Abstract
Siderophores and colicins enter bacterial cells through TonB-dependent outer membrane proteins. Using site-directed substitution mutagenesis, we studied ligand recognition by a prototypic Escherichia coli siderophore receptor, FepA, that binds the iron chelate ferric enterobactin and colicins B and D. These genetic experiments identified a common binding site for two of the three ligands, containing multiple positive charges, within cell surface residues of FepA. Elimination of single residues in this region did not impair the adsorption or transport of ferric enterobactin, but double mutagenesis in the charge cluster identified amino acids (Arg-286 and Arg-316) that participate in siderophore binding and function in FepA-mediated killing by colicins B and D. Ferric enterobactin binding, furthermore, prevented covalent modification of FepA within this domain by either a fluorescent probe or an arginine-specific reagent, corroborating the involvement of this site in ligand recognition. These results identify, for the first time, residues in a TonB-dependent outer membrane protein that participate in ligand binding. They also explain the competition between ferric enterobactin and the colicins on the bacterial cell surface: all three ligands interact with the same arginine residues within FepA during their penetration through the outer membrane.
Keywords: ferric enterobactin, siderophore, colicin, TonB, outer membrane
Siderophores are small organic molecules that chelate iron and supply it to microorganisms (1). Siderophore receptor proteins form TonB-dependent, ligand-gated channels in the outer membrane (OM) that transport metals into Gram-negative bacteria (2–4). Little information exists about how such proteins recognize and internalize their ligands. In Escherichia coli, for example, seven different OM proteins discriminate, bind, and transport at least seven different siderophores (1, 5), even though all the iron chelates have approximately the same size (700 Da) and shape (a hexadentate complex). FepA is one such OM protein that binds and transports the native E. coli siderophore, ferric enterobactin. FepA is also multifunctional: besides the siderophore, it also serves as the surface receptor for two lethal agents, colicins B and D (6–8). All three ligands bind with high affinity [the Kd of the ferric enterobactin–FepA equilibrium is 10−8 M (9, 10)] in a centrally located surface loop proposed to lie between residues 255 and 336 (11). FepA has been crystallized (12) but not structurally solved, and the amino acids that participate in these adsorption events are not known. Subsequent to binding on the cell surface, the siderophore and the colicins penetrate the OM bilayer by an uncharacterized mechanism that requires the participation of another cell envelope protein, TonB (6, 13).
FepA recognizes the iron center of ferric enterobactin, Fe3+ complexed by three identical catechol groups. The organic platform from which these catechols arise does not influence the recognition events; a variety of chemically different structures can substitute for the macrocyclic ring of ferric enterobactin (14). The chemistry of the siderophore provides a rationale for understanding its adsorption to proteins. Its catechol groups form a right-handed propeller around ferric iron (Fig. 1), and various left-handed analogs of ferric enterobactin do not supply iron to E. coli expressing FepA (15). Another distinguishing feature of the siderophore is its charge: chelation of Fe3+ by six catechol oxygens creates a net charge of −3. Thus, the ferric enterobactin iron complex presents a distinctive three dimensional structure, an outlying, chiral sphere of aromaticity, surrounding a negatively charged metal center.
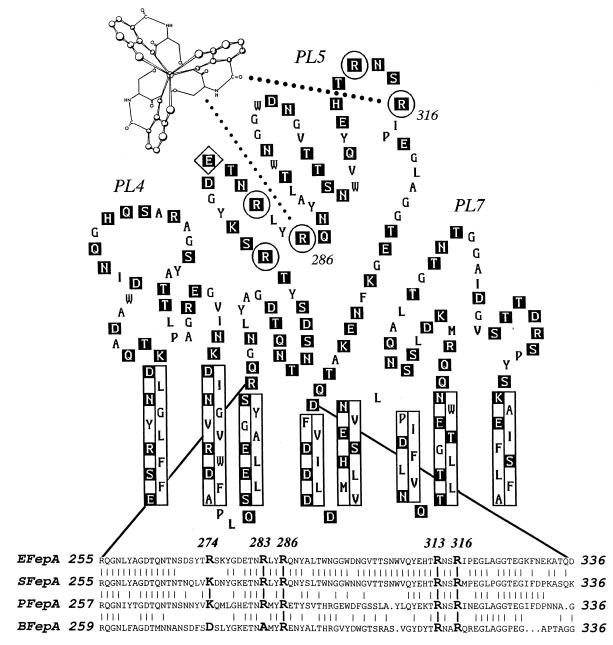
Figure 1.
Location of site-directed substitution mutations within the proposed structure of FepA. The predicted structural and topological features of three proposed surface loops (PL4, PL5, and PL7) of the E. coli ferric enterobactin receptor (11) are shown, including hydrophilic residues (boxed in black), transmembrane strands (enclosed in rectangles), target residues for mutagenesis (circled; Arg-274, -283, -286, -313, and -316), and a target residue for covalent modification (enclosed in a diamond; Glu-280). Below is a sequence comparison of four FepA proteins from E. coli (EFepA), S. typhimurium (SFepA), P. aeruginosa (PFepA), and B. pertussis (BFepA), that illustrates the conserved basic amino acids in the fifth proposed external loop of FepA (PL5).
A systematic genetic approach to membrane protein structure and function is germane to the understanding of bacterial transport. A large number of OM proteins are known and partially characterized, but few structures are crystallographically defined, and fewer mechanisms are well understood (16–18). However, the elucidation of a potentially multivalent binding interaction by site-directed mutagenesis is problematical, because single-residue substitutions may not sufficiently impair avidity to alter the transport phenotype. The binding of ligands by FepA illustrates this premise. Replacement of single amino acids in its proposed binding site did not diminish either adsorption or transport of ferric enterobactin. Double substitutions, on the other hand, among five candidate arginine residues, pinpointed critical amino acids (Arg-286 and Arg-316) in the binding event. Double mutagenesis in the region of interest recognized probable ligand contact residues and a chemical basis for ligand interaction with FepA. These experiments illustrate a general, abbreviated rationale for the genetic identification of ligand-binding residues in proteins.
MATERIALS AND METHODS
Sequence Comparisons.
Sequences of the FepA proteins from E. coli (19), Salmonella typhimurium (20), Pseudomonas aeruginosa (21), and Bordetella pertussis (22) were aligned by the pileup algorithm (Genetics Computer Group, Madison, WI).
Site-Directed Mutagenesis.
The fepA structural gene was subcloned from the pUC19 plasmid pITS449 (23) into M13 mp19, using the PstI and SacI sites of the vectors. Site-directed substitutions were generated by Kunkel mutagenesis (24, 25). After verification of correct mutagenesis by sequencing the single-stranded M13 constructs (Sequenase; United States Biochemical), we reversed the cloning procedure, returning the mutant fepA genes, under control of their natural promoter, to pITS449. Finally, all mutant constructions in pUC19 were resequenced to confirm their authenticity. Mutations were designated as follows: XnY, where X represents the one-letter abbreviation for the wild-type residue at position n, and Y represents the one-letter abbreviation for the substituted amino acid.
Siderophore Binding and Transport.
59Fe-enterobactin binding experiments were performed by a modification of prior methods (26, 27). Bacteria harboring fepA+ or mutant fepA genes on pUC19 were grown in Luria–Bertani broth with ampicillin (100 μg/ml) overnight, subcultured into Mops minimal medium with ampicillin (10 μg/ml), and grown for 5.5–6 hr at 37°C with vigorous aeration. Six 10-ml aliquots were transferred into culture tubes and incubated on ice for 1 hr. Appropriate volumes of freshly prepared, purified, ice-cold 59Fe-siderophore (26) were mixed in the six tubes. After 1 and 6 min, 5-ml aliquots were withdrawn and filtered through glass fiber filters, and the filters were counted. The absence of a differential in the siderophore associated with the cells at 1 and 6 min confirmed the inability of the bacteria to transport under these conditions. An aliquot of the cell suspension was analyzed at 600 nm to determine the cell density, and 2 × 108 bacteria were lysed and subjected to Western immunoblot to quantitate FepA expression. For transport experiments, bacteria were grown and quantitated in the same way, and the uptake of 59Fe-enterobactin was measured (26) at 37°C.
Colicin Binding and Killing.
Colicins B and D were purified (28) and labeled with 125I using Iodo Beads (Pierce). Their adsorption to KDF541 (fepA; ref. 2)-harboring derivatives of plasmid pITS449 carrying the different mutations was measured. Bacteria were grown as for ferric enterobactin binding, pelleted, and resuspended in cold 40 mM Mops (pH 6.9) containing 0.9% NaCl, at 4 × 109 cells per ml. Aliquots of 50 μl were incubated with 10 μl of colicin B (containing 2.6% 125I-colicin B) for 1 hr on ice. The mixture was diluted with 500 μl of Mops buffer and spun at 14,000 rpm for 2 min at 4°C. The radioactivity of the pellets was compared with that of the colicin solutions to calculate the amount bound. Incubation times (with FepA and selected mutants) were varied from 5 min to 3 hr, and the level of tracer 125I-colicin B varied from 1.3% to 7.5% without any alteration in results. Colicin D binding was performed as with colicin B, except that, to achieve saturation for some of the higher concentrations, we added 50-μl volumes of colicin D, giving a final assay volume of 100 μl.
Chemical Modifications.
7-Diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin (CPM) shows a large increase in fluorescent intensity upon coupling to a protein (29–31). Wild-type FepA and the mutant FepAE280C were purified (3) and reacted with a 5-fold molar excess of CPM in 50 mM Mops, pH 6.9/60 mM NaCl/1 mM dodecyl β-d-maltoside at 25°C, in the absence or presence of ferric enterobactin, and the time course of the fluorescence emission of the labeled proteins was recorded at 471 nm on an SLM-8000 fluorimeter (SLM–Aminco, Rochester, NY).
For modification of FepA with the arginine-specific reagent (32) phenyl glyoxal (PG; Sigma), cells were grown as for siderophore binding and suspended in 40 mM Mops (pH 8), containing 0.9% NaCl and 1.5% PG, for 1 hr at 25°C, in the absence and presence of ferric enterobactin, and then washed by centrifugation and resuspended in ice-cold Mops medium.
RESULTS
Candidate Residues for Mutagenesis.
The possibility of ionic interactions with the negatively charged molecule ferric enterobactin suggested the involvement of basic residues within FepA. The receptor protein contains nine candidate residues in the previously implicated region of ligand binding: five arginines, three lysines, and one histidine. Comparison of known FepA sequences, however, showed that only arginines are conserved in region 255–336. Alignment of the E. coli, S. typhimurium, P. aeruginosa, and B. pertussis FepA proteins revealed four centrally located arginines (Fig. 1), fulfilling the expectation of positive counterions to bond with the siderophore. We mutagenized these conserved basic residues, singly and in combinations, to test their participation in the interaction with ligands.
Effects of Positive Charge Replacement on Siderophore Binding and Transport.
We initially replaced Arg at positions 283, 286, 313, and 316 with Ala (abbreviated as substitutions A, B, C, and D, respectively). Three of these arginines (B, C, and D) are fully conserved in all the FepA sequences (Fig. 1); the fourth (A) changes to Ala in B. pertussis. These single mutations had little or no impact on the binding or transport of the siderophore (Table 1); they only slightly changed the biochemical characteristics of the ferric enterobactin–FepA interaction (binding Kd and transport Km and Vmax), and they did not affect siderophore-dependent growth. But combinations of certain mutations that eliminated two (or more) positive charges did diminish the recognition and transport of ligands by FepA: residues 286 (B) and 316 (D) were essential to siderophore binding and transport, whereas residues 283 (A) and 313 (C) were not essential. Single mutations were phenotypically silent, unless combined with a second substitution of an essential amino acid. For example, the binding of ferric enterobactin to B was indistinguishable from its binding to wild-type FepA, when analyzed individually or in combination with A or C. But when combined with D, with or without additional substitutions, a 30-fold reduction in affinity for the siderophore occurred (Fig. 2; Table 1). ABD, BCD, and ABCD showed even lower affinity for ferric enterobactin, while other multiple mutations (that did not consolidate B and D) were similar to wild type. For ABCD, these effects were difficult to quantitatively characterize, because the binding-impaired mutant protein was only marginally different from the fepA negative control strain. We used double mutagenesis to study a fifth, nonconserved arginine residue within PL5, close to the other arginines of interest at position 274 (Fig. 1). Mutations combining R274A (abbreviated E in Table 1) with A, B, C, and D showed no effects on siderophore binding and transport. Thus, among the five positively charged residues that we studied, Arg-286 and Arg-316 were essential and may create contact points for the siderophore, while Arg-274, Arg-283, and Arg-313 were not essential, and probably do not interact with ferric enterobactin.
Table 1.
Phenotypic properties of FepA charge substitution mutants
| FepA mutant | Name | Ferric enterobactin
|
Colicin B
|
Colicin D
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Binding* and transport†
|
Killing,‡ ++ | Binding§Kd | Killing,‡ ++ | Binding§Kd | |||||
| Kd | Km | Vmax | Nutrition | ||||||
| ++ | FepA | 22 | 155 | 85 | 20 | 100 | 145 | 100 | 169 |
| R283A | A | 23 | 253 | 97 | 20 | 100 | 210 | 100 | 184 |
| R286A | B | 19 | 571 | 98 | 22 | 100 | 140 | 20 | 188 |
| R313A | C | 26 | 522 | 109 | 20 | 100 | 190 | 100 | 187 |
| R316A | D | 27 | 309 | 97 | 21 | 20 | 180 | 4 | 173 |
| R274A | E | 21 | 152 | 79 | 20 | 100 | 108 | 100 | 177 |
| R283/286A | AB | 28 | 524 | 70 | 21 | 100 | 220 | 80 | 133 |
| R283/313A | AC | 25 | 186 | 73 | 20 | 100 | 290 | 100 | 174 |
| R283/316A | AD | 27 | 161 | 64 | 21 | 10 | 250 | 2 | 312 |
| R274/283A | AE | 20 | 287 | 96 | 20 | 100 | 142 | 100 | 227 |
| R286/313A | BC | 21 | 335 | 87 | 21 | 100 | 226 | 20 | 192 |
| R274/286A | BE | 29 | 108 | 74 | 20 | 100 | 148 | 100 | 152 |
| R313/316A | CD | 23 | 268 | 101 | 21 | 20 | 170 | 80 | 303 |
| R274/313A | EC | 32 | 132 | 105 | 20 | 100 | 510 | 100 | 215 |
| R274/316A | DE | 26 | 145 | 76 | 20 | 20 | 142 | 2 | 232 |
| R/283/286/313A | ABC | 12 | 1676 | 77 | 21 | 25 | 120 | 10 | 215 |
| R283/313/316A | ACD | 44 | 316 | 93 | 20 | 100 | 200 | 12.5 | 408 |
| R274/R313/R316A | ECD | 22 | 354 | 150 | 20 | 10 | 462 | 2.5 | 275 |
| R286/316A | BD | 701 | 2,745 | 24 | 17 | 2 | 220 | 0.2 | 517 |
| R283/286/316A | ABD | 1,586 | 2,834 | 18 | 15 | 5 | 210 | 2 | 865 |
| R286/313/316A | BCD | 1,708 | >10,000 | 37 | 14 | 2 | 280 | 1 | 1,065 |
| R233/286/313/316A | ABCD | >5,000 | >10,000 | 0 | 9 | 0.1 | 145 | 0.2 | 1,699 |
Kd values (nM) were determined from the concentration-dependence of 59Fe-enterobactin binding; the mean values from four independent experiments were plotted with grafit (Elsevier), using the “Bound versus Total” equation. The mean SE of the fitted curves that produced the Kd values was 32%, with a SD of 8.6%.
The Km (nM) and Vmax (pmol/min per 109 cells) of transport were determined from 59Fe-enterobactin uptake assays (26); mean values from two independent experiments were plotted with grafit to yield the kinetic parameters. The mean SE of the fitted curves that produced the Km values was 46%, with a SD of 17%. The diameters of growth halos in siderophore nutrition assays are reported in millimeters. The tabulated values represent a single experiment, but little variation was observed among several trials.
Bacteria were plated in top agar, and serial dilutions of colicin B or D were spotted onto the plates to determine the colicin titer on the various mutants, relative to wild-type FepA. The tabulated values represent a single experiment, but little variation was observed among several independent trials.
Combined data from two independent binding experiments were fit with grafit, using the Bound versus Total equation. For colicin B, the mean SE of the fitted curves that produced the Kd values (nM) was 30%, with a SD of 6%, whereas for colicin D, the mean SE was 21%, with a SD of 7.5%.
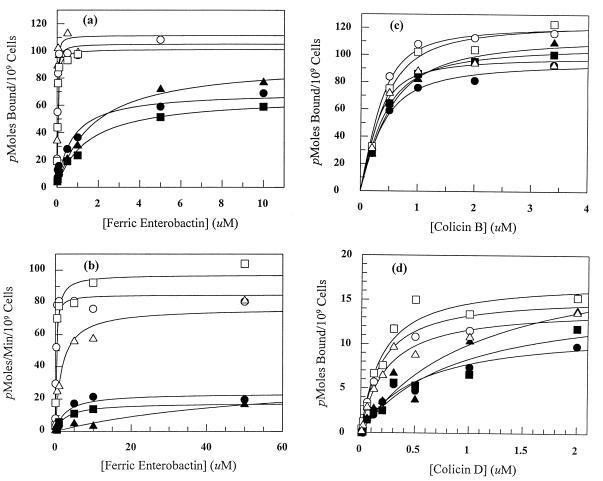
Figure 2.
Concentration dependence of ligand binding and transport by site-directed substitution mutants. (a) Ferric enterobactin binding; (b) ferric enterobactin transport; (c) colicin B binding; and (d) colicin D binding. ○, ++; □, D; ▵, ABC; •, BD; ▪, ABD; and ▴, BCD. Data points are the mean values of at least two experiments.
The fundamental importance of Arg-286 and Arg-316 appeared again in ferric enterobactin transport. Only constructions combining the B and D mutations affected the concentration dependence of siderophore uptake or nutrition (Fig. 3; Table 1). Among the 21 charge replacement mutations we analyzed, only consolidations of B and D significantly increased the siderophore transport Km (20-fold). Ferric enterobactin transport and nutrition were insensitive to 2- to 3-fold decreases in siderophore binding affinity, but both were impaired by 20-fold or greater reductions, which was consistent with the observation that binding to wild-type FepA occurred at lower concentrations (Kd = 20 nM) than ferric enterobactin transport (Km = 200 nM). The level of FepA expression in these experiments, either calculated from autoradiographic measurements of FepA expression (Fig. 3) or derived from ferric enterobactin binding capacity, was 80,000–90,000 monomers per cell, which translated to a maximum ferric enterobactin transport rate of approximately one molecule per FepA monomer per minute.
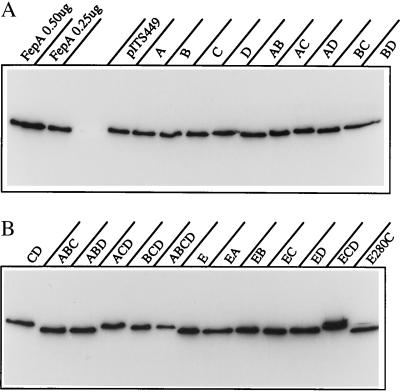
Figure 3.
Expression of FepA mutants. Samples from one experiment (2 × 108 bacteria) were lysed and subjected to Western immunoblot with anti-FepA mAb 41 (11) and 125I-protein A. The paper was subjected to autoradiography and FepA expression quantitated on a Packard Instant Imager.
Effects of Arginine Replacement on Colicin Binding and Killing.
Investigation of colicin adsorption and killing in the same set of mutants strengthened conclusions about the influence of Arg-286 and Arg-316 in the FepA ligand-binding site. As in siderophore transport, the BD double mutation profoundly reduced susceptibility to colicin B or colicin D (Table 1). It was noteworthy that, for colicin B, the reduction in killing did not originate from impaired binding to FepA. We did not find significant differences in the affinities of the various mutants for colicin B, including the BD double mutant and its siblings (Fig. 2; Table 1). For colicin D, on the other hand, the reduced killing of FepA BD mutants did originate, at least in part, from deficiencies in cell surface binding, in an identical manner to the binding defects observed with ferric enterobactin (Fig. 2; Table 1). Susceptibility to colicin D, furthermore, showed more dependence (10-fold) on the presence of Arg-286 and Arg-316 than susceptibility to colicin B (Table 1).
Inhibition of Covalent Modification in the FepA Ligand-Binding Domain by Ferric Enterobactin.
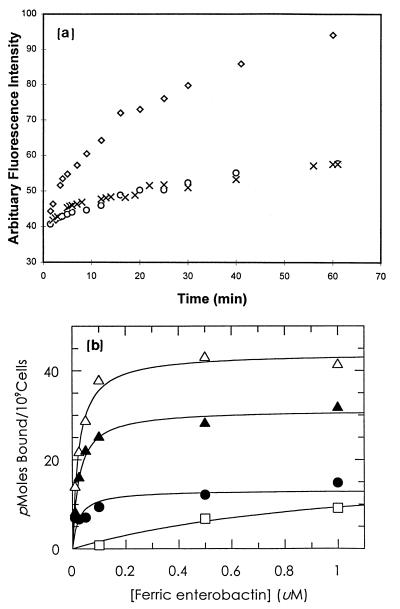
To further test the idea that ferric enterobactin physically contacts the positive charge cluster in region 255–336, we studied the chemical susceptibility of the site-directed substitution E280C, six residues upstream from Arg-286. In vivo and in vitro, FepAE280C bound and transported ferric enterobactin like wild-type FepA (ref. 33; data not shown). The engineered cysteine in purified FepAE280C was readily modified by the fluorescent probe CPM, unless ferric enterobactin was included in the labeling reaction (Fig. 4). Ferric enterobactin protected residue 280, located between Arg-274 and Arg-283 in the positive charge cluster of PL5. Finally, modification of wild-type FepA with the arginine-specific reagent PG prevented siderophore binding, and siderophore binding blocked FepA inactivation by PG (Fig. 4). These data confirm by chemical methods the involvement of arginines in siderophore recognition and demonstrate that ferric enterobactin binding shields functionally important arginines within the ligand-binding site.
Figure 4.
(a) Covalent modification of FepA with CPM in the absence and presence of ferric enterobactin. Purified wild-type FepA (○) or FepAE280C (◊) was incubated with the thiol-specific probe CPM, and the time course of fluorescence was recorded. FepAE280C was also incubated with CPM in the presence of 10 μM ferric enterobactin (×). (b) Effect of PG modification on ferric enterobactin binding by FepA. Bacteria were grown and tested for siderophore binding as described in Fig. 2, either with (filled symbols) or without (open symbols) prior modification by PG. The binding of 59Fe-enterobactin to RWB18-60 (fepA, □) and RWB18-60/pITS449 (fepA+), preincubated with PG (•), 10 μM ferric enterobactin and PG (▴), or 10 μM ferric enterobactin alone (▵), was measured. PG modification blocked ferric enterobactin binding, but preincubation with ferric enterobactin inhibited the PG-induced inactivation of FepA, protecting 70% of its binding capacity. The graph depicts mean values from two independent experiments.
DISCUSSION
The x-ray crystallographic solution of general (OmpF) and specific (LamB) porin folding explained the foundations of OM protein structure (16–18), but the TonB-dependent gated porins, like FepA, present a formidable next step in the understanding of OM protein architecture. A multitude of iron-transporting OM proteins exist among commensal and pathogenic Gram-negative bacteria alike, and ample evidence exists for the relationship between iron absorption and bacterial virulence (1, 5). Yet the instability, low abundance, energy dependence, and requirements for other proteins (i.e., TonB) increase the difficulty of relevant physical studies on siderophore receptors (34). The chemical lability of ferric enterobactin further complicates a crystallographic localization of its binding site in FepA, and to circumvent this problem, we used a noncrystallographic approach that analyzed ligand binding in vivo. Other investigators have successfully used alanine-scanning mutagenesis of charged residues to analyze protein structures (35) and double mutations to study interactions between proteins (36); our experiments applied double mutagenesis as a streamlined site-directed genetic method for identification of crucial residues in a multivalent binding event.
Multiple chemical interactions between the siderophore and the receptor likely explain why Arg → Ala substitutions were transparent individually but detrimental together. Single mutations in essential residues did not obstruct ferric enterobactin adsorption, ostensibly because sufficient residual charge remained in the recognition site to support binding. Double mutations that combined an essential residue with a nonessential residue also did not prevent binding, but double mutations in two essential residues severely impaired the binding event. These findings illustrate the logic and the advantages of double mutagenesis. Single site-directed substitutions may not abrogate or even reduce ligand binding or transport. At the other extreme, exhaustive combinatorial replacement is excessively arduous: the number of such constructions increases steeply with increasing numbers of target residues. For example, the generation of all possible substitutions at 5, 6, and 7 sites requires 31, 63, and 127 unique combinations, respectively [according to the formula C = n!/r!(n − r)!, where C is the total possible combinations of n sites taken r at a time]. The production of all double mutants from 5, 6, and 7 targets, on the other hand, involves only 10, 15, and 21 constructions, respectively, a more readily achievable objective.
A concern of protein analysis by mutagenesis is the differentiation of general structural perturbations from local, ligand-specific defects. Global conformational changes in OM proteins usually result in improper localization, degradation, and inviability. The site-directed substitutions in FepA did not exhibit these effects (Fig. 3). All the mutants, including ABCD, were active in transport and/or nutrition (albeit in some cases at much reduced levels), indicating that FepA assembled normally and functioned in the OM. Furthermore, structural deformities in the mutants were not detectable by cytofluorimetric analyses (2, 3) of antibody binding to FepA surface epitopes (data not shown). Perhaps most important, the impaired siderophore transport phenotype created by BD and its combinations was consistently traced to defects in ferric enterobactin binding. Varying degrees of binding impairment led to proportional reductions in siderophore transport Km. This quantitative correlation between loss of binding and loss of transport supports the idea that the mutated residues lie in a primary ligand-binding domain. Ultimately, three different approaches, site-directed mutagenesis, protection of specific residues by siderophore binding, and arginine-specific chemical inactivation, independently implicated Arg-286 and Arg-316 in siderophore recognition. These results establish the location of, and identify critical amino acids within, the cell surface siderophore binding site of FepA. They clarify the chemical basis (ionic bonds) of siderophore adsorption to a TonB-dependent OM protein. Although BD mutants showed minimal ferric enterobactin uptake over the 5-min assay period, they used the siderophore in nutrition assays (Table 1). This residual transport suggests that other ferric enterobactin binding determinants may also exist in FepA, which persist in the absence of Arg-286 and Arg-316.
Guterman (6) initially described the competition between ferric enterobactin and colicin B for binding to FepA on the E. coli cell surface. Our results explain her observations in molecular terms: the sensitivity of all three ligands to alterations in the Arg-286 and Arg-316 side chains demonstrates the commonality of these amino acids in siderophore transport and colicin killing. Every charge replacement in FepA that impaired ferric enterobactin uptake also decreased killing by colicins B and D. Furthermore, colicin D susceptibility was the most sensitive phenotypic trait to mutations in FepA, and the only one affected by single substitutions in the charge cluster (R316A). As with the siderophore, for colicin D, we measured decreases in binding affinity that accompanied the reductions in killing, indicating that this toxin also uses Arg-283 and Arg-316 as essential elements of productive binding. Yet none of the mutations reduced the binding of colicin B, which distinguished it from colicin D and reiterated that FepA surface topology was not generally disrupted by the mutations that impaired ferric enterobactin and colicin D adsorption. Bacteriocins are much larger molecules than siderophores [the molecular weights of colicins B, D, and ferric enterobactin are 58,000 (37), 75,000 (38), and 716 Da (1), respectively], which likely interact with OM proteins over a larger surface area involving more contact residues (23). The independence of colicin B from Arg-286 and Arg-316 during binding, combined with its dependence on these residues for killing, intimates that these amino acids function in the internalization of the bacteriocin through the OM bilayer. This may also be true for ferric enterobactin, as evidenced by the effects of the BD mutation on its transport Vmax.
Acknowledgments
We thank Dr. Tom Ferenci for helpful discussions, Marjorie A. Montague for expert technical assistance, and Drs. Leon T. Rosenberg, Paul F. Cook, J.B. Neilands, and Chris Murphy for their comments on the manuscript. This work was supported by National Science Foundation Grant MCB 9212070 and National Institutes of Health Grants GM53836 and RR1182, to P.E.K.
ABBREVIATIONS
- OM
outer membrane
- CPM
7-diethylamino-3-(4′-maleimidylphenyl)-4-methylcoumarin
- PG
phenyl glyoxal
References
- 1.Neilands J B. J Biol Chem. 1993;270:26723–26726. doi: 10.1074/jbc.270.45.26723. [DOI] [PubMed] [Google Scholar]
- 2.Rutz J M, Liu J, Lyons J A, Goranson S K, McIntosh M A, Feix J B, Klebba P E. Science. 1992;258:471–475. doi: 10.1126/science.1411544. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Rutz J M, Feix J B, Klebba P E. Proc Natl Acad Sci USA. 1993;90:10653–10657. doi: 10.1073/pnas.90.22.10653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Killmann H, Benz R, Braun V. EMBO J. 1993;12:3007–3016. doi: 10.1002/j.1460-2075.1993.tb05969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerinot M L. Annu Rev Microbiol. 1993;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 6.Guterman S. Biochem Biophys Res Commun. 1971;44:1149–1155. doi: 10.1016/s0006-291x(71)80206-1. [DOI] [PubMed] [Google Scholar]
- 7.Wayne R R, Frick K, Neilands J B. J Bacteriol. 1976;126:7–12. doi: 10.1128/jb.126.1.7-12.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugsley A P, Reeves P. J Bacteriol. 1976;127:218–228. doi: 10.1128/jb.127.1.218-228.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollifield W C, Jr, Fiss E H, Neilands J B. Biochemistry. 1978;17:1922–1928. doi: 10.1021/bi00603a019. [DOI] [PubMed] [Google Scholar]
- 10.Fiss E H, Stanley-Samuelson P, Neilands J B. Biochemistry. 1982;21:4517–4522. doi: 10.1021/bi00261a050. [DOI] [PubMed] [Google Scholar]
- 11.Murphy C K, Kalve V I, Klebba P E. J Bacteriol. 1990;172:2736–2346. doi: 10.1128/jb.172.5.2736-2746.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jalal M A F, van der Helm D. FEBS Lett. 1989;243:366–370. doi: 10.1016/0014-5793(89)80163-2. [DOI] [PubMed] [Google Scholar]
- 13.Wang C C, Newton A. J Bacteriol. 1969;98:1142–1150. doi: 10.1128/jb.98.3.1142-1150.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raymond K N. Pure Appl Chem. 1994;4:773–781. [Google Scholar]
- 15.Neilands J B, Erickson T J, Rastetter W H. J Biol Chem. 1981;256:3831–3832. [PubMed] [Google Scholar]
- 16.Weiss M S, Abele U, Weckesser J, Welte W, Schiltz E, Schulz G E. Science. 1991;254:1627–1630. doi: 10.1126/science.1721242. [DOI] [PubMed] [Google Scholar]
- 17.Cowan S W, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit R A, Jansonius J M, Rosenbusch J P. Nature (London) 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 18.Schirmer T, Keller T A, Wang Y-F, Rosenbusch J P. Science. 1995;267:512–514. doi: 10.1126/science.7824948. [DOI] [PubMed] [Google Scholar]
- 19.Lundrigan M D, Kadner R J. J Biol Chem. 1986;261:10797–10801. [PubMed] [Google Scholar]
- 20.Tumumuru M K R, Armstrong S K, McIntosh M A. Abstracts of the American Society for Microbiology Annual Meeting. Washington, DC: Am. Soc. Microbiol.; 1990. p. 234. [Google Scholar]
- 21.Dean C R, Poole K. J Bacteriol. 1993;175:317–324. doi: 10.1128/jb.175.2.317-324.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beall B, Sanden G. Microbiology. 1995;141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong S A, Francis C A, McIntosh M A. J Biol Chem. 1990;265:14536–14543. [PubMed] [Google Scholar]
- 24.Messing J. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rutz J M, Abdullah T, Kalve V I, Singh S P, Klebba P E. J Bacteriol. 1991;173:5964–5974. doi: 10.1128/jb.173.19.5964-5974.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ecker D, Matazanke B, Raymond K N. J Bacteriol. 1986;167:666–673. doi: 10.1128/jb.167.2.666-673.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pugsley A P, Reeves P. Antimicrob Agents Chemother. 1977;11:345–353. doi: 10.1128/aac.11.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman H A, Leonard K. Biochemistry. 1990;29:10640–10649. doi: 10.1021/bi00499a010. [DOI] [PubMed] [Google Scholar]
- 30.Sippel T O. J Histochem Cytochem. 1981;29:1377–1381. doi: 10.1177/29.12.7320496. [DOI] [PubMed] [Google Scholar]
- 31.Sippel T O. J Histochem Cytochem. 1981;29:314–316. doi: 10.1177/29.2.7019305. [DOI] [PubMed] [Google Scholar]
- 32.Means G E, Feeney R E. Chemical Modification of Proteins. San Francisco: Holden–Day; 1971. p. 196. , 224. [Google Scholar]
- 33.Liu J, Rutz J M, Klebba P E, Feix J B. Biochemistry. 1994;33:13274–13283. doi: 10.1021/bi00249a014. [DOI] [PubMed] [Google Scholar]
- 34.Klebba P E, Rutz J M, Liu J, Murphy C K. J Bioenerg Biomembr. 1993;25:603–611. doi: 10.1007/BF00770247. [DOI] [PubMed] [Google Scholar]
- 35.Wertman K F, Drubin D G, Botstein D. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pohlschroder M, Murphy C K, Beckwith J. J Biol Chem. 1996;271:19908–19914. doi: 10.1074/jbc.271.33.19908. [DOI] [PubMed] [Google Scholar]
- 37.Schram E, Mende J, Braun V, Kamp R M. J Bacteriol. 1987;169:3350–3357. doi: 10.1128/jb.169.7.3350-3357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timmis K. J Bacteriol. 1972;109:12–20. doi: 10.1128/jb.109.1.12-20.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]