support of this notion, several reports from the literature indicate that some metastatic prostate and breast cancer cells have a tendency to lose IRS-1 (18, 38, 39). One could ask why malignant cells down-regulate a molecule which should confer a growth advantage. Our speculation is that if proliferating cancer cells do not phosphorylate IRS-1, the fraction of hypophosphorylated IRS-1 may inhibit Rad51 nuclear translocation. Since DSBs have to be repaired for cell survival, the selection pressure may favor loss of IRS-1 to facilitate the release of Rad51 and to repair DSBs. In a different scenario, unscheduled activation of NHEJ during the S phase of the cell cycle could still rescue the cells (26); however, the price to be paid may involve an increased rate of mutations.
FIG. 8.
IGF-IR signaling and DNA repair. The findings presented in this study allowed us to postulate a sequence of molecular events in which IGF-IR triggers multiple signaling events that lead to synchronized activation of: (i) cell proliferation (SHC- or IRS-1-mediated activation of Ras-mitogen-activated protein kinase [MAPK] pathways), (ii) protection from apoptosis (IRS-1-amplified activation of Akt), and (iii) enhanced DNA repair by homologous recombination. IGF-I-mediated phosphorylation of IRS-1 plays a pivotal role in this model by facilitating the recruitment of Rad51 into DSBs (see last paragraph in Discussion).
Acknowledgments
We gratefully acknowledge the comments and editorial assistance of Sidney Croul.
This work was supported by grants from NIH (RO1CA095518-01 [to K.R.] and PO1 NS36466-06 [to K.K. and K.R.]) and from the Children's Brain Tumor Foundation (to K.R.). T.S. is a Scholar of the Leukemia and Lymphoma Society and is a recipient of grant R01CA89052.
REFERENCES
- 1.Bailey, S. M., J. Meyne, D. J. Chen, A. Kurimasa, G. C. Li, B. E. Lehnert, and E. H. Goodwin. 1999. DNA double-strand break repair proteins are required to cap the ends of mammalian chromosomes. Proc. Natl. Acad. Sci. USA 96:14899-14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, J., J. P. Liu, E. J. Robertson, and A. Efstratiadis. 1993. Role of insulin-like growth factors in embryonic and postnatal growth. Cell 75:73-82. [PubMed] [Google Scholar]
- 3.Baserga, R., P. Porcu, M. Rubini, and C. Sell. 1993. Cell cycle control by the IGF-1 receptor and its ligands. Adv. Exp. Med. Biol. 343:105-112. [DOI] [PubMed] [Google Scholar]
- 4.Baserga, R., C. Sell, P. Porcu, and M. Rubini. 1994. The role of the IGF-I receptor in the growth and transformation of mammalian cells. Cell Prolif. 27:63-71. [DOI] [PubMed] [Google Scholar]
- 5.Baumann, P., and S. C. West. 1998. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23:247-251. [DOI] [PubMed] [Google Scholar]
- 6.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462-42467. [DOI] [PubMed] [Google Scholar]
- 7.Chen, G., S. S. Yuan, W. Liu, Y. Xu, K. Trujillo, B. Song, F. Cong, S. P. Goff, Y. Wu, R. Arlinghaus, D. Baltimore, P. J. Gasser, M. S. Park, P. Sung, and E. Y. Lee. 1999. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J. Biol. Chem. 274:12748-12752. [DOI] [PubMed] [Google Scholar]
- 8.d'Adda di Fagagna, F., M. P. Hande, W. M. Tong, D. Roth, P. M. Lansdorp, Z. Q. Wang, and S. P. Jackson. 2001. Effects of DNA nonhomologous end-joining factors on telomere length and chromosomal stability in mammalian cells. Curr. Biol. 11:1192-1196. [DOI] [PubMed] [Google Scholar]
- 9.D'Ambrosio, C., B. Valentinis, M. Prisco, K. Reiss, M. Rubini, and R. Baserga. 1997. Protective effect of the insulin-like growth factor I receptor on apoptosis induced by okadaic acid. Cancer Res. 57:3264-3271. [PubMed] [Google Scholar]
- 10.Davies, A. A., J. Y. Masson, M. J. McIlwraith, A. Z. Stasiak, A. Stasiak, A. R. Venkitaraman, and S. C. West. 2001. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 7:273-282. [DOI] [PubMed] [Google Scholar]
- 11.Decraene, D., P. Agostinis, R. Bouillon, H. Degreef, and M. Garmyn. 2002. Insulin-like growth factor-1-mediated AKT activation postpones the onset of ultraviolet B-induced apoptosis, providing more time for cyclobutane thymine dimer removal in primary human keratinocytes. J. Biol. Chem. 277:32587-32595. [DOI] [PubMed] [Google Scholar]
- 12.Del Valle, L., S. Enam, A. Lassak, J. Y. Wang, S. Croul, K. Khalili, and K. Reiss. 2002. Insulin-like growth factor I receptor activity in human medulloblastomas. Clin. Cancer Res. 8:1822-1830. [PubMed] [Google Scholar]
- 13.Essers, J., A. B. Houtsmuller, L. van Veelen, C. Paulusma, A. L. Nigg, A. Pastink, W. Vermeulen, J. H. Hoeijmakers, and R. Kanaar. 2002. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 21:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferguson, D. O., and F. W. Alt. 2001. DNA double strand break repair and chromosomal translocation: lessons from animal models. Oncogene 20:5572-5579. [DOI] [PubMed] [Google Scholar]
- 15.Flygare, J., F. Benson, and D. Hellgren. 1996. Expression of the human RAD51 gene during the cell cycle in primary human peripheral blood lymphocytes. Biochim. Biophys. Acta 1312:231-236. [DOI] [PubMed] [Google Scholar]
- 16.Heron-Milhavet, L., and D. LeRoith. 2002. Insulin-like growth factor I induces MDM2-dependent degradation of p53 via the p38 MAPK pathway in response to DNA damage. J. Biol. Chem. 277:15600-15606. [DOI] [PubMed] [Google Scholar]
- 17.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411:366-374. [DOI] [PubMed] [Google Scholar]
- 18.Jackson, J. G., M. F. White, and D. Yee. 1998. Insulin receptor substrate-1 is the predominant signaling molecule activated by insulin-like growth factor-I, insulin, and interleukin-4 in estrogen receptor-positive human breast cancer cells. J. Biol. Chem. 273:9994-10003. [DOI] [PubMed] [Google Scholar]
- 19.Jayanth, V. R., C. A. Belfi, A. R. Swick, and M. E. Varnes. 1995. Insulin and insulin-like growth factor-1 (IGF-1) inhibit repair of potentially lethal radiation damage and chromosome aberrations and alter DNA repair kinetics in plateau-phase A549 cells. Radiat. Res. 143:165-174. [PubMed] [Google Scholar]
- 20.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247-254. [DOI] [PubMed] [Google Scholar]
- 21.Labhart, P. 1999. Nonhomologous DNA end joining in cell-free systems. Eur. J. Biochem. 265:849-861. [DOI] [PubMed] [Google Scholar]
- 22.Lassak, A., L. Del Valle, F. Peruzzi, J. Y. Wang, S. Enam, S. Croul, K. Khalili, and K. Reiss. 2002. Insulin receptor substrate 1 translocation to the nucleus by the human JC virus T-antigen. J. Biol. Chem. 277:17231-17238. [DOI] [PubMed] [Google Scholar]
- 23.Lavin, M. F., and Y. Shiloh. 1997. The genetic defect in ataxia-telangiectasia. Annu. Rev. Immunol. 15:177-202. [DOI] [PubMed] [Google Scholar]
- 24.LeRoith, D., R. Baserga, L. Helman, and C. T. Roberts, Jr. 1995. Insulin-like growth factors and cancer. Ann. Intern. Med. 122:54-59. [DOI] [PubMed] [Google Scholar]
- 25.Ludwig, T., J. Eggenschwiler, P. Fisher, A. J. D'Ercole, M. L. Davenport, and A. Efstratiadis. 1996. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev. Biol. 177:517-535. [DOI] [PubMed] [Google Scholar]
- 26.Lundin, C., K. Erixon, C. Arnaudeau, N. Schultz, D. Jenssen, M. Meuth, and T. Helleday. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 22:5869-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myers, M. G., Jr., X. J. Sun, and M. F. White. 1994. The IRS-1 signaling system. Trends Biochem. Sci. 19:289-293. [DOI] [PubMed] [Google Scholar]
- 28.Paull, T. T., E. P. Rogakou, V. Yamazaki, C. U. Kirchgessner, M. Gellert, and W. M. Bonner. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886-895. [DOI] [PubMed] [Google Scholar]
- 29.Peretz, S., R. Jensen, R. Baserga, and P. M. Glazer. 2001. ATM-dependent expression of the insulin-like growth factor-I receptor in a pathway regulating radiation response. Proc. Natl. Acad. Sci. USA 98:1676-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peruzzi, F., M. Prisco, A. Morrione, B. Valentinis, and R. Baserga. 2001. Anti-apoptotic signaling of the IGF-I receptor through mitochondrial translocation of c-Raf and Nedd4. J. Biol. Chem. 276:25990-25996. [DOI] [PubMed] [Google Scholar]
- 31.Petrini, J. H. 2000. The Mre11 complex and ATM: collaborating to navigate S phase. Curr. Opin. Cell Biol. 12:293-296. [DOI] [PubMed] [Google Scholar]
- 32.Pierce, A. J., and M. Jasin. 2001. NHEJ deficiency and disease. Mol. Cell 8:1160-1161. [DOI] [PubMed] [Google Scholar]
- 33.Pierce, A. J., R. D. Johnson, L. H. Thompson, and M. Jasin. 1999. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13:2633-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raderschall, E., E. I. Golub, and T. Haaf. 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc. Natl. Acad. Sci. USA 96:1921-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reiss, K. 2002. Insulin-like growth factor-I receptor—a potential therapeutic target in medulloblastomas. Expert Opin. Ther. Targets 6:539-544. [DOI] [PubMed] [Google Scholar]
- 36.Reiss, K., W. Cheng, A. Ferber, J. Kajstura, P. Li, B. Li, G. Olivetti, C. J. Homcy, R. Baserga, and P. Anversa. 1996. Overexpression of insulin-like growth factor-1 in the heart is coupled with myocyte proliferation in transgenic mice. Proc. Natl. Acad. Sci. USA 93:8630-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiss, K., B. Valentinis, X. Tu, S. Q. Xu, and R. Baserga. 1998. Molecular markers of IGF-I-mediated mitogenesis. Exp. Cell Res. 242:361-372. [DOI] [PubMed] [Google Scholar]
- 38.Reiss, K., J. Y. Wang, G. Romano, F. B. Furnari, W. K. Cavenee, A. Morrione, X. Tu, and R. Baserga. 2000. IGF-I receptor signaling in a prostatic cancer cell line with a PTEN mutation. Oncogene 19:2687-2694. [DOI] [PubMed] [Google Scholar]
- 39.Reiss, K., J. Y. Wang, G. Romano, X. Tu, F. Peruzzi, and R. Baserga. 2001. Mechanisms of regulation of cell adhesion and motility by insulin receptor substrate-1 in prostate cancer cells. Oncogene 20:490-500. [DOI] [PubMed] [Google Scholar]
- 40.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubini, M., A. Hongo, C. D'Ambrosio, and R. Baserga. 1997. The IGF-I receptor in mitogenesis and transformation of mouse embryo cells: role of receptor number. Exp. Cell Res. 230:284-292. [DOI] [PubMed] [Google Scholar]
- 42.Sell, C., R. Baserga, and R. Rubin. 1995. Insulin-like growth factor I (IGF-I) and the IGF-I receptor prevent etoposide-induced apoptosis. Cancer Res. 55:303-306. [PubMed] [Google Scholar]
- 43.Sigurdsson, S., S. Van Komen, G. Petukhova, and P. Sung. 2002. Homologous DNA pairing by human recombination factors Rad51 and Rad54. J. Biol. Chem. 277:42790-42794. [DOI] [PubMed] [Google Scholar]
- 44.Skorski, T., M. Nieborowska-Skorska, K. Campbell, R. V. Iozzo, G. Zon, Z. Darzynkiewicz, and B. Calabretta. 1995. Leukemia treatment in severe combined immunodeficiency mice by antisense oligodeoxynucleotides targeting cooperating oncogenes. J. Exp. Med. 182:1645-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Slupianek, A., C. Schmutte, G. Tombline, M. Nieborowska-Skorska, G. Hoser, M. O. Nowicki, A. J. Pierce, R. Fishel, and T. Skorski. 2001. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol. Cell 8:795-806. [DOI] [PubMed] [Google Scholar]
- 46.Strauss, B. S. 2000. The stability of the genome and the genetic instability of tumors. Perspect. Biol. Med. 43:286-300. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, L. H., and D. Schild. 2002. Recombinational DNA repair and human disease. Mutat. Res. 509:49-78. [DOI] [PubMed] [Google Scholar]
- 48.Tombline, G., and R. Fishel. 2002. Biochemical characterization of the human RAD51 protein. I. ATP hydrolysis. J. Biol. Chem. 277:14417-14425. [DOI] [PubMed] [Google Scholar]
- 49.Tombline, G., K. S. Shim, and R. Fishel. 2002. Biochemical characterization of the human RAD51 protein. II. Adenosine nucleotide binding and competition. J. Biol. Chem. 277:14426-14433. [DOI] [PubMed] [Google Scholar]
- 50.Tu, X., P. Batta, N. Innocent, M. Prisco, I. Casaburi, B. Belletti, and R. Baserga. 2002. Nuclear translocation of insulin receptor substrate-1 by oncogenes and Igf-I. Effect on ribosomal RNA synthesis. J. Biol. Chem. 277:44357-44365. [DOI] [PubMed] [Google Scholar]
- 51.Turner, B. C., B. G. Haffty, L. Narayanan, J. Yuan, P. A. Havre, A. A. Gumbs, L. Kaplan, J. L. Burgaud, D. Carter, R. Baserga, and P. M. Glazer. 1997. Insulin-like growth factor-I receptor overexpression mediates cellular radioresistance and local breast cancer recurrence after lumpectomy and radiation. Cancer Res. 57:3079-3083. [PubMed] [Google Scholar]
- 52.Valentinis, B., A. Morrione, F. Peruzzi, M. Prisco, K. Reiss, and R. Baserga. 1999. Anti-apoptotic signaling of the IGF-I receptor in fibroblasts following loss of matrix adhesion. Oncogene 18:1827-1836. [DOI] [PubMed] [Google Scholar]
- 53.Valentinis, B., K. Reiss, and R. Baserga. 1998. Insulin-like growth factor-I-mediated survival from anoikis: role of cell aggregation and focal adhesion kinase. J. Cell. Physiol. 176:648-657. [DOI] [PubMed] [Google Scholar]
- 54.Valle, L. D., J. Y. Wang, A. Lassak, F. Peruzzi, S. Croul, K. Khalili, and K. Reiss. 2002. Insulin-like growth factor I receptor signaling system in JC virus T antigen-induced primitive neuroectodermal tumors-medulloblastomas. J. Neurovirol 8(Suppl. 2):138-147. [DOI] [PubMed] [Google Scholar]
- 55.Van Komen, S., G. Petukhova, S. Sigurdsson, and P. Sung. 2002. Functional cross-talk among Rad51, Rad54, and replication protein A in heteroduplex DNA joint formation. J. Biol. Chem. 277:43578-43587. [DOI] [PubMed] [Google Scholar]
- 56.Venkitaraman, A. R. 2001. Chromosome stability, DNA recombination and the BRCA2 tumour suppressor. Curr. Opin. Cell Biol. 13:338-343. [DOI] [PubMed] [Google Scholar]
- 57.White, M. F. 1997. The insulin signalling system and the IRS proteins. Diabetologia 40(Suppl. 2):S2-S17. [DOI] [PubMed] [Google Scholar]
- 58.White, M. F. 1994. The IRS-1 signaling system. Curr. Opin. Genet. Dev. 4:47-54. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347-352. [DOI] [PubMed] [Google Scholar]