Abstract
While a number of proteins are involved in elongation processes, the mechanism for action of most of these factors remains unclear primarily because of the lack of suitable in vivo model systems. We identified in yeast several genes that contain internal poly(A) sites whose full-length mRNA formation is reduced by mutations in RNA polymerase II subunit RPB2, elongation factor SPT5, or TFIIS. RPB2 and SPT5 defects also promoted the utilization of upstream poly(A) sites for genes that contain multiple 3′ poly(A) signaling sequences, supporting a role for elongation in differential poly(A) site choice. Our data suggest that elongation defects cause increased transcriptional pausing or arrest that results in increased utilization of internal or upstream poly(A) sites. Transcriptional pausing or arrest can therefore be visualized in vivo if a gene contains internal poly(A) sites, allowing biochemical and genetic study of the elongation process.
The regulation of eucaryotic gene expression can occur at different and multiple levels. Control of the elongation phase of transcription has been found to be important for a number of genes, most notably those in higher eucaryotes (14, 53). It is expected, therefore, that numerous biological controls are in place to ensure that elongation occurs to the extent and degree necessary to result in the proper levels of mRNA for the different genes in the cell. Most importantly, recent evidence has suggested that the elongation process is linked and critical to other posttranscriptional processes such as mRNA capping, splicing, polyadenylation and cleavage, and transport (7, 42, 48). Elongation can therefore be viewed as the center through which the whole quality control of mRNA formation can be integrated (33, 36, 42).
Recently a number of factors have been identified in yeast and other organisms as either components of the elongating polymerase or as possible modulators of the process of elongation. Biochemical and in vitro studies have demonstrated or suggested a requirement for several of these factors in elongation. As a result of the RNA Pol II structure being recently determined, it is clear that the RPB2 subunit of RNA Pol II may play multiple roles during elongation (16). In vitro evidence confirms a key role for RPB2 because RNA polymerase II (Pol II) containing either the rpb2-10 or rpb2-4 protein fails to elongate well in vitro (41). In particular, the rpb2-10 protein causes arrest at known mammalian pause sites and requires elongation factor TFIIS to overcome the arrest. Moreover, the rpb2-10 allele in combination with a dst1 (encoding TFIIS) deletion causes severe defects in the synthesis of most mRNA in vivo (29) and reduces the induction of a number of other genes (56). These observations suggest that in vivo rpb2-10 actually causes promiscuous arrest of RNA Pol II that requires TFIIS to suppress and overcome.
Other important factors involved in elongation are the yeast proteins SPT5 and SPT4, whose higher eucaryotic orthologs are components of the DSIF complex. SPT5 and SPT4 have been found to be both activators and repressors of elongation. The DSIF complex is known to be important in repressing elongation in vitro and appears to do so by its interaction with the hypophosphorylated form of the RNA Pol II carboxy-terminal domain (54, 55, 59, 60). In the control of hsp70, Drosophila SPT5 is known to be recruited to the transcribing polymerase, although it may also have a role in forming the paused RNA Pol II complex at hsp70 (1, 27). Also, depleting SPT5 in a TAT-dependent system promotes pausing and transcriptional termination, indicating that SPT5/SPT4 can have positive roles on elongation in addition to previously described negative roles (6). It has also recently been shown that SPT4 plays a positive role in elongation in yeast (43). Various genetic analyses have implicated SPT5 and SPT4 in controlling elongation in vivo (22) and initiation (50). In addition, certain spt5 alleles can be suppressed by rpb2-10 or by the presumed slowing of elongation (22). The physical association of the SPT5/SPT4 complex with RNA Pol II further confirms its importance in RNA Pol II function (22).
In the yeast Saccharomyces cerevisiae, the principal limitation in characterizing the action of factors involved in elongation has been the lack of identification of specific genes whose expression is clearly affected by defects in these elongation factors. For instance, no particular gene whose elongation is affected by these factors has been identified (29, 45, 56), although it has been suggested that transcription of long genes and genes with high G+C content is defective with certain elongation defects (9, 43). Similarly, whole genome microarray analysis with dst1 and rpb9 alleles did not yield particular genes controlled at the level of elongation (23). Moreover, chromatin immunoprecipitation analysis was unable to verify in vivo that deletion of TFIIS resulted in increased RNA Pol II occupancy even with a gene whose transcriptional elongation was apparently impaired (28). These results suggest that the pausing that is presumed to be occurring in vivo with elongation defects is difficult to detect either as effects upon gene expression or in enhanced RNA Pol II occupancy. Because many putative elongation factors (SPT5/SPT4, PAF1 complex, TFIIE/IIF/IIH, RPB subunits, TFIIS, and CCR4-NOTs) (13, 14, 17, 19, 45, 49-51) can also play roles in affecting initiation, it is of paramount importance to identify genes in yeast that are regulated at the level of elongation.
Previously, the lacZ gene, albeit an Escherichia coli gene, has been found to be regulated at the level of elongation when expressed in yeasts that are defective in the HPR1 gene (8). The hpr1 deletion reduces transcription through lacZ, apparently because of both its high G+C content and its extreme length (9). We have consequently examined whether other defects in elongation factors also affected expression through lacZ. Our results show that defects in SPT5 and RNA Pol II subunit RPB2 impair transcription through the E. coli lacZ gene but by a different mechanism than that found for hpr1. We show that transcription through genes containing internal polyadenylation sequences is particularly sensitive to spt5-4 and rpb2-10 defects. Most importantly, we identify several bona fide yeast genes containing naturally occurring internal poly(A) sites whose elongation is impaired by rpb2-10, spt5-4, and dst1 defects. Our model is that spt5-4, rpb2-10, and dst1 alleles, which would be expected to cause pausing or arrest during elongation, result in increased usage of internal poly(A) sites. These results are consistent with those of other studies linking downstream pause sites to poly(A) site utilization (2, 5, 61, 62) and imply that elongation pausing or arrest can be studied biochemically and genetically in vivo by using genes containing internal poly(A) sites.
MATERIALS AND METHODS
Yeast strains, growth conditions, and enzyme assays.
The yeast strains used are listed in Table 1. Strains containing spt5 or spt4 alleles are all isogenic to FY1642 (wild type) except as indicated (22). The Z96 set of strains has been described previously (41). Yeast were grown on medium containing 1% yeast extract-2% Bacto Peptone, minimal medium, or CAA-U− medium (31) supplemented with an appropriate carbon source, as indicated in the figures. β-Galactosidase activities were determined as described previously (31).
TABLE 1.
Yeast strains used
| Strain | Genotype |
|---|---|
| FY1642 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 SPT5-FLAG |
| FY1668-uH | MATa his4-912δ lys2-128δ spt5-4 ura3::HIS3 |
| FY1635 | MATα his4-912δ lys2-128δ leu2Δ1 ura3-52 spt5-242 |
| L615 | MATa his4-912δ lys2-128δ ura3-52 ade2-1 trp5 can1-100 spt5-25 |
| FY300 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 spt5-194 |
| GHY180 | MATα ura3-52 leu2Δ1 his4-912δ lys2-128δ spt4Δ2::HIS3 |
| FY276-uT | MATa leu2Δ1 ura3::TRP1 his4-9128 lys2-128δ spt5-8 |
| AYW3-1B1 | MATa leu2 trp1 ura3 his3 |
| AYW3-3D1 | MATa leu2 his3 ura3 leu2-k::ADE2-URA3-leu2-k hpr1Δ3::HIS3 |
| Z96 | MATa ura3-52 leu2-3,112 his3Δ200 rpbΔ297::HIS3 [pRP214 (LEU2 RPB2)] |
| Z100 | Isogenic to Z96 except [pRP2-4L (LEU2 rpb2-4)] |
| Z106 | Isogenic to Z96 except [pRP2-10L (LEU2 rpb2-10)] |
| Z103 | Isogenic to Z96 except [pRP2-7L (LEU2 rpb2-7)] |
| DY106-u | Isogenic to Z96 except dstl::hisG |
| DY108 | Isogenic to Z106 except dst1::hisG |
RNA analyses.
Quantitative S1 nuclease protection assays were conducted as previously described (13) using the oligonucleotides listed in Table 2. Control experiments in each case indicated that at the concentration of S1 nuclease used no radioactively labeled oligonucleotide remained if no RNA was present and that the S1 nuclease assay was linear over the concentration of RNAs used.
TABLE 2.
Characteristics of oligonucleotide probes used
| Probe | Sequence | Location (nt) |
|---|---|---|
| ADH2 | ||
| 5′ | 5′-GTATTACGATATAGTTAATAGTTGATAGTTGATTGTATGCTTTTTGTAGCTTGATATTCTATTTA CCAAGAAGAAAC-3′ | −80 to −4 |
| 3′ | 5′-GGCATACTTGATAATGAAAACTATAAATCGTAAAGACATAAG-3′ | +1126 to +1167 |
| NOT1 | ||
| 5′ | 5′-GCGGCCTGTTTTTCCTTTGATGTTTCTAAATTAGATGCTGTGTTCAAATCACGGGTCC-3′ | +18 to +71 |
| 3′ | 5′-GGCGGATTGGTCATCTTGTTCACTGGTTGTGTATTTTTTGG-3′ | +6245 to +6285 |
| YAT1 | ||
| 5′ | 5′-GGCGCTCGTCCTGCAGGGGTTCCACGCGTGCCAGGTAGCGG-3′ | +51 to +91 |
| 3′ | 5′-GCGGTCGATCTCCAGCAGCGACTTTTCCATGAGCGACGCAAACCGAGCAGTCTGGCG-3′ | +1876 to +1932 |
| RNA14 | ||
| 5′ | 5′-GGCTCTGCGACTTTGTCCGCAGAGGGATATAGTAAATCAGGAGTCGTAGAG-3′ | +5 to +59 |
| 3′ | 5′-GCGCATCGAGTAAATTTGTATTAAAATATTGACGTTTTGG-3′ | +1927 to +1966 |
| CBP1 | ||
| 5′ | 5′-CCCTGCTGCTGTGGTTGATTCGTCGCAAGGTCCTGGTAGGTACCATTTTTATAAACCTCTCGG-3′ | +32 to +94 |
| 3′ | 5′-GCCGTTCATCTTAAGTAACGTTTGACAGCCGACACACCATGC-3′ | +1929 to +1970 |
| AEP2 | ||
| 5′ | 5′-CGGGGATAGACAGAATGACATATTGTGCTATTCGGCAGTACACCAAATTCACGCAGCG-3′ | +86 to +143 |
| 3′ | 5′-CCCCGGTTTGAAACTCCTTAAAACATCAATTCCAAGCGGG-3′ | +1401 to +1440 |
| lacZ | ||
| 5′ | 5′-GGATCCGGTCATTATTAATTTAGTGTGTGTATTTGTGTTTGCGTGTCTATAGAAGTATAGTA-3′ | −49 to +12 |
| 0.5-kb | 5′-GCGCTCAGGTCAAATTCAGACGGCAAACGACTGTCCTGGCC-3′ | +487 to +527 |
| 1.5-kb | 5′-CGGGAAGGGCTGGTCTTCATCCACGCGCGCGTACATCGGGC-3′ | +1502 to 1542 |
| 2.3-kb | 5′-CGCCAATGTCGTTATCCAGCGGTGCACGGGTGAACTGATCGC-3′ | +2345 to +2386 |
| 3′ | 5′-CCGCGTGCAGCAGATGGCGATGGCTGGTTTCCATCAGTTGC-3′ | +2898 to +2938 |
Total RNA and poly(A) mRNA were purified and Northern blotting was conducted as described previously (15, 18). mRNA was verified as being enriched in polyadenylated RNA because only 1% of the total RNA was recovered, rRNA was substantially reduced as evidenced by ethidium bromide staining, and one-fifth the abundance of poly(A) was required to give a much stronger mRNA signal following Northern analysis than that required for total RNA. Oligonucleotides were radiolabeled at their 5′ end with T4 polynucleotide kinase as previously described (10).
The rates of degradation for GAL1-lacZ, RP51-lacZ, RP51-ADH2, and GAL1 RNA were determined following growth of yeast on galactose-containing medium for 3 h (15 min for GAL1) and shifting to medium containing glucose, as described previously (10). For the analysis of the polyadenylated species of GAL1 mRNA, GAL1 gene expression was induced for 15 min by shifting yeast from raffinose-containing medium to galactose-containing medium. The newly synthesized GAL1 mRNA was detected by using an RNase H assay in which an 18-nt DNA probe (5′-GCCATTTGGGCCCCCTGG-3′) complementary to the sequences 133-bp upstream of the GAL1 translation stop codon was hybridized to total yeast RNA prior to RNase H cleavage (52). The resultant GAL1 3′ polyadenylated species were detected by Northern analysis by using a probe that was complementary to the 3′ end of GAL1 (5′-GCCCAATGCTGGTTTAGAGACGATGATAGCATTTTCTAGCTCAGCATCAGTGATCTTAGGG-3′).
RESULTS
The rpb2-10, rpb2-4, spt5-4, and spt4 alleles affect lacZ reporter expression irrespective of the promoter.
A previous study of the hpr1 deletion showed that it affected expression of the PHO5-lacZ or GAL1-lacZ gene in yeast although hpr1 had no effect on endogenous PHO5 or GAL1 expression (8). These and other results led to the conclusion that HPR1 was required for efficient transcriptional elongation through the lacZ gene (8). Based on the observation that the lacZ gene might contain specific sequences or structures that interfered with elongation, we conducted an analysis to determine whether defects in other factors known or presumed to play roles in transcriptional elongation also failed to properly express lacZ. We used three lacZ reporter constructs, each of which contained a different promoter: ADH2-lacZ, FKS1-lacZ, or GAL1-lacZ (31). These promoters were chosen to represent genes that are constitutively expressed (FKS1), inducible (GAL1), or subject to derepression by nonfermentative growth (ADH2). We subsequently analyzed the effects of defects in transcription elongation and initiation factors with each of these three reporters. As was found for hpr1, the rpb2-10, rpb2-4, spt5-4, and spt4 alleles conferred large decreases in lacZ expression for all reporters tested (Table 3). These effects were not likely due to effects on the respective promoters, as spt5-4 does not reduce GAL1 or ADH2 expression (data not shown; see Fig. 4A). rpb2-10 is known to reduce GAL1 steady-state mRNA but did so by only 1.4-fold (56), and it had no effect on ADH2 derepression (see Fig. 4A).
TABLE 3.
Effects of elongation defects on lacZ-reporter activities
| Strain | Relative β-galactosidase activity witha:
|
||
|---|---|---|---|
| FKS1-lacZ | ADH2-lacZ | GAL1-lacZ | |
| WT | 100 | 100 | 100 |
| hpr1 | 2.3 | 1.6 | 2.2 |
| rpb2-4 | 20 | 12 | 5.5 |
| rpb2-10 | 24 | 13 | 15 |
| rpb2-7 | ND | 290 | 56 |
| spt5-4 | 23 | 2.9 | 4.1 |
| spt5-8 | 65 | 35 | 56 |
| spt5-242 | ND | 59 | 51 |
| spt4 | 1.5 | 4.9 | 0.50 |
β-Galactosidase activities were determined for each allele and its corresponding isogenic wild-type parent containing the lacZ reporter as indicated. In order to ease comparison between strains, all wild-type parent values were set to 100 for each reporter. FKS1-lacZ was assayed on medium containing 4% glucose, ADH2-lacZ was assayed on 2% ethanol-2% glycerol, and GAL1-lacZ was assayed on 2% galactose-2% raffinose. All values represent the averages of the results for five separate transformants. The SEMs were less than 20%. ND, no data.
FIG. 4.
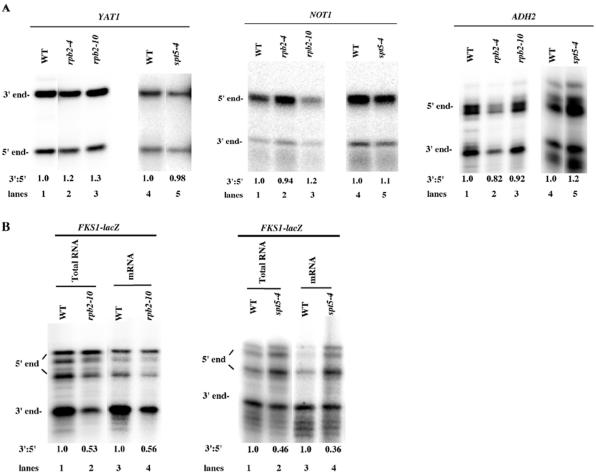
spt5 and rpb2 alleles do not affect genes lacking internal poly(A) sites and do affect lacZ containing internal poly(A) sites. (A) spt5 and rpb2 alleles do not affect elongation through the ADH2, NOT1, and YAT1 genes. Cells were grown on 1% yeast extract-2% Bacto Peptone medium containing 4% glucose (for NOT1), 2% galactose-2% raffinose (for YAT1), or 2% ethanol (for ADH2). 5′- and 3′-end RNA levels were quantitated as described for Fig. 1 by using 5′ and 3′ probes specific to ADH2, NOT1, and YAT1, respectively (Table 2). To quantitate YAT1 5′ and 3′ RNA levels, yeast strains were transformed with a GAL1-YAT1 plasmid (9). The values represent the data as presented, but repetitions showed no significant differences. (B) rpb2-10 and spt5-4 alleles reduce full-length lacZ mRNA formation by increasing shortened lacZ mRNA formation. Both total RNA (left panel, lanes 1 and 2; right panel, lanes 1 and 2) and poly(A) RNA (left panel, lanes 3 and 4; right panel, lanes 3 and 4) were extracted and analyzed by using an S1 nuclease protection assay. S1 nuclease protection assays, growth conditions, and calculations were as described for Fig. 1. The values represent the data as presented. Shown are wild-type (WT) strain Z96 (left panel, lanes 1 and 3), rpb2-10 strain Z106 (lanes 2 and 4), wild-type strain FY1642 (right panel, lanes 1 and 3), and spt5-4 strain FY1668-uH (lanes 2 and 4).
Another rpb2-7 allele, which like rpb2-4 and rpb2-10 confers 6AU sensitivity (41), did not display a consistent major effect on the lacZ reporters (Table 3). Other alleles of SPT5 (spt5-8 and spt5-242) had much less effect on lacZ expression (Table 3). Moreover, defects in the elongation factor ELP1 (38) or SPT16 (37) did not affect lacZ expression (data not shown). Similarly, defects in a number of transcription initiation factors, such as CCR4-NOT complex components, RNA Pol II holoenzyme components SRB9, SRB10, SRB11, SIN4, and GAL11, and SAGA components ADA2 and GCN5, did not display decreases in lacZ gene expression for each reporter plasmid (4, 11, 12, 31; data not shown). We conclude that the rpb2-10, rpb2-4, spt5-4, and spt4 alleles are affecting an apparently postinitiation step in the expression of the lacZ gene. Importantly, the rpb2-4- and rpb2-10-containing RNA Pol IIs have been shown in vitro to display increased transcriptional arrest, whereas rpb2-7-RNA Pol II did not (41).
The rpb2-10, rpb2-4, spt5-4, and spt4 alleles reduce the ability of RNA Pol II to form full-length lacZ mRNA.
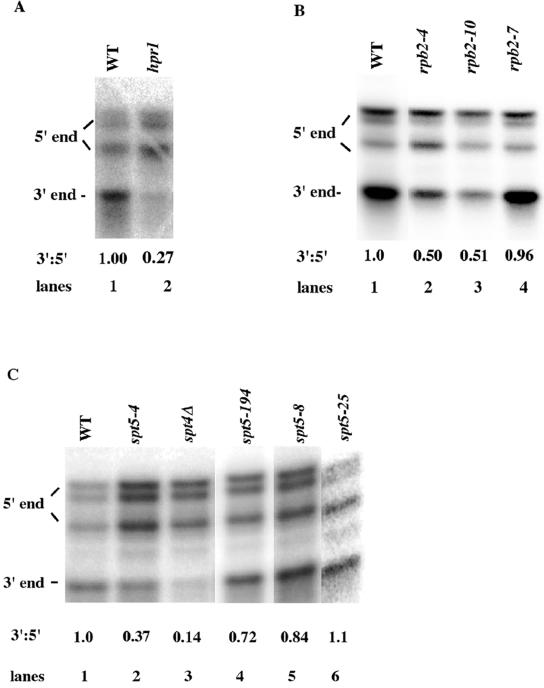
To address whether transcriptional elongation through the lacZ gene was being impaired by the above-described rpb2, spt5, and spt4 alleles, we used the quantitative S1 nuclease protection assay (13) to identify the abundance of lacZ transcripts that were full length. To do this, we compared the abundance of total lacZ mRNA to the abundance of full-length lacZ mRNA. Previously, it was shown by using Northern analysis that an hpr1 deletion blocked the synthesis of lacZ mRNA (8). In our assay system, hpr1 similarly displayed an inability to form full-length lacZ mRNA regardless of the promoter. Comparing an hpr1 strain to its isogenic parent, we observed fourfold less full-length lacZ mRNA (corresponding to 3′-end bands) than total lacZ RNA (corresponding to 5′-end bands) (Fig. 1A and data not shown).
FIG. 1.
S1 nuclease protection analysis of the effects of HPR1, RPB2, SPT4 and SPT5 defects on full-length lacZ mRNA formation. lacZ 5′- and 3′-end RNA levels were quantified by using an S1 nuclease protection assay with probes directed against the 5′ end of lacZ RNA and the site 2.9 kb downstream of the lacZ initiation site, respectively (Table 2). Total RNA was extracted, and the ratios of 3′ to 5′ RNA levels were quantified by using a phosphorimager. The values were normalized based on the ratio for the wild type, and the average ratio is given below each panel except as indicated. The multiple 5′ends that are visualized resulted from multiple initiation sites. Changes in the ratios of these 5′ band intensities appear to vary somewhat with the strain background. (A) hpr1 reduces full-length lacZ mRNA formation. Strains AYW3-1B1 (WT, lane 1) and AYW3-3D1 (hpr1, lane 2) containing plasmids expressing FKS1-lacZ were grown on 4% glucose-containing CAA-U− medium. The values represent the averages of the results of five determinations, and the SEM for hpr1 was 20%. (B) rpb2-4 and rpb2-10 alleles reduce full-length lacZ mRNA formation. The growth conditions with FKS1-lacZ and the calculations for the results were as described for panel A. Lane 1, WT strain Z96; lane 2, rpb2-4 strain Z100; lane 3, rpb2-10 strain Z106; and lane 4, rpb2-7 strain Z103. The values represent the averages of the results of five determinations for rpb2-4 with an SEM of 4%, four determinations for rpb2-10 with an SEM of 7%, and a single determination for rpb2-7. Similar results for the analysis of ADH2-lacZ mRNA expression were observed for rpb2-7. (C) spt5-4 and spt4 alleles reduce full-length lacZ mRNA formation. The growth conditions with FKS1-lacZ and the calculations for the results were as described for panel A. Lane 1, WT strain FY1642; lane 2, spt5-4 strain FY1668-uH; lane 3, spt4 strain GHY180; lane 4, spt5-194 strain FY300; lane 5, spt5-8 strain FY276-uT; and lane 6, spt5-25 strain L615. The spt5-4 value represents the average of the results for eight determinations, with an SEM of 8%. The other values represent the results for single determinations, although similar results were obtained for these defects with the analysis of ADH2-lacZ mRNA expression.
As shown in Fig. 1B and C, the rpb2-10, rpb2-4, spt5-4, and spt4 alleles which displayed consistent reductions in overall lacZ reporter expression concomitantly displayed reduced levels of full-length lacZ mRNA formation compared to the quantity of total lacZ mRNA that was present (Fig. 1B and C). In contrast, rpb2-7 (Fig. 1B), spt5-8, spt5-25, and spt5-194 (Fig. 1C) had no significant effect or less effect on formation of full-length lacZ mRNA. The same results obtained with FKS1-lacZ (Fig. 1) were obtained with ADH2-lacZ (data not shown), indicating that these effects were independent of the promoter. These data confirm a direct correlation between the effect these elongation factors have on lacZ reporter expression and the formation of full-length lacZ mRNA. Importantly, only the rpb2 alleles which displayed reduced ability to transcribe through elongation blocks in vitro (41) failed to form full-length lacZ mRNA in vivo.
Note that, in agreement with the results of other studies (32, 58), these S1 analyses indicate that, due to differences in length and sequence, oligonucleotide probes can display different degrees of stable binding to the same mRNA and hence different apparent levels of mRNA. Because of these inherent differences in stability of binding between the individual probes and a given mRNA, only the ratios of 5′ to 3′ mRNA levels can be compared between a particular mutant and its isogenic parent. Importantly several of these mutants can affect the overall abundance of a particular mRNA that results in either an increased or reduced level of total mRNA levels relative to that of the wild type (for example, see spt5-4, Fig. 1C and 3B). The rpb2-4 and rpb2-10 alleles also reduced total FKS1-lacZ expression (Fig. 1B). However, as we were interested in comparing the relative abilities of different elongation defects to reduce the formation of the full-length mRNA, alterations in the level of initiation of the transcript did not affect these comparisons.
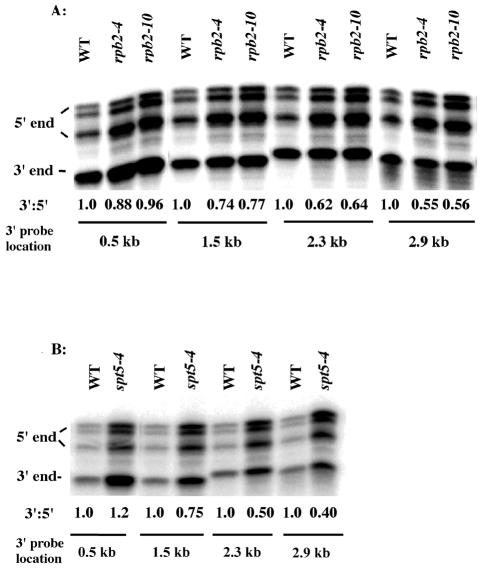
FIG. 3.
spt5-4 and rpb2 alleles affect full-length lacZ RNA formation at multiple sites. The 3′ lacZ probes for conducting the S1 nuclease protection assays are listed in Table 2. In all cases, the 5′ probe corresponded to sequences at the initiation codon of lacZ. Growth conditions, assays, and analyses were as described for Fig. 1. The rpb2 (A) and spt5-4 (B) strains contained FKS1-lacZ. The values represent the averages of two experiments, and the SEMs were 10% or less. Note that the rpb2-4 and rpb2-10 alleles resulted in less total FKS1-lacZ gene expression than for the wild type, which necessitated loading fourfold more rpb2-4 and rpb2-10 RNA than for the wild type in panel A. Equivalent amounts of total RNA were analyzed in panel B for the wild type and spt5-4.
The effect on lacZ mRNA formation is not due to increased mRNA 3′-end degradation.
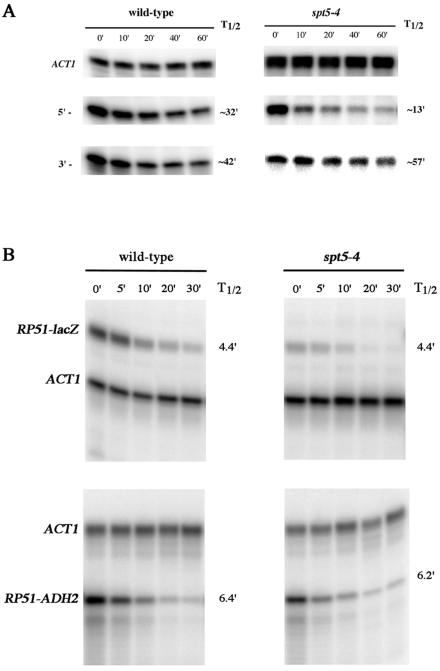
The reduced level of formation of full-length lacZ mRNA could be attributed either to a block in expression through the lacZ gene or to increased degradation of the 3′ end of the mRNA in comparison to the degradation of the 5′ end. To address this latter possibility, we utilized the GAL1-lacZ reporter to determine the rate of degradation of the 5′ and 3′ ends of the lacZ mRNA following the shutting off of GAL1-lacZ transcription by growth on glucose-containing medium. As shown in Fig. 2A, the spt5-4 allele had little effect on the stability of the full-length lacZ mRNA (3′-end probe). Note that with the 5′ probe, in the spt5-4 strain, the rate of degradation of the lacZ RNA was actually enhanced relative to that in the wild-type strain. This finding may be the result of higher rates of degradation for RNAs that are not full length, as identified with the 5′ probe, and which are in greater abundance in the spt5-4 background. A similar analysis showed that in the rpb2-10 background full-length lacZ RNA had a half-life of 50 min, whereas the 5′-end RNA had a half-life of 20 min, and that in the rpb2-4 background full-length RNA had a half-life of 41 min, while the 5′-end RNA had a half-life of 26 min. These data indicate that the spt5-4, rpb2-10, and rpb2-4 alleles are not reducing full-length lacZ RNA expression by enhancing the degradation rate of the full-length mRNA relative to all RNAs containing the 5′ end.
FIG. 2.
spt5-4 does not preferentially enhance full-length lacZ and RP51-lacZ mRNA half-lives. mRNA half-life determinations were conducted as described in Materials and Methods (10). (A) GAL1-lacZ mRNA was detected at the 5′ end with the 0.5-kb lacZ probe and at the 3′ end with the 2.3-kb lacZ probe (Fig. 3). (B) RP51-lacZ and RP51-ADH2 RNAs were detected, respectively, with the 0.5-kb lacZ probe 2 (Fig. 5A) and with the probe 1 corresponding to the RP51-ADH2 5′ intron junction (Fig. 5A). Quantitation of ACT1 RNA levels was used to standardize for loadings. The wild-type strain was FY1642, and the spt5-4 strain was FY1668.
The block to transcriptional elongation in lacZ occurs at multiple sites.
We subsequently examined where in the lacZ gene the block to elongation was occurring by using the S1 nuclease protection assay with probes spaced across the lacZ gene. In backgrounds with mutations in spt5-4, rpb2-4, and rpb2-10, lacZ mRNA synthesis appeared unimpeded through the first 500 bp of the lacZ gene (Fig. 3). However, with probes at 1.5, 2.3, and 2.9 kb, decreased levels of lacZ 3′-end mRNA formation became apparent. In moving from 1.5 to 2.9 kb, at each step an additional decrease in lacZ 3′-end mRNA formation was observed, suggesting the existence of multiple sites for blockage of mRNA synthesis (Fig. 3).
spt5 and rpb2 defects may enhance use of cryptic poly(A) sites in lacZ.
The above observations indicate that lacZ mRNA formation is particularly sensitive to SPT5 and RPB2 defects. One model to explain these data is that transcriptional pausing or arrest caused by the spt5-4, spt4, rpb2-10, and rpb2-4 alleles results in increased utilization of known cryptic poly(A) sites located within the lacZ gene (8, 44). An alternative hypothesis for how hpr1 affects lacZ has been suggested: that the long length of lacZ or its high G+C content contributes to its impaired transcription (9). spt5-4 and rpb2-10 had no effect on transcription through the native YAT1 gene, which is extremely G+C rich (Fig. 4A). hpr1 and spt4 have both been shown to reduce overall YAT1 gene expression, presumably by effects on elongation (9, 43). We also found that spt5-4 reduced the overall expression of YAT1 relative to that of the wild type by about threefold (data not shown). Our results (Fig. 4A), in contrast, indicate that the spt5-4 allele does not specifically reduce the abundance of full-length YAT1 mRNA relative to the levels of the initiated mRNA. Likewise, spt5-4 and rpb2-10 did not affect transcription through NOT1, a 6.5-kb transcript (Fig. 4A), suggesting that it is not the length per se of lacZ which impedes expression. Finally, rpb2-4, rpb2-10, and spt5-4 did not affect full-length ADH2 expression (Fig. 4A), although they all affected ADH2-lacZ expression (Table 3 and data not shown).
If the poly(A) usage model were correct, the incompletely formed lacZ RNA would be polyadenylated. We would expect, therefore, that when polyadenylated enriched RNA is analyzed, both spt5-4 and rpb2-10 would reduce the formation of full-length lacZ mRNA to the same extent that they reduce full-length lacZ RNA formation when total RNA is analyzed. As shown in Fig. 4B, rpb2-10 and spt5-4 affected the formation of full-length lacZ polyadenylated RNA to the same degree as they affected the formation of full-length lacZ RNA isolated from total RNA. These results support the model that spt5-4 and rpb2-10 enhance utilization of the cryptic poly(A) sites in lacZ (44) and do not simply cause RNA Pol II to cease transcription, resulting in shortened RNAs lacking poly(A) tails. Northern analysis, however, was unable to detect discrete shortened RNAs, probably because of multiple diffuse ends (data not shown).
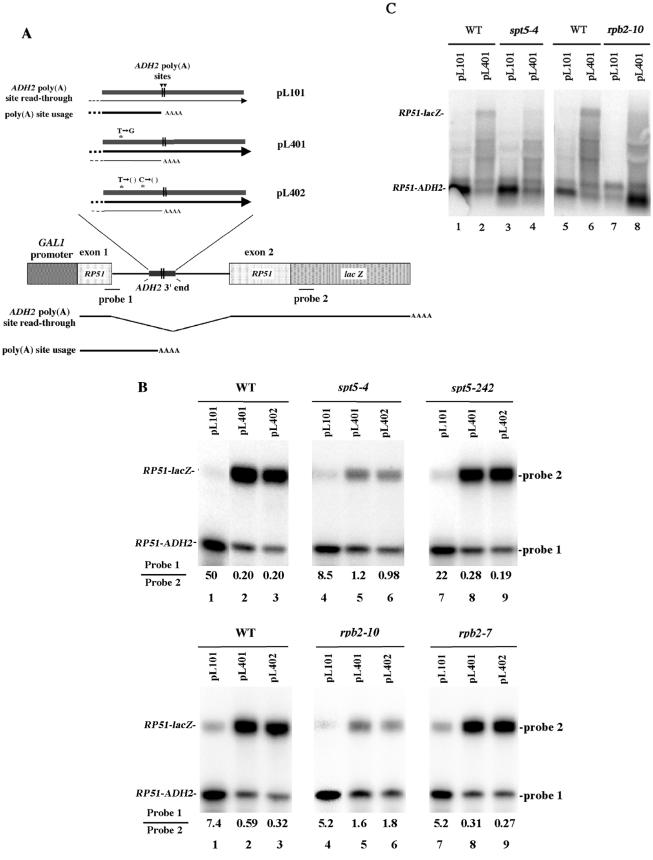
spt5-4 and rpb2-10 enhance utilization of defective ADH2 poly(A) sites.
If spt5-4 and rpb2-10 were causing increased pausing or arrest that results in subsequent upstream poly(A) site utilization, then it would be expected that these mutations should be able to enhance the use of known defective poly(A) sites internal to genes. To test this hypothesis, we utilized the previously described ADH2 defective poly(A) sites that have been used in a poly(A) site usage assay (26). For this assay, as shown in Fig. 5A, the 3′ end of the ADH2 gene containing its poly(A) signaling sequences was embedded within the RP51 intron. Usage of the ADH2 poly(A) cleavage site promotes the formation of a short mRNA that does not include downstream RP51 sequences that are fused to lacZ (26). Read-through of the poly(A) site, on the other hand, promotes splicing of the intron and synthesis through lacZ. We monitored these two alternative events by quantitating with an S1 nuclease protection assay the relative amount of mRNA that is expressed by using either probe 1, which overlaps the 5′ RP51 junction of the exon and intron [thus measuring mRNA resulting from ADH2 poly(A) site usage], or probe 2, which is at 150 bp within the lacZ gene [thus measuring RNA in which the poly(A) site has not been used and splicing has occurred]. As shown in Fig. 3, transcription through the first 500 bp of lacZ is unaffected by spt5-4 or rpb2-10 alleles.
FIG.5.
rpb2-10 and spt5-4 enhance utilization of defective ADH2 poly(A) sites embedded within the RP51 gene. (A) Results of the internal ADH2 poly(A) site assay as previously described (26). Plasmid pL101 contains the RP51 locus fused to the lacZ gene in which the 3′ end of ADH2 [containing the signals for poly(A) cleavage and adenylation] is inserted into the RP51 intron. ADH2 poly(A) site usage results in a short transcript ending in the intron that will be detected with probe 1 (5′-GCCTCCTTTAGTCCATATTAACATACCATTTTGTTATTGC-3′; see panel B, lane 1). Plasmids pL401 and pL402 contain the same ADH2 sequence inserted into the intron of RP51 except that each contains mutations in the ADH2 poly(A) recognition sequences resulting in substantial read-through and the formation of a full-length mRNA transcript that is detectable with probe 2 (5′-GCGCCATTCGCCATTCAGGCTGCGCAACTGTTGGGAAGGGCGATCGGTGCGGGCCAGAA-3′; see panel B, lanes 2 and 3). (B) S1 nuclease protection analysis of the relative levels of read-through and short transcripts. Strains were pregrown in 4% glucose-containing CAA-U− medium at 30°C for 20 h and then shifted to CAA-U− medium with 2% galactose-2% raffinose at 30°C for 4 h. RNA was subjected to S1 nuclease protection assays as described for Fig. 1 using probes 1 and 2 (as described for panel A). The values represent the data as presented. Other repetitions resulted in the same trends. Note that because of differential effects of the spt5-4 and rpb2-10 alleles on RP51-lacZ gene expression from the pL401 and pL402 plasmids compared to those from pL101, only conclusions about the relative abundances of RP51-lacZ and RP51-ADH2 RNA can be made. (C) Northern analysis of the effect of elongation defects on full-length RP51-lacZ and truncated RP51-ADH2 RNA levels. Total RNA was isolated as described for panel B, and probe 1 and RP51 probe 2 were used to detect RNA. The RNA band just above the RP51-ADH2 RNA species most likely represents the native RP51 RNA, and the two faint bands above that represent nonspecific hybridization to the 18S and 25S rRNA species.
Three plasmids containing this setup were used: pL101, which has a wild-type ADH2 3′-end sequence; pL401, which contains a T to G alteration upstream of the ADH2 poly(A) site that blocks ADH2 poly(A) site usage by 10-fold; and pL402, which contains two single nucleotide deletions upstream of the ADH2 poly(A) site that also block poly(A) site usage (Fig. 5A). As shown in Fig. 5B (upper and lower panels, lane 1), in a wild-type strain, with pL101 the ADH2 poly(A) site is used and very little read-through mRNA is made. With both pL401 and pL402 (lanes 2 and 3, respectively), poly(A) site usage is substantially weakened at ADH2 and corresponding read-through into the lacZ gene occurs. These results are the same as those previously obtained by Hyman et al. (26).
In an spt5-4 strain, however, the defective poly(A) sites in pL401 and pL402 display increased usage. The ratio of ADH2 poly(A) site usage RNA (probe 1) to that of read-through RNA (probe 2) increased about five- to sixfold in both cases relative to the ratio observed in the wild type (Fig. 5B, upper panel; compare lanes 5 and 6 to lanes 2 and 3). In contrast, the spt5-242 allele, which does not affect lacZ expression, did not have a corresponding effect on poly(A) site usage in pL401 and pL402 (Fig. 5B, upper panel, lanes 8 and 9). Exactly the same results were obtained for the rpb2 alleles as were observed for spt5. rpb2-10, which reduces lacZ expression, increased the ratio of poly(A) site usage RNA (probe 1) to that of read-through RNA (probe 2) by about three- and fivefold relative to that for the wild-type ratio in plasmids pL401 and pL402, respectively (Fig. 5B, lower panel; compare lanes 5 and 6 to lanes 2 and 3). rpb2-7, which does not affect lacZ, also did not affect pL401 and pL402.
Northern analysis was used to confirm this increased utilization of the ADH2 poly(A) sites in rpb2-10 and spt5-4 backgrounds (Fig. 5C). The spt5-4 and rpb2-10 alleles resulted in increased utilization of the internal ADH2 poly(A) site for pL401 relative to that observed for the wild type (Fig. 5C; compare lanes 4 and 8 to lanes 2 and 6, respectively). As shown in Fig. 5C, lanes 4 and 8, the full-length RP51-lacZ RNA becomes diminished relative to that of the wild type without the formation of any smaller RNA products other than those of the RP51-ADH2 species. These data indicate that the spt5-4 and rpb2-10 alleles enhance utilization of an upstream, partially defective poly(A) site.
One alternative explanation for these results with the ADH2 poly(A) usage assay is that spt5-4 and rpb2-10 decrease the rate of degradation of the short RP51-ADH2 mRNA that terminates in the intron relative to that of the read-through RP51-lacZ mRNA. This result is unlikely because, as observed in Fig. 5B, upper and lower panels, lane 4, spt5-4 and rpb2-10 do not cause a corresponding augmentation of the levels of the short mRNA relative to the those of the long mRNA. We did, however, determine the mRNA half-lives for the short and long mRNA and found that spt5-4 had no effect on the half-life of either mRNA (Fig. 2B).
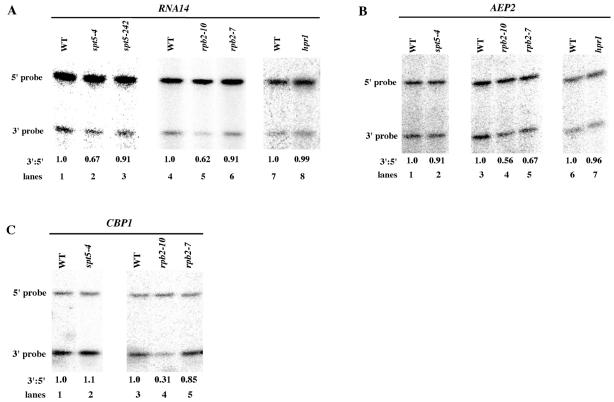
rpb2-10 reduces full-length mRNA formation from yeast genes that contain internal poly(A) sites.
Since rpb2-10 has been shown to increase pausing or arrest in vitro, the most likely interpretation of the above results is that increases in pausing downstream of the ADH2 poly(A) site caused by the rpb2-10 allele promote greater utilization of the upstream ADH2 poly(A) sites. These results also imply that rpb2-10 pausing will become manifest in reduced full-length gene expression of native yeast genes if partially functional internal poly(A) sites are present. In yeast, three genes, RNA14, AEP2, and CBP1, have been shown to contain internal inefficiently utilized poly(A) sites (46, 47). RNA14 contains two such sites, whereas AEP2 and CBP1 each contain one. We therefore assayed the effects of rpb2-10 and rpb2-7 on expression through RNA14, AEP2, and CBP1 (Fig. 6A through C). With 5′ and 3′ probes to the RNA14 mRNA, rpb2-10 reduced the level of the 3′ RNA relative to the amount of 5′ RNA (Fig. 6A, compare lanes 4 and 5). Over seven such experiments, the average drop in the 3′/5′ ratio in an rpb2-10 mutant was 0.62 ± 0.045 (mean ± standard error of the mean [SEM]). The rpb2-7 allele again displayed much less of an effect (Fig. 6A, lane 6), with an average decline in the 3′/5′ ratio for five determinations of 0.91 ± 0.082.
FIG. 6.
rpb2-10 and spt5-4 reduce full-length expression of yeast genes containing internal poly(A) sites. (A through C) Cells were grown on minimal medium with 4% glucose overnight and shifted to 2% glycerol-containing minimal medium for 6 h. S1 nuclease protection assays were conducted as described for Fig. 1 using probes specific to the 5′ and 3′ ends of RNA14, CBP1, and AEP2 (Table 2). The values represent the averages of the results for three to seven repetitions except for the values for hpr1 and the rpb2-7 effect on CBP1, which are the results for single experiments. SEMs not indicated in the text were less than 10% except that for rpb2-10 on CBP1, which was 20%. (A) S1 analysis of RNA14. (B) S1 analysis of AEP2. (C) S1 analysis of CBP1.
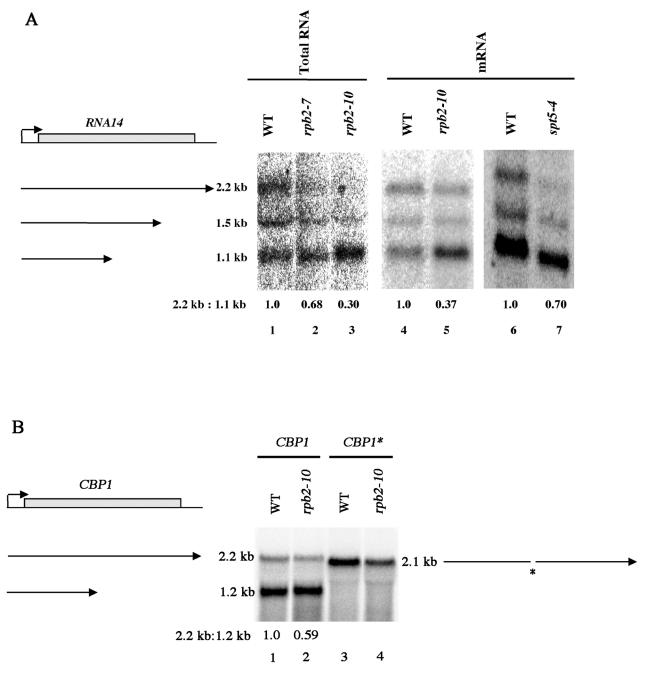
We reexamined these results using Northern analysis to verify whether one or both of the RNA14 internal poly(A) sites were being preferentially affected. As shown in Fig. 7A, lanes 3 and 5, rpb2-10 resulted in diminished full-length mRNA formation and augmented levels of the shortest (1.1-kb) mRNA. This result agrees with the S1 analysis shown in Fig. 6A and establishes that the rpb2-10 allele is directly enhancing upstream poly(A) usage. It suggests further that the two internal poly(A) sites in RNA14 might behave differently or that there are specific sequences across the RNA14 gene that display different levels of responsiveness to rpb2-10. In addition, rpb2-10 also reduced CBP1(Fig. 6C, lane 4) and AEP2 (Fig. 6B, lane 4) full-length formation. As mentioned above, rpb2-10 did not affect expression through several other genes analyzed, ADH2, NOT1, and YAT1, indicating that it specifically affects expression through genes containing internal poly(A) sites. However, hpr1 had no effect on RNA14 full-length expression (Fig. 6A, lane 8) or that of AEP2 (Fig. 6B), confirming that hpr1 operates by a different mechanism than rpb2-10 in affecting elongation. Note that rpb2-7 did display some enhanced internal poly(A) site utilization at RNA14 (Fig. 7A, lane 3) and at AEP2 (Fig. 6B, lane 5). While these effects are consistently less than those observed with rpb2-10, they do indicate that the rpb2-7 allele, which confers a 6AU phenotype in vivo (41) consistent with an elongation defect, can affect elongation under certain in vivo conditions.
FIG. 7.
(A) Northern analysis of rpb2-10 and spt5-4 effects on full-length formation of RNA14 RNA. Cells were grown as described for Fig. 6A. Both total RNA (lanes 1 to 3) and poly(A) RNA (lanes 4 to 7) were extracted and subjected to Northern analysis using the 5′ probe from RNA14. The values represent the data as presented. Repeat experiments gave similar data. (B) Effect of deleting the CBP1 internal poly(A) site on rpb2-10-enhanced usage of upstream poly(A) sites. Northern analysis of CBP1 RNA levels was conducted as described for Fig. 6. Plasmid pG::-26 (CBP1) contains the CBP1 gene under the control of the GAL10 promoter (47); plasmid pdeltaGM (CBP1*) is the same as pG::-26 except that the 98-bp region encompassing the internal poly(A) site of CBP1 has been removed (47). An asterisk indicates the location of the internal poly(A) sites. The apparently reduced abundance of CBP1* RNA in lane 4 (rpb2-10) compared to that in lane 3 (wild type) was not borne out by other analyses.
spt5-4 also resulted in decreased full-length formation for RNA14 (Fig. 6A, lane 2) with an average decrease in the 3′/5′ ratio of 0.67- ± 0.046-fold for six experiments. In contrast, spt5-242 had little apparent effect on full-length RNA14 expression (Fig. 6A, lane 3) with an average drop in the 3′/5′ ratio of 0.91 ± 0.12 for four experiments. These results were also confirmed by Northern analysis (Fig. 7A). spt5-4, however, displayed no effect on CBP1 or AEP2 expression (Fig. 6B and C), suggesting that there is sequence specificity to its pausing, its effects are weaker than rpb2-10, or the multiple poly(A) sites within RNA14 allow greater amplification of its putative pausing.
We further tested the model that rpb2-10 enhances utilization of upstream poly(A) sites by examining the effect of deleting the internal poly(A) sites present in the CBP1 gene on CBP1 RNA formation. When the internal poly(A) region of the CBP1 gene was deleted (47), only full-length CBP1 RNA was formed in both wild-type and rpb2-10 backgrounds (Fig. 7B, lanes 3 and 4). These data establish that the rpb2-10 allele enhances upstream poly(A) site usage and does not just result in a blockage to elongation.
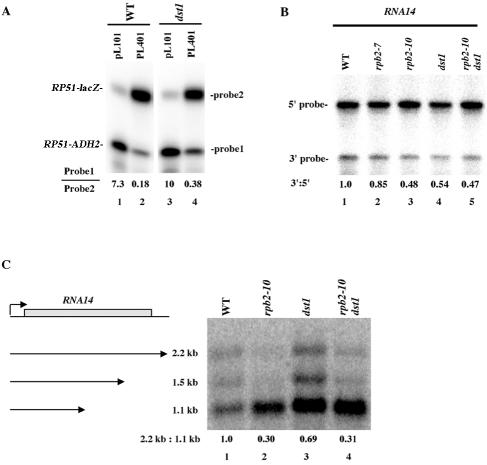
Deletion of TFIIS also reduces full-length expression of RNA14.
The above results indicate that defects in transcriptional elongation in vivo can be visualized by monitoring the use of internal poly(A) sites. It had already been shown previously that a dst1 deletion blocks GAL1-lacZ expression, presumably by affecting its elongation (28). We subsequently examined the effect of dst1 by using the defective ADH2 poly(A) signal inserted into the RP51 gene. As shown in Fig. 8A (compare lanes 2 and 4), dst1, like rpb2-10 and spt5-4 (Fig. 5B), caused the formation of increased truncated RP51-ADH2 RNA (for dst1, lane 4, about a twofold-more-shortened transcript relative to the read-through transcript than that found in the wild type, shown in lane 2), consistent with dst1 augmenting internal ADH2 poly(A) site utilization. To further test this hypothesis, we subsequently examined the effect of a TFIIS deletion on RNA14 expression. As shown in Fig. 8B, a dst1 deletion reduced full-length RNA expression by 1.6-fold. This result was confirmed by Northern analysis, in which the shortest transcript was increased in abundance relative to the full-length transcript by twofold (Fig. 8C). Combining a dst1 deletion with an rpb2-10 allele did not result, however, in any significantly worse effects than those observed with the rpb2-10 allele alone (Fig. 8C). These observations identify the first native gene in vivo whose elongation is impaired by deletion of TFIIS.
FIG. 8.
dst1 has effects on full-length RNA formation similar to those for rpb2-10. (A) Wild-type (WT) strain Z96 (lanes 1 and 2) and dst1 strain DY106-u (lanes 3 and 4) were analyzed for their effects on ADH2 poly(A) site usage as described for Fig. 5B. Plasmids pL101 and pL401 are the same as for Fig. 5. (B) RNA14 full-length RNA expression was analyzed as described for Fig. 5A. Total RNA was extracted from strains Z96 (WT), Z103 (rpb2-7), Z106 (rpb2-10), DY106-u (dst1), and DY108 (dst1 rpb2-10). The values represent the data as presented. Repeat experiments gave similar data. (C) Northern analysis of RNA14 mRNA levels in the strain backgrounds described above was conducted as described for Fig. 7A. The values represent the data as presented. Repeat experiments gave similar data.
rpb2-10 and spt5-4 affect poly(A) site usage at the 3′ end of genes.
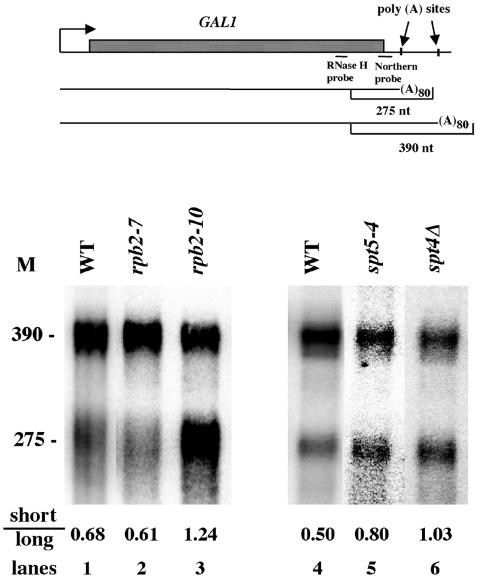
Differential poly(A) site choice at the 3′ ends of genes can be regulated in response to a variety of biological influences (20). The above effects of elongation defects on internal upstream poly(A) site utilization suggest that elongation may also play a role in regulating poly(A) site choice. To test this prediction, we analyzed poly(A) site utilization for the GAL1 gene, which contains two poly(A) sites about 50 and 160 bp downstream of its translation stop codon (21, 35). Newly synthesized GAL1 polyadenylated mRNA was created by inducing GAL1 expression for 15 min in galactose-containing medium followed by repression of GAL1 mRNA synthesis with the addition of glucose. The 3′ ends of GAL1 mRNA were detected by using an RNase H assay (52) and a DNA probe that was complementary to sequences present in both species (Fig. 9, top panel). Two polyadenylated species migrating at about 380 and 275 nucleotides (nt) that corresponded to poly(A) sites at about 160 bp and 50 bp, respectively, downstream of the GAL1 stop codon were identified (Fig. 9, bottom panel). Each mRNA species contained 80 nt of poly(A), as determined by a deadenylation assay (52; data not shown). As shown in Fig. 9, lanes 1 and 4, the downstream site is preferred by about twofold over the upstream site in the wild-type strain. In a rpb2-10 background, use of the GAL1 upstream poly(A) site was increased by about twofold (Fig. 9, compare lanes 1 and 3), whereas the rpb2-7 allele had no effect on GAL1 poly(A) site utilization (Fig. 9, lane 2). Similarly, in an spt5-4 or spt4 strain background, the use of the upstream site is increased by 1.6-fold and twofold, respectively (Fig. 9, lanes 5 and 6). The spt5-242 allele, which does not affect internal poly(A) site utilization (see above), had no effect on augmenting GAL1 upstream poly(A) site usage (data not shown). rpb2-10, spt5-4, and spt4 also did not enhance the stability of the short GAL1 mRNA relative to that of the long GAL1 mRNA (Table 4). These results confirm our prediction that elongation defects enhance utilization of upstream poly(A) sites and indicate that regulating the elongation process will be critical to 3′-end poly(A) site choice.
FIG. 9.
rpb2-10, spt5-4 and spt4 affect 3′-end poly(A) site choice at GAL1. (Top panel) Diagram of GAL1 gene RNA. The two poly(A) sites located 50 and 160 bp downstream of the stop codon are indicated, as are the RNase H probe and the Northern probe. (Bottom panel) Northern analysis of GAL1 mRNA. Yeast were grown on galactose-containing medium for 15 min, followed by glucose addition, upon which total RNA was extracted. Newly synthesized GAL1 polyadenylated mRNA species were detected by using an RNase H assay (52). GAL1-specific transcripts were identified by Northern analysis. The larger RNA species (migrating at about 390 nt) corresponds to a poly(A) cleavage site about 160 bp downstream of the GAL1 gene translation stop codon, and the smaller RNA species (migrating at about 275 nt) corresponds to a cleavage site about 50 bp downstream of the stop codon. Each mRNA species carries a poly(A) tail measured at about 80 nt by a deadenylation assay (data not shown). The values represent the average ratios of the results for the 275-nt species to those for the 390-nt species of two to five determinations, with SEMs less than 20%.
TABLE 4.
Half-lives of GAL1 mRNAa
The half-lives of GAL1 RNAs were determined after yeast was grown on galactose-containing medium for 15 min, followed by the addition of glucose and harvesting of RNA for times up to 60 min. The two GAL1 RNA species were detected as described in the legend to Fig. 9. The SEMs were less than 15%. The value for rpb2-10 is the result for a single determination.
Strain FY1642, which is isogenic to FY1668 (spt5-4) and GHY180 (spt4), except for the indicated allele.
Strain Z96, which is isogenic to Z106 (rpb2-10), except as indicated.
DISCUSSION
In this report, we have identified native yeast genes whose full-length mRNA formation is significantly reduced by defects in the elongation factors RPB2, SPT5,and TFIIS. These experiments represent the first demonstration in vivo of genes whose transcriptional elongation is regulated by these elongation factors, as previous studies, including microarray analyses, were unsuccessful in clearly identifying such genes (23, 45, 56). In addition, we have shown that expression through other chimeric yeast gene constructs and that of the E. coli lacZ gene are also affected by RPB2, SPT5,and TFIIS defects. The common feature of these genes and constructs is that they contain internal poly(A) sites. These observations should aid the study of elongation in vivo and allow the development of genetic assays for the identification of novel elongation factors and new relationships among known factors.
It has previously been shown in vitro and in vivo that transcriptional pausing downstream to a poly(A) site promotes polyadenylation and cleavage of the RNA in both yeast and mammalian cells (2, 5, 40, 61). These observations are consistent with several observations for the requirement of RNA Pol II in 3′ processing (24, 34, 42). We have established in this report that defects in SPT5, SPT4, RPB2, and TFIIS, all factors involved in transcriptional elongation, affect mRNA formation in vivo of genes containing internal poly(A) sites. We postulate, therefore, that transcriptional pausing caused by defects in elongation factors enhances internal poly(A) site usage and consequently increases truncated mRNA formation relative to that of full-length mRNA.
Several pieces of evidence support our hypothesis. First, mutations in SPT5, SPT4, and RPB2 (such as spt5-4, spt4, rbp2-4, and rpb2-10 alleles) reduced full-length RNA formation in lacZ, a gene known to contain cryptic poly(A) sites (44). Other SPT5 or RPB2 mutations had no effect on full-length lacZ RNA formation, indicating that this effect was allele specific. Second, the truncated lacZ RNAs were polyadenylated. Third, these SPT5 and RPB2 defects did not affect full-length mRNA formation for genes such as YAT1 or NOT1 containing high G+C content or excessive length that lacked internal poly(A) sites, indicating that their effects on elongation occurred by a mechanism different from or in addition to that ascribed to HPR1 (9). Fourth, we showed that spt5-4, rpb2-10, and dst1 alleles can enhance the usage of defective ADH2 poly(A) sites embedded within the RP51 gene. The other SPT5 or RPB2 alleles, which did not affect lacZ expression, also had no effect on ADH2-defective poly(A) site usage. Importantly, the rpb2-10 allele, which is known to cause increased pausing in vitro and to block in vitro transcriptional elongation, resulted in enhanced internal poly(A) site usage whereas the rpb2-7 allele, which does not block in vitro elongation, did not display the same effects. Fifth, we showed that several yeast genes with known internal poly(A) sites displayed decreased full-length mRNA expression and increased internal poly(A) site usage with rpb2-10 and to a lesser extent with spt5-4 and dst1. Importantly, deleting the internal poly(A) site for CBP1 in an rpb2-10 background resulted in only full-length CBP1 mRNA being visualized. Finally, the rpb2-10, spt5-4, and spt4 defects affected 3′-end poly(A) site choice in which the usage of the upstream poly(A) site became preferred in strains carrying these defects.
That the usage of the upstream poly(A) site is always enhanced relative to that of the downstream site with these elongation mutations suggests that RNA Pol II pausing occurs throughout the gene and that poly(A) site usage becomes favored at the first available poly(A) signal. Our results also imply that at normal genes such as ADH2, NOT1, and YAT1, where rpb2-10 does not have an apparent effect on 3′-end formation, or the first 1.0 kb of lacZ, rpb2-10-induced RNA Pol II pausing probably still occurs but is not significant enough in vivo to cause reduced levels of 3′ RNA formation relative to that of 5′ RNA levels. Only when a cryptic or defective poly(A) site is present can the pausing be visualized as reduced normal 3′-end formation due to increased poly(A) site usage. This interpretation is consistent with the observation that rpb2-10 did not appear to affect transcription in vivo even when a known in vitro arrest site was introduced into a gene (57). Relatedly, dst1 did not affect RNA Pol II occupancy at the lacZ gene (28), implying that enhanced pausing caused by dst1 or rpb2-10 may not have noticeable effects on measurable RNA Pol II association with the gene. The known effects of rpb2-10, dst1, spt5-4, or spt4 on many genes' expression, either positive or negative (50, 56), must therefore be interpreted carefully as to whether they are due to transcriptional initiation or elongation defects. The observation that these defects did not result in reductions in full-length mRNA formation for long genes such as NOT1 or genes with a very high G+C content such as YAT1 does not imply that spt5, rpb2, or dst1 strains were not impaired in transcribing through these genes. For example, YAT1 total mRNA expression was decreased in an spt5-4 or rpb2-10 background, but we are unable to ascertain whether this effect is at the level of transcriptional initiation or not.
Interestingly, it has been shown that nonfermentative growth conditions promote increased poly(A) site cleavage at the RNA14 1.1-kb site and enhanced internal poly(A) site cleavage at CBP1 and AEP2 (46). These effects of nonfermentative growth on RNA14, CBP1, and AEP2 expression are similar to those observed for rpb2-10 and suggest that they occur by a similar mechanism, that is, by causing increased RNA Pol II pausing. Several stress conditions have also been observed to enhance SUA7 upstream poly(A) site utilization (25). Nonfermentative growth and other stress conditions may impair elongation and thereby alter poly(A) site utilization. However, the effects of the several elongation defects on internal poly(A) site usage that we observed would occur in addition to the nonfermentative growth effect since all of our experiments were conducted under glycerol growth conditions.
While rpb2-10 can result in decreased full-length RNA formation for several yeast genes containing internal poly(A) sites, such as RNA14, CBP1, and AEP2 (46, 47), and differential 3′-end poly(A) usage at GAL1, spt5-4 clearly affected only full-length RNA14 RNA formation and 3′-end choice of GAL1. This difference in behavior may be due to sequence differences between RNA14, CBP1, and AEP2. While CBP1 and AEP2 each contain one internal poly(A) site, RNA14 contains two internal poly(A) sites, which may allow a greater amplification of the effect caused by pausing or arrest. RNA14 and the 3′ end of GAL1 may also contain specific sequences that can result in more pausing. Alternatively, the spt5-4 allele may have a weaker effect on elongation than rpb2-10.
It could be argued that the spt5, spt4, rpb2, and dst1 alleles affect full-length RNA formation not by blocking RNA elongation but by directly enhancing the polyadenylation and cleavage process at upstream poly(A) sites. While we cannot formally exclude this possibility, several factors suggest otherwise. First, these alleles have displayed a number of effects on elongation and are known to be involved in elongation. Second, in vitro evidence has shown that rpb2-10 causes RNA Pol II to pause, while rpb2-7 had no effect (41). Similarly, rpb2-10 can strongly decrease full-length lacZ RNA formation and enhance defective ADH2 poly(A) site usage, whereas rpb2-7 had no effect in our experiments. Although rpb2-7 did reduce RNA14, CBP1, and AEP2 full-length RNA formation to a limited extent, rpb2-10 displayed much stronger effects. In addition, while SPT5 has been indicated to play a role in transcription initiation, elongation, and mRNA capping (22, 39, 49, 50, 54, 55, 59, 60), it has not been identified as affecting polyadenylation or cleavage directly, although through its contacts to RNA Pol II it can immunoprecipitate with polyadenylation or cleavage factors (30). Finally, known defects in poly(A) cleavage or adenylation factors that reduce poly(A) site utilization have not been shown to enhance upstream poly(A) site use as presented herein (C. Moore, personal communication). However, in mammalian systems alternations in the activity or abundance of the 64-kDa subunit of CstF polyadenylation factor can in some circumstances influence poly(A) site choice (20). It remains possible, therefore, that the rpb2-10, spt5-4, and dst1 defects could exert indirect effects on the activities of poly(A) cleavage or adenylation factors.
Many genes with alternative poly(A) sites have been identified and characterized in mammalian cells, yeast, and several types of viruses. Differential poly(A) site choice of some genes is regulated by development stages or in different tissues in mammalian cells (20). More than 5,000 human and 1,000 mouse genes with two or more poly(A) sites have been identified by using expressed sequence tag data (3). Therefore, regulation of poly(A) site choice is an important method for regulating certain mRNA levels in different cellular environments. The results presented herein indicate that the elongation process, through effects on transcriptional pausing or arrest, may also regulate mRNA levels by affecting poly(A) site choice. Since the same mRNA with different poly(A) site ends can display vastly different deadenylation rates and mRNA stabilities, altering poly(A) site choice could greatly influence protein translation and abundance in the cell. In addition, more than 1,000 yeast genes could contain internal poly(A) sites (21). It is obvious, therefore, that the proper regulation of pausing or arrest in vivo will be extremely important in maintaining both the formation and fidelity of full-length mRNA.
Not only do our results confirm an in vivo connection between transcription elongation and polyadenylation or cleavage, but they also provide a method to examine transcriptional elongation in vivo. The enhanced utilization of internal poly(A) sites that occurs as a result of defects in elongation factors can be used to develop appropriate genetic assays for identifying novel elongation factors and elucidating their mechanisms in transcriptional elongation and for analyzing the types of DNA sites that affect transcriptional elongation in vivo.
Acknowledgments
We thank A. Aguilera for the GAL1-YAT1 plasmid, C. Dieckmann for the GAL10-CBP1 plasmids, and Claire Moore for the RP51-ADH2-lacZ plasmids. We also thank D. Reines and R. Parker for discussions related to this project.
This research was supported by NIH grant GM41215 to C.L.D. and by HATCH project H291.
Footnotes
Scientific contribution no. 2087 from the New Hampshire Agriculture Experiment Station.
REFERENCES
- 1.Andrulis, E. D., E. Guzman, P. Doring, J. Werner, and J. T. Lis. 2000. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 14:2635-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda, A., and N. J. Proudfoot. 1999. Definition of transcriptional pause elements in fission yeast. Mol. Cell. Biol. 19:1251-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaudoing, E., and D. Gautheret. 2001. Identification of alternate polyadenylation sites and analysis of their tissue distribution using EST data. Genome Res. 11:1520-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 5.Birse, C. E., B. A. Lee, K. Hansen, and N. J. Proudfoot. 1997. Transcriptional termination signals for RNA polymerase II in fission yeast. EMBO J. 16:3633-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourgeois, C. F., Y. K. Kim, M. J. Churcher, M. J. West, and J. Karn. 2002. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 22:1079-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calvo, O., and J. Manley. 2001. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell 7:1013-1023. [DOI] [PubMed] [Google Scholar]
- 8.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavez, S., M. Garcia-Rubio, F. Prado, and A. Aguilera. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7054-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, J., Y. C. Chiang, and C. L. Denis. 2002. CCR4, a 3′-5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J. 21:1414-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, J., J. Rappsilber, Y. C. Chiang, P. Russell, M. Mann, and C. L. Denis. 2001. Purification and characterization of the 1.0 MDa CCR4-NOT complex identifies two novel components of the complex. J. Mol. Biol. 314:683-694. [DOI] [PubMed] [Google Scholar]
- 12.Chiang, Y. C., P. Komarnitsky, D. Chase, and C. L. Denis. 1996. ADR1 activation domains contact the histone acetyltransferase GCN5 and the core transcriptional factor TFIIB. J. Biol. Chem. 271:32359-32365. [DOI] [PubMed] [Google Scholar]
- 13.Collart, M. A., and K. Struhl. 1993. CDC39, an essential nuclear protein that negatively regulates transcription and differentially affects the constitutive and inducible HIS3 promoters. EMBO J. 12:177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 15.Cook, W. J., and C. L. Denis. 1993. Identification of three genes required for the glucose-dependent transcription of the yeast transcriptional activator ADR1. Curr. Genet. 23:192-200. [DOI] [PubMed] [Google Scholar]
- 16.Cramer, P., D. A. Bushnell, J. Fu, A. L. Gnatt, B. Maier-Davis, N. E. Thompson, R. R. Burgess, A. M. Edwards, P. R. David, and R. D. Kornberg. 2000. Architecture of RNA polymerase II and implications for the transcription mechanism. Science 288:640-649. [DOI] [PubMed] [Google Scholar]
- 17.Denis, C. L., Y. C. Chiang, Y. Cui, and J. Chen. 2001. Genetic evidence supports a role for the yeast CCR4-NOT complex in transcriptional elongation. Genetics 158:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denis, C. L., J. Ferguson, and E. T. Young. 1983. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J. Biol. Chem. 258:1165-1171. [PubMed] [Google Scholar]
- 19.Denis, C. L., and T. Malvar. 1990. The CCR4 gene from Saccharomyces cerevisiae is required for both nonfermentative and spt-mediated gene expression. Genetics 124:283-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edwalds-Gilbert, G., K. L. Veraldi, and C. Milcarek. 1997. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 25:2547-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graber, J. H., G. D. McAllister, and T. F. Smith. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res. 30:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemming, S. A., D. B. Jansma, P. F. Macgregor, A. Goryachev, J. D. Friesen, and A. M. Edwards. 2000. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem. 275:35506-35511. [DOI] [PubMed] [Google Scholar]
- 24.Hirose, Y., and J. L. Manley. 1998. RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395:93-96. [DOI] [PubMed] [Google Scholar]
- 25.Hoopes, B. C., G. D. Bowers, and M. J. DiVisconte. 2000. The two Saccharomyces cerevisiae SUA7 (TFIIB) transcripts differ at the 3′-end and respond differently to stress. Nucleic Acids Res. 28:4435-4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hyman, L. E., S. H. Seiler, J. Whoriskey, and C. L. Moore. 1991. Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3′-end formation. Mol. Cell. Biol. 11:2004-2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaplan, C. D., J. R. Morris, C. Wu, and F. Winston. 2000. Spt5 and Spt6 are associated with active transcription and have characteristics of general elongation factors in D. melanogaster. Genes Dev. 14:2623-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulish, D., and K. Struhl. 2001. TFIIS enhances transcriptional elongation through an artificial arrest site in vivo. Mol. Cell. Biol. 21:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lennon, J. C., III, M. Wind, L. Saunders, M. B. Hock, and D. Reines. 1998. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:5771-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 23:1368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, H. Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann, and C. L. Denis. 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17:1096-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luebke, K. J., R. P. Balog, and H. R. Garner. 2003. Prioritized selection of oligodeoxyribonucleotide probes for efficient hybridization to RNA transcripts. Nucleic Acids Res. 31:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maquat, L. E., and G. G. Carmichael. 2001. Quality control of mRNA function. Cell 104:173-176. [DOI] [PubMed] [Google Scholar]
- 34.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miyajima, A., N. Nakayama, I. Miyajima, N. Arai, H. Okayama, and K. Arai. 1984. Analysis of full-length cDNA clones carrying GAL1 of Saccharomyces cerevisiae: a model system for cDNA expression. Nucleic Acids Res. 12:6397-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 37.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 38.Otero, G., J. Fellows, Y. Li, T. de Bizemont, A. M. Dirac, C. M. Gustafsson, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 1999. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol. Cell 3:109-118. [DOI] [PubMed] [Google Scholar]
- 39.Pei, Y., and S. Shuman. 2002. Interactions between fission yeast mRNA capping enzymes and elongation factor Spt5. J. Biol. Chem. 277:19639-19648. [DOI] [PubMed] [Google Scholar]
- 40.Peterson, M. L., S. Bertolino, and F. Davis. 2002. An RNA polymerase pause site is associated with the immunoglobulin μs poly(A) site. Mol. Cell. Biol. 22:5606-5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell, W., and D. Reines. 1996. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem. 271:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 43.Rondon, A. G., M. Garcia-Rubio, S. Gonzalez-Barrera, and A. Aguilera. 2003. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 22:612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russnak, R., K. W. Nehrke, and T. Platt. 1995. REF2 encodes an RNA-binding protein directly involved in yeast mRNA 3′-end formation. Mol. Cell. Biol. 15:1689-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sparks, K. A., and C. L. Dieckmann. 1998. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 26:4676-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sparks, K. A., S. A. Mayer, and C. L. Dieckmann. 1997. Premature 3′-end formation of CBP1 mRNA results in the downregulation of cytochrome b mRNA during the induction of respiration in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4199-4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strasser, K., S. Masuda, P. Mason, J. Pfannstiel, M. Oppizzi, S. Rodriguez-Navarro, A. G. Rondon, A. Aguilera, K. Struhl, R. Reed, and E. Hurt. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417:304-308. [DOI] [PubMed] [Google Scholar]
- 49.Swanson, M. S., E. A. Malone, and F. Winston. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11:3009-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson, M. S., and F. Winston. 1992. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan, S., R. C. Conaway, and J. W. Conaway. 1995. Dissection of transcription factor TFIIF functional domains required for initiation and elongation. Proc. Natl. Acad. Sci. USA 92:6042-6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 53.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66:117-172. [DOI] [PubMed] [Google Scholar]
- 54.Wada, T., T. Takagi, Y. Yamaguchi, A. Ferdous, T. Imai, S. Hirose, S. Sugimoto, K. Yano, G. A. Hartzog, F. Winston, S. Buratowski, and H. Handa. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wada, T., T. Takagi, Y. Yamaguguhi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effects of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wind-Rotolo, M., and D. Reines. 2001. Analysis of gene induction and arrest site transcription in yeast with mutations in the transcription elongation machinery. J. Biol. Chem. 276:11531-11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wind-Rotolo, M., and D. Reines. 2000. Transcription elongation factor SII. Bioessays 22:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woolf, T. M., D. A. Melton, and C. G. Jennings. 1992. Specificity of antisense oligonucleotides in vivo. Proc. Natl. Acad. Sci. USA 89:7305-7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 60.Yamaguchi, Y., T. Wada, D. Watanabe, T. Takagi, J. Hasegawa, and H. Handa. 1999. Structure and function of the human transcription elongation factor DSIF. J. Biol. Chem. 274:8085-8092. [DOI] [PubMed] [Google Scholar]
- 61.Yonaha, M., and N. J. Proudfoot. 1999. Specific transcriptional pausing activates polyadenylation in a coupled in vitro system. Mol. Cell 3:593-600. [DOI] [PubMed] [Google Scholar]
- 62.Yonaha, M., and N. J. Proudfoot. 2000. Transcriptional termination and coupled polyadenylation in vitro. EMBO J. 19:3770-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]