Abstract
The replication checkpoint is a dedicated sensor-response system activated by impeded replication forks. It stabilizes stalled forks and arrests division, thereby preserving genome integrity and promoting cell survival. In budding yeast, Tof1 is thought to act as a specific mediator of the replication checkpoint signal that activates the effector kinase Rad53. Here we report studies of fission yeast Swi1, a Tof1-related protein required for a programmed fork-pausing event necessary for mating type switching. Our studies have shown that Swi1 is vital for proficient activation of the Rad53-like checkpoint kinase Cds1. Together they are required to prevent fork collapse in the ribosomal DNA repeats, and they also prevent irreversible fork arrest at a newly identified hydroxyurea pause site. Swi1 also has Cds1-independent functions. Rad22 DNA repair foci form during S phase in swi1 mutants and to a lesser extent in cds1 mutants, indicative of fork collapse. Mus81, a DNA endonuclease required for recovery from collapsed forks, is vital in swi1 but not cds1 mutants. Swi1 is recruited to chromatin during S phase. We propose that Swi1 stabilizes replication forks in a configuration that is recognized by replication checkpoint sensors.
Replication of a eukaryotic genome is a challenging task that is often made more difficult by conditions that interfere with replisome progression. These circumstances include DNA lesions that obstruct replicative polymerases, drugs that target DNA polymerases or enzymes required to synthesize deoxynucleoside triphosphates (dNTPs), and protein complexes bound to DNA (7, 36). Stalled replication forks are prone to collapse, regression, and recombination (32). Collapsed forks are among the most serious forms of DNA damage and as such pose a grave threat to cell survival and genome integrity (28). Discovering how cells cope with aberrant replication forks is therefore essential for understanding mechanisms of genome maintenance.
Studies of budding and fission yeasts have uncovered a network of proteins that form the replication checkpoint (7, 36). Central to this network are protein kinases of the ATM/ATR family, such as Mec1 in the budding yeast Saccharomyces cerevisiae and Rad3 in the fission yeast Schizosaccharomyces pombe. In fission yeast, Rad3 forms a complex with Rad26 and functions together with a trimeric checkpoint clamp (Rad1-Rad9-Hus1) and five-subunit checkpoint clamp loader (Rad17-RFC2-RFC3-RFC4-RFC5) to sense stalled replication forks and transmit a checkpoint signal. These proteins act together with Mrc1, a mediator of the replication checkpoint, to activate the replication checkpoint effector Cds1, a protein kinase homologous to Rad53 in budding yeast and Chk2 in humans (1, 46).
Budding yeast rad53 mutants starved of dNTPs will arrest in S phase and accumulate aberrant DNA structures such as regressed forks or hemireplicated regions, and they are unable to fully resolve replication intermediates when dNTP levels are restored (30, 44, 47). How Rad53 preserves stalled forks is unknown, but there is accumulating evidence that it and Cds1 control phosphorylation of several replication and recombination proteins (7, 36). These proteins include Mus81, a subunit of the Mus81-Eme1 DNA endonuclease complex. Mus81-Eme1 is required for recovery from collapsed forks and is thought to be a component of Holliday junction resolvase in fission yeast. Mus81 physically interacts with Cds1 homologs in budding yeast, fission yeast, and humans (5, 6, 11, 22). Mammalian Chk2 plays an important role in controlling the apoptotic response to DNA damage, apparently through its ability to control the p53 tumor suppressor, and recent evidence has indicated that Chk2 is a bona fide tumor suppressor (21, 45).
In some situations, stalled forks are important for cell vitality, as is the case for cell type switching in fission yeast (12, 13). Mating type in fission yeast is determined by the expressed gene cassette at the mat1 locus. Mating type switching occurs when the DNA cassette at mat1 is replaced by one of two silent donor cassettes. Switching requires a strand-specific imprinting event that occurs when mat1 is replicated in a specific direction. The direction of replication of mat1 is determined by polar replication fork pausing and termination sites located near mat1 and is dependent on several proteins. One of these proteins is Swi1, a 971-amino acid (aa) protein that has ∼25% sequence identity to Drosophila melanogaster Timeless, mammalian Tim1, and budding yeast Tof1 (13). Timeless controls circadian rhythms in Drosophila (35), whereas mouse Tim1 is an essential nuclear protein that does not regulate circadian rhythms but whose function is unknown (19). Tof1, first identified as topoisomerase 1-associated factor in a two-hybrid screen, is involved in DNA damage responses during S phase (16). Mutant tof1 cells exhibit no obvious phenotypes and are not sensitive to genotoxic agents, but deletion of TOF1 enhances the genotoxic-sensitive phenotype of rad9 mutants. Rad9 is a mediator protein that facilitates activation of Rad53 by Mec1 (18, 42). Deletion of TOF1 exacerbated the Rad53 activation defect of rad9 mutants, suggesting that Tof1 and Rad9 act in redundant pathways to control Rad53 activation (16).
Fission yeast swi1 mutants have additional phenotypes that are unconnected to mating type switching (13). They have a reduced growth rate that is exacerbated by mutations in top1, which encodes topoisomerase I. Swi1 is essential for viability in a mutant that is partially defective for DNA polymerase alpha. These studies indicated that Swi1 might have a more general role in DNA replication. Here we report that Swi1 is crucial for survival of replication fork arrest. Swi1 has both Cds1-dependent and -independent functions. Swi1 appears to act early in the response to fork arrest, perhaps as a replisome component, to stabilize forks in a configuration that is recognized by the replication checkpoint sensors.
MATERIALS AND METHODS
General techniques.
Methods for genetic and biochemical analyses of fission yeast have been described previously (2, 33). For immunoblotting, extracts from ∼108 cells were made by glass bead disruption in lysis buffer A (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% NP-40, 10% glycerol, 50 mM NaF, 1 mM Na3VO4, 5 mM EDTA, 5 mM N-methylmaleimide, 1 μM microcystin, 0.1 μM okadaic acid, and 1 mM dithiothreitol) supplemented with protease inhibitors (0.2 mM para-(4-amidoinophenyl)-methane sulfonyl fluoride hydrochloride monohydrate) and 5 μg each of leupeptin, pepstatin, and aprotinin per ml). Protein extracts were clarified by centrifugation at 15,000 × g for 5 min at 4°C. Immunoblotting and Cds1 kinase assays have been described previously (29, 46). In situ chromatin binding assays were carried out as previously described (25). Mating type switching assays were performed as previously described (20). h90 strains were plated on sporulation medium and incubated for 6 to 7 days at 25°C. Plates were exposed to iodine vapors. Colonies that have efficient mating type switching stain darkly with iodine vapors, whereas inefficient strains show mottled staining. For UV sensitivity assays, cells were exposed to a short wavelength (254 nm) in a Stratalinker machine (Stratagene, La Jolla, Calif.).
2D gel electrophoresis.
Two-dimensional (2D) gel electrophoresis was performed as previously described (8, 26). Cells grown in YES medium (yeast extract with supplements) at 30°C were arrested with 0.1% sodium azide and chilled on ice for 5 min. Approximately 2 × 109 cells were harvested, washed in ice-cold water, and frozen at −80°C. Genomic DNA was purified as described previously (43). Cell pellets were resuspended in 5 ml of nuclear isolation buffer containing 17% glycerol, 50 mM morpholinepropanesulfonic acid (MOPS) (pH 7.2), 150 mM potassium acetate, 2 mM MgCl2, 500 μM spermidine, and 150 μM spermine. Cells were treated with 1 mg of Zymolyase 100T per ml at 37°C for 20 min and then lysed by the addition of 4 volumes of ice-cold water. Nuclei were collected by centrifugation and resuspended in 3 ml of TEN buffer containing 50 mM Tris (pH 8.0), 50 mM EDTA, and 100 mM NaCl. The nuclei were treated with 0.3 mg of proteinase K per ml in 1.5% N-laurylsarcosine at 37°C for 1 h. Cell debris were removed by centrifugation at 3,000 × g. CsCl and Hoechst 33258 were added to final concentrations of 1.05 g/ml and 0.02 mg/ml, respectively. Samples were centrifuged at 45,000 rpm in a 65VTi rotor at 20°C for 16 h. The genomic DNA band was collected and precipitated twice with ethanol. For analysis of ars3001, 5 μg of DNA was digested with 30 U each of HindIII and KpnI. Precipitated DNA was run on a 0.4% agarose gel for the first dimension and a 1.0% agarose gel for the second dimension. For the analysis of the ars3001 5′ region, DNA was digested with HindIII and BamHI. Precipitated DNA was run on a 1.0% agarose gel for the first dimension and a 2.0% agarose gel for the second dimension. Gels were transferred to Hybond N+ membranes. After hybridization, radioactive signals were detected with a Molecular Dynamics Storm 840 machine. The ars3001 gels were probed with the 3-kb HindIII-KpnI ribosomal DNA (rDNA) fragment (48). The ars3001 5′ gels were probed with the 0.8-kb HindIII-BamHI fragment as shown in Fig. 3.
FIG. 3.
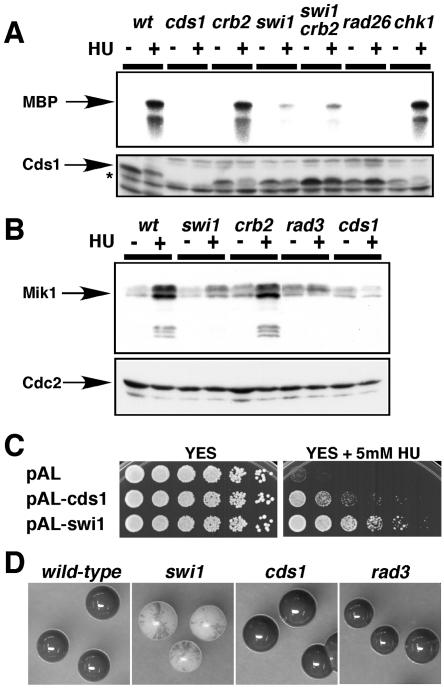
Deficient Cds1 activation in swi1 mutant. (A) Cds1 activation is strongly reduced in swi1 mutant cells. Cells of the indicated genotypes were incubated in YES liquid medium supplemented with 0 or 12 mM HU for 4 h at 30°C. Kinase activity of immunoprecipitated Cds1 was measured by using myelin basic protein (MBP) substrate (upper panel). A Cds1 immunoblot confirmed that approximately equal amounts of Cds1 (absent in cds1 mutant strain) were present in the samples (lower panel). The Cds1 polyclonal antisera cross-react with nonspecific proteins that migrate faster than Cds1 (asterisk). (B) HU induction of Mik1 accumulation is deficient in swi1 mutant cells. Cells of the indicated genotypes that contained genomic mik1-13myc were incubated with 0 or 12 mM HU for 4 h at 30°C. Mik1 was detected with anti-Myc monoclonal antibody (upper panel). A Cdc2 immunoblot was used as a loading control (lower panel). (C) HU sensitivity of swi1 mutant cells was partially suppressed by multicopy cds1+ plasmid. Fivefold serial dilutions of swi1 mutant cells transformed with the indicated plasmids were incubated on YES agar medium supplemented with the indicated amount of HU for 2 to 5 days at 30°C. (D) Colonies of homothallic wild-type and swi1, cds1, and rad3 mutant cells were grown in sporulation medium and then stained by iodine vapor to detect spores. The swi1 mutant colonies showed a mottled phenotype, indicating a defect in mating type switching, whereas wild-type and cds1 and rad3 mutant cells stained darkly with iodine, indicating proficient mating type switching.
Strains and plasmids.
The following strains were used for this study: PR109 (h−), BF2406 (h− rad3::ura4+ mik1-13myc:Kanr his7-366), BF2408 (h− cds1::ura4+ mik1-13myc:Kanr his7-366), BF2506 (h− mik1-13myc:Kanr his7-366), BF2115 (h+ cds1::ura4+ chk1::ura4+), BF2521 (h− chk1-9myc2HA6His:ura4+), EN3180 (h− rad26::ura4+), EN3181 (h+ chk1::ura4+), EN3182 (h− swi1::Kanr), EN3183 (h− swi1::Kanr cds1::ura4+), EN3184 (h+ swi1::Kanr chk1::ura4+), EN3185 (h− swi1::Kanr mik1-13myc:Kanr), EN3186 (h− crb2::ura4+ mik1-13myc:Kanr), EN3187 (h− crb2::ura4+ chk1-9myc2HA6His:ura4+), EN3188 (h− swi1::Kanr chk1-9myc2HA6His:ura4+), EN3189 (h− cds1::ura4+ chk1-9myc2HA6His:ura4+), EN3190 (h+ mus81::Kanr), EN3191 (h+ swi1-GFP:Kanr), EN3192 (h− swi1::Kanr mus81::Kanr), EN3193 (h− swi1::Kanr rhq1::ura4+), EN3194 (h+ cds1::ura4+ mus81::Kanr), EN3195 (h90 rad3::ura4+), EN3196 (h90 cds1::ura4+), EN3197 (h90 swi1::Kanr), EN3204 (h− swi1-GFP:Kanr rad26::ura4+), EN3220 (h+ rad22-YFP:Kanr cds1::ura4+), EN3221 (h+ rad22-YFP:Kanr swi1::Kanr), EN3222 (h− rad22-YFP:Kanr), EN3279 (h+ swi1::Kanr rad26::ura4+), EN3280 (h+ swi1::Kanr crb2::ura4+), EN3285 (swi1::Kanr rad13::ura4+ uve1::LEU2), EN3286 (rad13::ura4+ uve1::LEU2), EN3287 (h90 swi1-GFP:Kanr), FG2216 (h− crb2::ura4+), KT2751 (h− cds1::ura4+), PR636 (h90), PS2375 (h− rqh1::ura4+), and PS2383 (h− smt0 rhp51::ura4+). All strains are leu1-32 and ura4-D18 mutants. The 3.6-kb swi1+ genomic fragment was amplified by EXtaq polymerase (TaKaRa) and introduced into the SacI/BamHI site of pAL-sk (34), resulting in pAL-swi1. pAL-cds1 has been described previously (46).
RESULTS
Swi1 is important for survival of replication fork arrest.
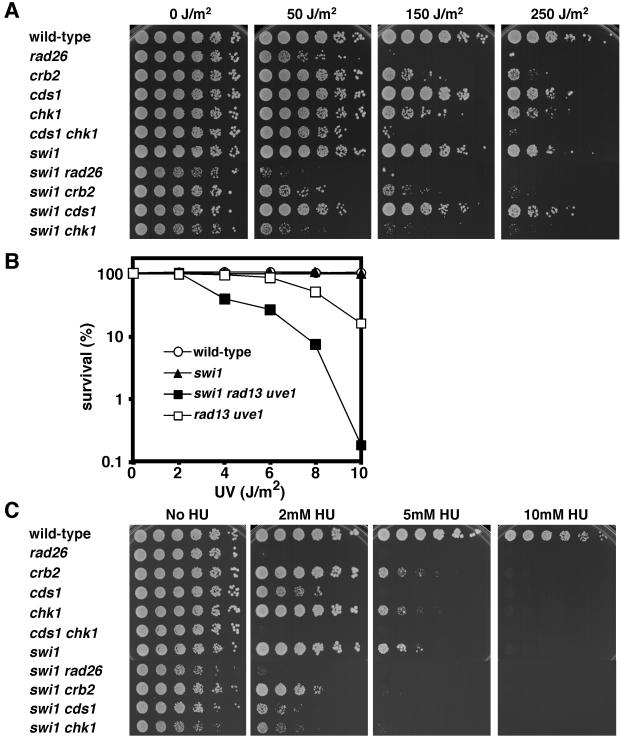
Exposure of DNA to UV irradiation causes the formation of cyclobutane pyrimidine dimers and other lesions that block the progression of replicative polymerases (17). To explore whether Swi1 is involved in tolerance of DNA lesions that block fork progression, survival assays were performed with a set of mutant strains exposed to UV (Fig. 1A). (All mutant strains used for this study had deletions of alleles.) Serial dilutions of cells plated on growth media were exposed to increasing doses of UV (Fig. 1A). The swi1 mutant was only weakly UV sensitive, indicating that Swi1 does not have a direct role in the repair of UV lesions. However, there was a strong synergistic interaction with inactivation of Chk1, the effector kinase of the DNA damage checkpoint. The swi1 chk1 mutant was much more sensitive to UV than either single mutant (Fig. 1A). The absence of Chk1 abolishes the G2-M DNA damage checkpoint, thereby greatly increasing the number of cells that enter S phase with unrepaired UV lesions. A similar synergistic interaction occurs between cds1 and chk1 mutations (Fig. 1A), as previously reported (29). Loss of Swi1 substantially increased UV sensitivity in a rad26 mutant that is defective in activation of both the Cds1 and Chk1 pathways (Fig. 1A), suggesting that Swi1 has a checkpoint-independent role in UV damage tolerance. This function may explain why swi1 chk1 mutant cells are more UV sensitive than rad26 or cds1 chk1 mutant cells (Fig. 1A). Curiously, the swi1 mutation displayed a somewhat stronger synergistic interaction with chk1 than with crb2, even though crb2 cells were more sensitive to UV than chk1 cells (Fig. 1A).
FIG. 1.
Replication checkpoint function of Swi1. (A) Synergistic interactions of swi1 and chk1 mutations in UV survival assays indicate that Swi1 is required for tolerance of UV damage during DNA replication. Fivefold serial dilutions of cells were plated on YES agar medium and then exposed to the indicated dose of UV. Agar plates were incubated for 2 to 5 days at 30°C. (B) Swi1 acts independently of the proteins that promote UV damage repair. Cells of the indicated genotypes were spread on YES agar medium and exposed to the indicated dose of UV. Agar plates were incubated for 3 days at 30°C to measure UV survival. (C) Swi1 is involved in survival of HU-induced replication arrest. Synergistic interactions of swi1 with cds1 and chk1 mutations indicate that Swi1 has an HU survival function that is at least partially independent of Cds1 and Chk1. Fivefold serial dilutions of cells were incubated on YES agar medium supplemented with the indicated amount of HU for 2 to 5 days at 30°C.
The strong synergistic interaction involving chk1 and swi1 mutations indicated that Swi1 is vital for tolerance of replication fork arrest caused by UV lesions. To test this hypothesis, we determined whether Swi1 contributed to survival in a mutant strain defective for nucleotide excision repair (NER) and UV damage excision repair (UVER). NER and UVER account for all detectable UV damage repair in fission yeast (49). NER was eliminated by inactivation of rad13+, which encodes a protein related to FEN-1 endonucleases. UVER was inactivated by mutation of uve1+, which encodes the UV damage endonuclease. The swi1 mutation substantially enhanced UV sensitivity in the rad13 uve1 double-mutant background (Fig. 1B). These data supported the proposition that Swi1 is important for UV damage tolerance.
Hydroxyurea (HU), a compound that inactivates ribonucleotide reductase, causes replication fork arrest by depleting dNTPs. Serial dilutions of cells plated on medium containing increasing concentrations of HU showed that Swi1 plays an important role in HU tolerance (Fig. 1C). Colony formation by swi1 mutant cells was severely compromised by 5 mM HU, whereas wild-type cells readily formed colonies under these conditions. As was the case for UV, there was a strong synergistic interaction between the swi1 and chk1 mutations (Fig. 1C). Replication fork arrest is thought to lead to DNA damage that is repaired in the subsequent G2 phase, explaining why chk1 mutants are sensitive to HU (9). Unlike the situation for UV, there was a reproducible synergistic interaction between swi1 and cds1 mutations. This interaction indicated that Swi1's role in tolerating HU-induced replication arrest might be partially independent of Cds1. As was the case for the UV sensitivity assay, swi1 crb2 mutant cells displayed weaker HU sensitivity than did swi1 chk1 mutant cells (Fig. 1C). The potential meaning of this finding is explored in Discussion.
Replication checkpoint and Cds1 activation defects in swi1 mutant cells.
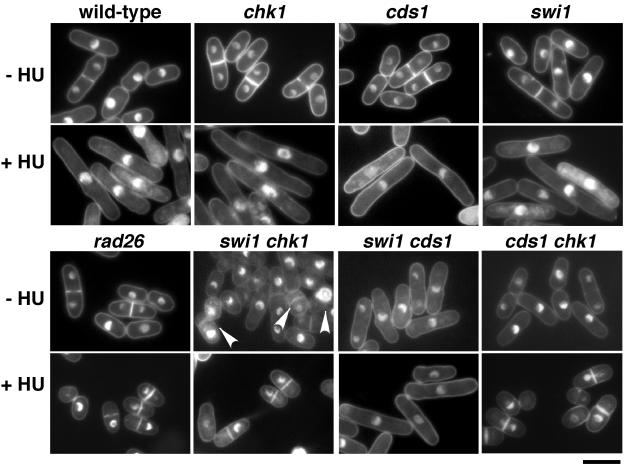
We next assessed Swi1's role in HU-induced division arrest. Cds1 and Chk1 have overlapping roles in this arrest (4, 29); therefore, Swi1's role was evaluated in a cds1 or chk1 background. Wild-type and swi1, cds1, and chk1 mutant strains arrested division in the presence of HU, whereas rad26 mutant cells, which are unable to activate Chk1 and Cds1, displayed the mitotic catastrophe or “cut” phenotype that is indicative of checkpoint failure. Double-mutant swi1 chk1 cells failed to arrest division in HU, whereas arrest was proficient in swi1 cds1 mutant cells (Fig. 2). These experiments showed that Swi1 is required for HU-induced cell cycle arrest enforced by Cds1.
FIG. 2.
Swi1 is required for Cds1 pathway of replication checkpoint arrest. The indicated strains were incubated in YES liquid medium supplemented with 0 or 12 mM HU for 6 h at 30°C and then stained with DAPI to visualize nuclear DNA. Wild-type and chk1, cds1, swi1, and swi1 cds1 mutant cells treated with HU underwent checkpoint arrest, as indicated by the appearance of elongated, uninucleate cells without septa. In contrast, rad26, cds1 chk1, and swi1 chk1 mutant cells treated with HU failed to undergo cell cycle arrest and instead displayed aberrant mitosis as indicated by a cut phenotype. The cut phenotype was also observed in ∼12% of septated swi1 chk1 mutant cells grown in the absence of HU (arrowheads). Bar, 10 μm.
These findings prompted analysis of Swi1's role in Cds1 activation. Cds1 was immunoprecipitated and assayed using myelin basic protein as a substrate. HU caused potent activation of Cds1 in wild-type or chk1 mutant cells, whereas Cds1 activation was eliminated in rad26 mutant cells (Fig. 3A), as previously reported (4, 29). Very weak Cds1 activation occurred in swi1 mutant cells (Fig. 3A). This defect was reflected in immunoblot analysis of Mik1 (Fig. 3B), a mitotic inhibitor kinase that accumulates in HU-treated cells via a Cds1-dependent mechanism (4). Residual activity of Cds1 seen in swi1 mutant cells is consistent with the observation that swi1 chk1 mutant cells are less HU sensitive than rad26 or cds1 chk1 mutant cells (Fig. 1A).
In budding yeast, Tof1 and Rad9 are thought to have redundant roles in activating Rad53 (16). Therefore, we examined whether Crb2, a Rad9-related protein, had any roles in Cds1 activation in swi1 mutant cells. We observed that swi1 crb2 double-mutant cells had no further decrease in Cds1 activation than swi1 single-mutant cells in response to HU (Fig. 3A). Together with the finding that crb2 mutant cells had a wild-type level of Cds1 activation (Fig. 3A), these experiments showed that Crb2 was not involved in activating Cds1.
To explore whether the HU-sensitive phenotype of swi1 mutant cells can be attributed to a defect in Cds1 activation, we tested whether Cds1 overexpression rescued swi1 mutants. A multicopy plasmid containing cds1+ substantially improved the growth of swi1 mutant cells in medium containing 5 mM HU (Fig. 3C). These findings indicated that the HU-sensitive phenotype of swi1 mutant cells was at least partly attributable to a defect in Cds1 activation.
Replication fork collapse and arrest in swi1 and cds1 cells treated with HU.
Our studies indicated that Swi1 and Tof1 were functional homologs that controlled Cds1 and Rad53, respectively. Rad53 stabilizes replication forks in HU-treated cells, preventing formation of aberrant structures and maintaining forks in a replication-competent state (30, 44, 47). To investigate whether Cds1 and Swi1 participate in fork stabilization, 2D gel analysis was used to detect replication intermediates (RIs) from the rDNA repeats (41). Fission yeast has ∼150 tandem arrayed rDNA repeats at the ends of chromosome III. Each rDNA repeat has a single replication origin designated ars3001 located in the nontranscribed spacer (NTS) region (Fig. 4A). About half of the rDNA origins are activated during each cell cycle. A polar replication pause site located ∼2 kb to the left of ars3001 inhibits fork progression from entering the rDNA genes counter to the direction of transcription (41).
FIG.4.
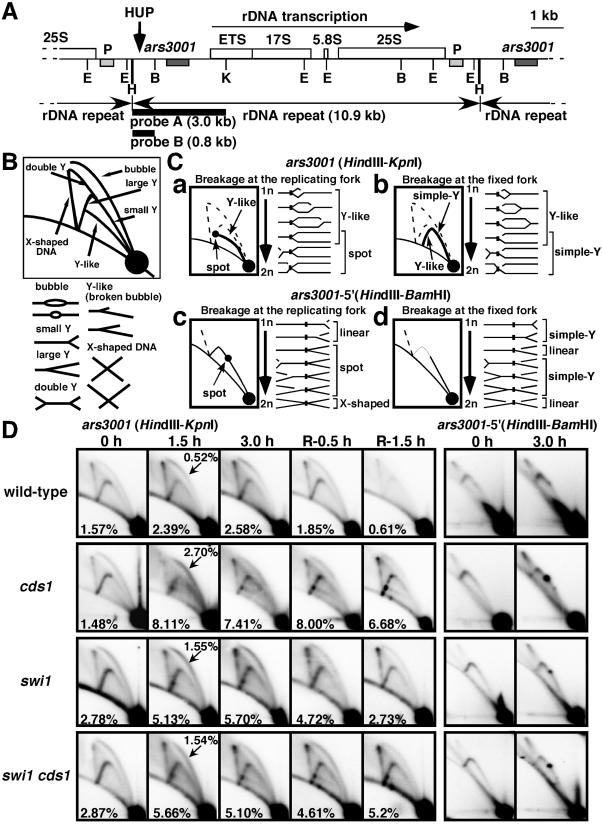
2D gel analysis of replication forks in swi1 and cds1 mutants. (A) Map of the rDNA repeats as reported by Sanchez et al. (41). The ars3001 box indicates the origin region and the P box indicates a pause site mapped by Sanchez et al. (41). The HUP site is indicated, as are the probes and restriction enzyme sites (E, EcoRI; H, HindIII; B, BamHI; K, KpnI). (B) Diagram of the migration pattern of replication intermediates that can be detected by 2D gel electrophoresis. (C) Diagrams of potential Y-like arc and pause site patterns. (D) Wild-type and cds1, swi1, and swi1 cds1 mutant cells were incubated in YES liquid medium supplemented with 12 mM HU for the indicated times at 30°C. Genomic DNA prepared from cells was analyzed by 2D gel electrophoresis. The five columns on the left show 2D gels of DNA digested with HindIII and KpnI and hybridized with probe A. Cells were incubated with HU for 0, 1.5, or 3 h and then HU was washed out and cells were incubated for a further 0.5 or 1.5 h. Numbers in the panels indicate fractions of RIs relative to total signal. Numbers with arrows in 1.5-h samples represent fractions of bubble structures relative to total signal. The two columns on the right show gels digested with HindIII and BamHI and hybridized with probe B. Cells were incubated with HU for 0 or 3 h.
The HindIII-KpnI fragment that encompasses ars3001 was analyzed in samples taken from wild-type and cds1, swi1, and swi1 cds1 mutant cells before, during, and after treatment with HU. Patterns from untreated cells were similar among all the strains (Fig. 4B to D). Bubbles, small and large Y structures, double-Y structures, and X-shaped DNA structures were detected. Bubbles arise from origin activation within the HindIII-KpnI region, as do the majority of large Y's, whereas small Y's and double-Y's are formed by passive replication. X-shaped structures are thought to arise from recombination associated with DNA replication and are more common in fission yeast than in budding yeast (43).
HU treatment led to an increase in bubbles in all strains, consistent with accumulation of cells in S phase (Fig. 4D). More bubbles appeared in the mutant strains and they were generally smaller than in the wild type. These observations indicated that latent origins were activated in the mutants but not in the wild type, consistent with previous studies of rad53 and cds1 mutants (26, 30).
HU treatment of cds1, swi1, or swi1 cds1 mutant cells led to the appearance of a novel Y-like arc that was not observed in the wild type (Fig. 4B to D). Intersection of this arc with the Y arc produced a distinctive spot that was faintly visible in HU-treated wild-type cells but was readily apparent in the mutants. The Y-like arc was similar to broken fork structures detected with simian virus 40 replicons (24), but it most closely resembled an arc detected with unidirectionally replicated ColE1 plasmids in bacteria (31). The ColE1 Y-like arc arose from breakage of the migrating replication fork, creating a series of asymmetrical Y structures that have two arms of fixed but unequal lengths and a third arm of variable length (Fig. 4C, panel a). The position of the spot on the Y arc indicated that the HU-induced pause (HUP) site was located ∼1kb away from ars3001 (Fig. 4A and D). We analyzed flanking fragments of ars3001 by 2D gel analyses (data not shown) and confirmed that the HUP site was located ∼1kb to the left of ars3001 and ∼1kb to the right of the previously mapped pause site (P) that inhibits leftward-moving forks (Fig. 4A) (41).
A different type of Y-like arc is predicted if breakage of a paused fork stops migration of the diverging fork (Fig. 4C, panel b). This Y-like arc intersects with the linear DNA line. This arc was not detected in the mutant strains, indicating preferential breakage of the moving fork. To substantiate this conclusion, we examined the 0.8-kb HindIII-BamHI fragment that contains the HUP site (Fig. 4D, right panels). Arrest at the HUP site without fork breakage should produce a distinctive spot on the Y arc, whereas fork breakage at the HUP site should produce an arc that fades at the HUP site (Fig. 4C, panels c and d). HU treatment of cds1 mutant cells created an intense spot on the Y arc, whereas a very weak spot was detected in the wild type (Fig. 4D). Intermediate intensity Y-arc spots were observed in HU-treated swi1 or swi1 cds1 mutant cells. These observations confirmed that Swi1 and Cds1 were required for efficient progression through the HUP site in HU-treated cells.
Samples taken after HU removal showed that Swi1 and Cds1 were important for recovery from HU arrest (Fig. 4D). RIs rapidly disappeared from wild-type cells but persisted in cds1, swi1, and swi1 cds1 mutant cells. These findings were consistent with studies of Rad53 (30, 47) and indicated that Swi1 and Cds1 were needed to maintain stalled replication forks in a replication-competent state. The Y-like arc appeared to persist longer in cds1 mutant cells than in swi1 or swi1 cds1 mutant cells after HU removal (Fig. 4D). swi1 mutant cells retained weak Cds1 activity (Fig. 3A), which might help to resolve broken forks in swi1 mutant cells. Double-mutant swi1 cds1 cells had inefficient replication (our unpublished results) and this might explain why the Y-like arc was weak in these cells relative to the cds1 single-mutant cells.
Swi1 has Cds1-independent functions.
The aforementioned studies indicated that Swi1 functions in a Cds1-dependent pathway, consistent with the model proposed for Tof1 and Rad53 in budding yeast (16). There were, however, strong indications that Swi1 also has significant Cds1-independent functions. Notably, swi1 and cds1 mutations synergistically interacted for survival in HU, showing that Swi1 contributed to survival in HU in the absence of Cds1 (Fig. 1C). We also noticed that chk1 displayed a stronger genetic interaction with swi1 than cds1 in UV survival assays (Fig. 1A). In addition, we found that swi1 rad26 mutant cells were more UV sensitive than were rad26 mutant cells (Fig. 1A). Furthermore, swi1 chk1 mutant cells displayed an elevated frequency of spontaneous aberrant mitoses when grown without HU (Fig. 2). Analyses showed that 12% (12 of 100) of septated swi1 chk1 mutant cells displayed a cut phenotype, whereas rad26 (0 of 100) or cds1 chk1 (0 of 100) mutant cultures contained no cut cells. These findings correlated with a reduced growth rate of swi1 chk1 mutant cells in medium lacking HU compared to those of other cells, including cds1 chk1 mutants (Fig. 1C). Moreover, swi1 mutant cells were moderately elongated, and this mitotic delay required Chk1 (Fig. 2). These observations demonstrate that Swi1 has activities that are independent of Cds1.
cds1 mutant cells are proficient in mating type switching.
To further explore the relationship between Swi1 and Cds1, we determined whether mating type switching required Cds1 or Rad3. Switching-proficient strains mate and form spores that can be stained with iodine vapors. As seen in Fig. 3D, wild-type and cds1 and rad3 mutant colonies produced uniformly intense iodine staining, whereas the swi1 mutant colonies had a mottled phenotype typical of switching-defective mutants. Therefore, Swi1's role in mating type switching does not involve Cds1 or Rad3, the latter of which is needed for both Cds1 and Chk1 activation.
Rad22 foci accumulate in swi1 mutant cells.
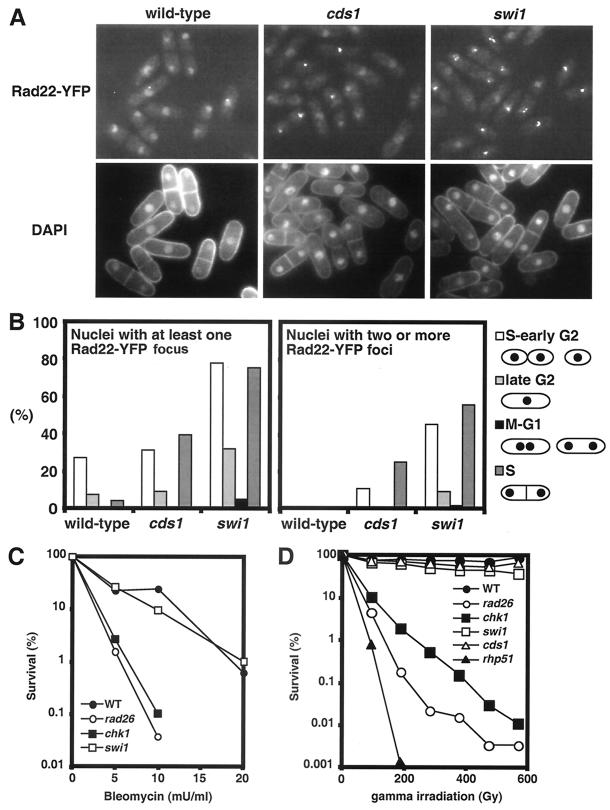
Genetic interactions involving swi1 and chk1 mutations indicated that swi1 mutant cells experience spontaneous DNA damage that triggers a Chk1-dependent mitotic delay. We investigated this possibility by analyzing localization of Rad22-yellow fluorescent protein (YFP) fusion protein. Rad22 is a Rad52 homolog that plays a central role in repair of DNA breaks (37, 38). Chromatin immunoprecipitation analysis showed that Rad22 is recruited to the double-strand break (DSB) at mat1 (27). We recently found that Rad22-YFP forms foci at DSBs in live cells (15). Rad22-YFP displayed diffuse nuclear fluorescence in wild-type cells (Fig. 5A). Quantitative analysis showed that 15.8% of the nuclei in these cells contained a single Rad22-YFP focus. No wild-type nuclei contained multiple Rad22-YFP foci. In contrast, 50.3% of the swi1 mutant nuclei had at least one Rad22-YFP focus and 28.3% had multiple foci (Fig. 5A). An intermediate phenotype was observed in nuclei of cds1 mutant cells, with 22.1% having at least one Rad22-YFP focus and 9.1% having multiple foci (Fig. 5A).
FIG. 5.
Spontaneous Rad22-YFP foci formation in swi1 mutant cells. (A) Cells that had genomic rad22-YFP were grown in EMM medium at 25°C until mid-log phase. DNA was stained with DAPI. Rad22-YFP foci formation was strikingly elevated in swi1 mutant cells. (B) Quantification of Rad22-YFP foci according to cell cycle stages. S and early G2 cells had the most Rad22-YFP foci. (C) Swi1 is not required for survival of DNA damage caused by bleomycin. Cells of the indicated genotypes were treated with the indicated concentrations of bleomycin for 2 h and then incubated on YES plates for 3 days to measure survival. (D) Swi1 is not required for survival of DNA damage caused by IR. Cells of the indicated genotypes were tested for resistance to IR. Irradiated cells were incubated on YES plates for 3 days to measure survival.
To explore whether Rad22 foci arose from replication abnormalities, the cell cycle position of cells containing Rad22-YFP foci was estimated by analyzing cell length, nuclei number and position, and the presence of a division plate (Fig. 5B). Nuclei with one or more Rad22-YFP foci were heavily skewed towards the S and early G2 phases, indicating that swi1 mutant cells frequently experience DNA damage during replication.
These observations suggested that swi1 mutant cells experience increased rates of fork collapse. An alternative explanation was that swi1 mutant cells were deficient in repairing DNA damage that normally occurs during S phase, leading to persistence of Rad22-YFP foci at DNA breaks. As mentioned above (Fig. 1A and B), swi1 mutant cells were only weakly sensitive to UV irradiation, indicating that Swi1 was not generally required for repair of DNA damage. To specifically address whether Swi1 was involved in DSB repair, swi1 mutant cells were tested for sensitivity to bleomycin, a radiomimetic drug that causes DSBs. Wild-type and swi1 mutant cells survived bleomycin treatment equally well, whereas rad26 and chk1 checkpoint mutants were profoundly sensitive to bleomycin (Fig. 5C). We also examined swi1 mutant cells for sensitivity to ionizing radiation (IR) that causes DSBs. Wild-type and cds1 and swi1 mutant cells had no obvious sensitivities to IR. This phenotype contrasted with the profound sensitivity of chk1, rad26, and rhp51 mutant cells to IR (Fig. 5D). These results show that Swi1 is not required for DSB repair and support the idea that swi1 mutant cells have increased Rad22-YFP foci because of elevated rates of fork collapse.
Mus81 is vital in the absence of Swi1.
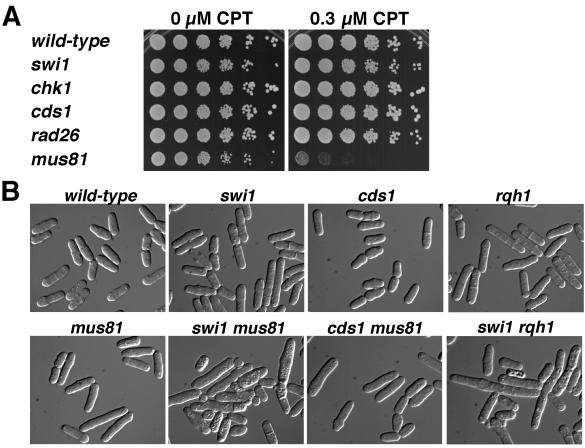
Mus81-Eme1 is crucial for recovery from replication fork collapse (5, 6), a fact made most evident by the sensitivity of mus81 mutant cells to camptothecin (CPT), a drug that traps topoisomerase I while it makes a single-strand nick in DNA (14). The replication fork breaks when it collides with the CPT-stabilized topoisomerase I-DNA complex (23, 39). Restoration of the fork necessarily forms a Holliday junction that must be resolved to segregate chromosomes in mitosis (32). We confirmed that mus81 mutants are profoundly sensitive to CPT, more so than even the rad26 mutant that lacks both the replication and DNA damage checkpoints (Fig. 6A). This relationship contrasts with that to IR, in which rad26 mutant cells are extremely sensitive to IR (Fig. 5D), whereas mus81 mutants are only slightly IR sensitive (3, 6). DSBs induced by IR in G2 are thought to be repaired by a synthesis-dependent strand annealing mechanism that does not require Holliday junction resolution (38). Hence, mutations that cause fork collapse should have strong genetic interactions with the mus81 mutation that causes a defect in processing collapsed forks, whereas mutations that cause DSBs to arise after replication should not display strong genetic interactions with mus81.
FIG. 6.
Genetic interactions involving Swi1 and Mus81. (A) Mus81 is vital for cell survival in CPT. Fivefold serial dilutions of cells were incubated for 2 to 3 days at 30°C on YES agar medium supplemented with 0 or 0.3 μM CPT. Growth of mus81 mutant cells was severely impaired by 0.3 μM CPT, whereas growth of wild-type and swi1 and checkpoint mutant strains was unaffected. The swi1 and checkpoint mutant strains were sensitive to higher concentrations of CPT (data not shown). (B) Mus81 and Rqh1 are vital in swi1 mutant background. Cells of indicated genotypes were grown in YES liquid medium and photographed. The swi1 mus81 and swi1 rqh1 double mutants showed a severe growth defect, with many elongated and vacuolated cells, whereas cds1 mus81 mutant cells and other single mutants grew relatively well.
Tetrad analysis revealed that most swi1 mus81 mutant spores failed to form colonies. The spores germinated and formed a small group of highly elongated and misshapen cells. A few swi1 mus81 mutant spores formed visible colonies that were much smaller than the corresponding single mutants. In liquid culture, these cells were elongated and vacuolated, and many cells were inviable (Fig. 6B). The swi1 mus81 mutant cells had a doubling time of ∼8 h compared to ∼2.5 h for each of the single mutants. These data further indicated that the frequent Rad22-YFP foci that appeared in swi1 mutant cells during S phase arose from replication fork collapse. In contrast to swi1 mus81 mutant cells, cds1 mus81 mutant cells grew as well as mus81 single-mutant cells (doubling time, ∼2.5 h) and few were elongated (Fig. 6B). This difference reflected the significantly fewer Rad22-YFP foci detected in cds1 mutant cells.
Rqh1 is a RecQ-like DNA helicase that is thought to be involved in stabilizing or processing stalled replication forks (6). Double-mutant rqh1 mus81 cells are inviable (6). Rqh1 and Mus81 might participate in separate pathways of restoring stalled replication forks that have regressed to form Holliday junctions, although it is likely that Rqh1 has multiple roles in DNA replication, processing of stalled forks, and recombinational repair (5). We investigated genetic interactions between swi1 and rqh1. Most swi1 rqh1 mutant spores failed to grow and a few formed tiny colonies. These cells were highly elongated and extremely sick, with a doubling time of ∼8 h (Fig. 6B). These findings show that Rqh1 is vital in the absence of Swi1 and suggest that the two proteins have independent functions.
Swi1 associates with chromatin in S phase.
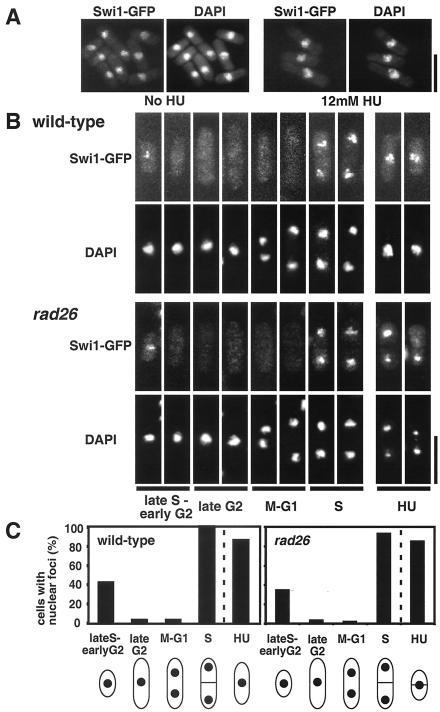
To further understand Swi1's role in fork stability and checkpoint signaling, we determined whether Swi1-green fluorescent protein (GFP) associated with chromatin. Cells that expressed Swi1-GFP from the genomic locus were insensitive to HU and proficient in mating type switching (data not shown), indicating that Swi1-GFP was functional. Swi1-GFP colocalized with DAPI (4′,6′-diamidino-2-phenylindole)-stained DNA in cells at all stages of the cell cycle and in HU-treated cells (Fig. 7A). In situ chromatin binding assays were used to determine whether Swi1-GFP interacts with chromatin (25). Strikingly, after Triton X-100 extraction of soluble nuclear proteins, Swi1-GFP was detected predominantly in septated cells and short cells corresponding to those that were in S or early G2 phase (Fig. 7B and C). Prior treatment with DNase I eliminated all Swi1-GFP (data not shown), confirming that detergent-insoluble Swi1-GFP was bound to chromatin. More than 85% of HU-treated cells retained Swi1-GFP after Triton X-100 extraction (Fig. 7B and C). These were also sensitive to DNase I treatment (data not shown). Interestingly, Swi1-GFP chromatin recruitment was proficient in rad26 mutant cells (Fig. 7B and C). These data indicate that Swi1's role in fork stability and checkpoint signaling involves direct interaction with chromatin.
FIG. 7.
Swi1-GFP is recruited to chromatin in S phase. (A) Cells that had genomic swi1-GFP were incubated in EMM liquid medium supplemented with 0 or 12 mM HU for 6 h at 25°C. Cells were modified into spheroplasts and fixed prior to microscopic analysis of Swi1-GFP fluorescence. All cells displayed nuclear Swi1-GFP. Essentially identical results were obtained with living cells (data not shown). (B) In situ chromatin binding assay of Swi1-GFP protein in wild-type and rad26 mutant backgrounds. The assay was the same as that described for panel A, except that spheroplasts were extracted with Triton X-100 to remove soluble nuclear protein and then were fixed for microscopic analysis. Representative patterns of fluorescence are shown. Swi1-GFP protein was detected predominantly in septated cells and unseptated small cells, which are in S phase or possibly early G2 phase. Representative photos of HU-treated cells are also shown. (C) Quantification of Triton X-100-resistant nuclear Swi1-GFP in wild-type and rad26 mutant cells according to cell morphology.
DISCUSSION
These studies establish that Swi1 is vital for Cds1 activation and that loss of either protein has severe consequences, including fork collapse and irreversible fork arrest, in HU-treated cells. These findings strongly suggest that Swi1 and Tof1 are functional homologs, indicating broad functional conservation of this family of proteins. These studies also show that Swi1 has important functions that are independent of Cds1, some of which are revealed in the absence of HU. They include enhanced sensitivity to UV lesions encountered during replication, elevated formation of Rad22 DNA repair foci during S phase, and striking genetic interactions involving Mus81 and Rqh1. Coupled with evidence that Swi1 associates with chromatin during S phase, these studies strongly suggest that Swi1 has a proximal role in stabilizing forks in a manner that is recognized by the replication checkpoint apparatus.
Swi1 and Tof1 are homologs.
One important conclusion of these studies is that Swi1 and Tof1 are functional homologs. This fact was not previously obvious because Swi1 and Tof1 were highly divergent and equally related to Drosophila Timeless, a protein that controls circadian rhythms. Moreover, the reported phenotypes of swi1 or tof1 mutants had little in common. Our studies establish that Swi1 is crucial for Cds1 activation in a manner that closely approximates the role of Tof1 in controlling Rad53 in rad9 mutant cells (16). RAD53 is essential, and therefore residual Cds1 activation in swi1 mutants is probably mirrored by weak Rad53 activity in rad9 tof1 mutant cells. Furthermore, it was reported that Crb2, the Rad9-like protein in fission yeast, is not involved in Cds1 regulation and instead is absolutely required for Chk1 function (40). Foss (16) proposed that Tof1 and Rad9 have overlapping roles in the activation of Rad53 in response to fork arrest, but are they involved in responding to precisely the same types of replication intermediates? Tof1 might act at the earliest stage of fork impediment, with Rad9 becoming involved when a fork has regressed, collapsed, or formed other abnormal structures. Curiously, we found that swi1 had a stronger synergistic interaction with chk1 than with crb2 in sensitivity to UV and HU (Fig. 1A and C). These facts suggest that Crb2 interferes with recovery from fork arrest in swi1 mutant cells. Interestingly, experiments performed with budding yeast strains synchronized to enter an HU-induced arrest showed that Rad53 activation was delayed in mrc1 mutants relative to the wild type (1). The activation of Rad53 in mrc1 mutants required Rad9, a relationship that is analogous to that between Tof1 and Rad9 (16). The delay of Rad53 activation in mrc1 mutant cells might be attributable to an undiscovered form of cell cycle regulation of Rad9, but an alternative possibility is that the delay is due to time-dependent collapse or rearrangement of the fork into a structure that is recognized by Rad9. In this model, Tof1 and Mrc1 act in a common pathway to sense stalled forks and activate Rad53, whereas Rad9-dependent activation of Rad53 occurs when the Tof1-Mrc1 pathway fails or after bulk DNA replication is complete.
We showed that Swi1 and Tof1 are structural and functional homologs, but what about homologs, such as Tim1, in mammals? Tim1 is an essential protein in mice (19). We speculate that the greater size and complexity of the mouse genome may lead to a higher incidence of fork stalling. These problems may create a greater demand for a Swi1/Tof1 homolog, explaining why Tim1 is an essential protein.
Swi1 and Cds1 protect impeded forks.
Detection of the ars3001-associated Y-like arc in HU-treated cds1 or swi1 mutants has provided unexpected insights into the fate of replication forks in checkpoint mutants. The Y-like arc formed when the leftward fork arrested at the HUP site and the rightward fork broke. Fork breakage without concomitant arrest of the diverging fork at a specific site would generate a continuous series of arcs that would form a shadow under the Y arc. Y-arc shadows are not a distinctive or indisputable feature of 2D gels, and thus their meaning has been uncertain. The existence of the ars3001 Y-like arc provides compelling physical evidence that replication forks break in fission yeast replication checkpoint mutants treated with HU.
The HUP site appears to be a novel genetic element. It does not correspond to the previously identified pause region that inhibits fork progression from entering rDNA genes counter to the direction of transcription (41). Persistence of the spot on the Y arc after removal of HU demonstrates that the paused fork is converted to a permanently arrested fork. Collapse of the fork converging into the HUP region from the adjacent rDNA repeat presumably contributes to persistence of the Y-like arc after HU removal. Why the fork arrests at HUP in cds1 or swi1 mutant cells is unknown. We speculate that a protein complex bound to HUP impedes fork progression when the replisome is slowed by limited availability of dNTPs. Cds1 activation seems to be crucial for maintaining the replisome in a competent state when it enters the HUP region.
It remains to be determined whether fork collapse and irreversible fork arrest during HU treatment occur throughout the genome in swi1 or cds1 mutants. It was recently found that chromosomes tend to break in ∼10-kb replication slow zones (RSZs) spaced at large chromosome intervals (∼50 kb) in budding yeast mec1 hypomorphic mutants that are presumably defective in dNTP synthesis and Rad53 activation (10). The relatively large RSZs seem to be significantly different from the narrowly defined HUP site, but the underlying mechanism of chromosome breakage in the RSZs may be similar to the fork breakage described in this report.
Detection of the Y-like arc in cds1 cells seems to contradict the notion that Cds1 protects stalled forks from breakage. Formation of the Y-like arc requires arrest but not breakage of the leftward fork at the HUP site. Perhaps forks collapse in HU-treated cds1 cells not because stalled forks are unstable but because leading- and lagging-strand DNA synthesis is uncoordinated. Budding yeast rad53 mutants treated with HU accumulate extensive single-stranded gaps near the fork, indicative of a lagging-strand synthesis defect (44). These structures may be the precursors to fork collapse, as opposed to forks that have coordinately stopped leading- and lagging-strand synthesis because they have encountered a replication obstacle.
The studies of Dalgaad and Klar (13) indicated that Swi1 was specifically required to initiate fork pausing, perhaps being directly involved as a protein that impedes replication fork progression near the mat1 locus. Our findings suggest a different model in which Swi1 acts after a fork has encountered a barrier. Blockage can be caused by a protein complex bound to DNA, as might be the case for the pausing and termination sites near the mat1 locus, or by a DNA lesion such as a cyclobutane pyrimidine dimer that is an impassable obstacle to the replisome. Failure to stabilize a stalled fork may have different consequences depending on the circumstances. In the case of a cyclobutane pyrimidine dimer lesion, failure of Swi1-dependent fork stabilization may result in fork collapse, as indicated by the synergistic interaction involving swi1 and chk1 mutations (Fig. 1A). In the situation of fork pausing near the mat1 locus, which apparently does not involve an impassable obstacle, the absence of Swi1 may lead to resumption of fork progression instead of fork collapse. This response may appear as a defect in pausing, but the defect may reside with events that occur after the fork has encountered a structure that impedes progression. The fact that Swi1 associates with chromatin only during S phase is more consonant with a model in which Swi1 is specifically recruited to a stalled fork or arrives there because of its association with the replisome, as opposed to a model in which Swi1 binds to chromosomes at specific sites and acts as a barrier to fork progression.
Cds1-independent roles of Swi1.
Our studies have revealed that Swi1 has Cds1-independent functions. This fact is evident from the mating type switching proficiency of cds1 mutant cells (Fig. 3D), but there is additional evidence more obviously related to genome integrity mechanisms. In the absence of Chk1, swi1 mutants display a high level of mitotic catastrophe that is not observed in cds1 chk1 or even rad26 mutant cells (Fig. 2). Coupled with the evidence that swi1 mutant cells undergo a Chk1-dependent mitotic delay when grown without genotoxic agents, these observations imply that swi1 mutant cells frequently suffer DNA damage that triggers a G2-M DNA damage checkpoint. This conclusion is supported by the large increase of spontaneous Rad22 foci in swi1 mutant cells (Fig. 5). Most of these foci appeared during S phase, indicating that DNA was damaged during replication and repaired in the G2 phase. The damage was apparently incompletely or improperly repaired in swi1 chk1 mutant cells that underwent mitotic catastrophe. Mus81 was vital for survival in swi1 but not cds1 mutant cells (Fig. 6). The genetic interaction of swi1 and mus81 is most straightforwardly explained by spontaneous fork collapse in the absence of Swi1 followed by replication restart leading to the formation of Holliday junctions.
Swi1's position in the checkpoint signaling network.
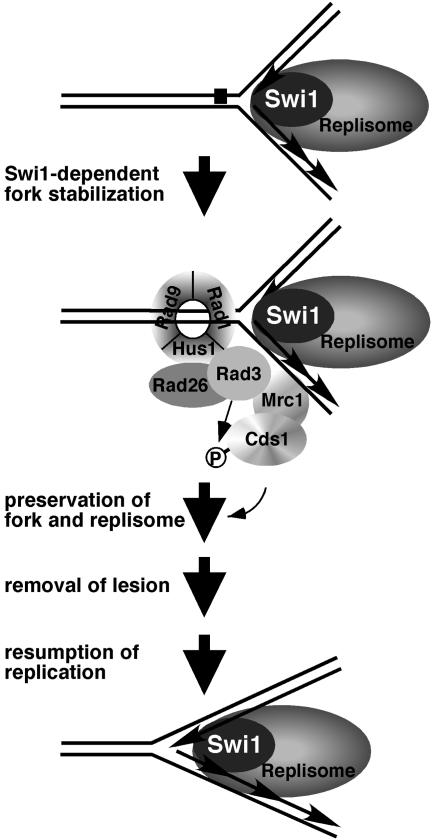
Foss (16) proposed that Tof1 acts as an intermediary in two redundant pathways that link the primary checkpoint sensors (e.g., Mec1) to the checkpoint effector Rad53. Our studies suggest a different model for Swi1's role in checkpoint signaling. Swi1 inactivation causes a range of phenotypes and genetic interactions that are not shared by mutations of checkpoint genes. It seems unlikely, therefore, that Swi1 acts downstream of the checkpoint sensors. Many of the properties associated with swi1 mutations can be explained by the appearance of DNA damage during S phase, suggesting a role for Swi1 in proficient replication. Swi1 is recruited to chromatin during S phase in rad26 mutant strains, further indicating that Swi1 functions independently of Rad26. We therefore favor a model in which Swi1 functions upstream or in parallel with Rad26 and the other checkpoint sensors to detect stalled forks (Fig. 8). In this model, Swi1 acts at the initial stages of the replication fork arrest to stabilize the replisome and protect the fork. Swi1 might be a component of the replisome and one of its roles may be to stabilize the fork and replication apparatus when it encounters obstacles.
FIG. 8.
Proposed model of Swi1's involvement in the response to fork arrest. Swi1 is recruited to chromatin during S phase, suggesting that it might be an ancillary component of the replisome. Swi1 is required for proficient activation of Cds1 in response to replication arrest, suggesting that it stabilizes the stalled fork in a conformation that is recognized by the checkpoint sensor proteins (Rad3-Rad26 and Rad9-Rad1-Hus1 complexes) and Mrc1. Mrc1 facilities Cds1 activation by recruiting it to Rad3 and/or mediating Cds1 autophosphorylation. Cds1 enhances fork and replisome stabilization while the lesion is removed.
Swi1's association with chromatin during S phase does not appear to be increased further by HU treatment. This observation suggests that Swi1's association with chromatin is not triggered by replication arrest but instead is a normal consequence of DNA replication. It does seem reasonable to propose that the replisome has components that are not essential for DNA synthesis per se but are important for stabilizing the replisome when it encounters obstacles. Components such as these would not have been detected by mutant hunts that discovered essential replication proteins, but nevertheless could be vital for preserving genome integrity.
Acknowledgments
We thank J. A. Huberman and M. A. Marchetti for instruction in the technique of 2D gel analysis, T. Wang for providing Cds1 polyclonal antisera, and M. N. Boddy for helpful comments. Members of the Scripps Cell Cycle Groups are thanked for their support and encouragement.
E.N. was supported by the Human Frontier Science Program. L.-L.D. is a fellow of the Leukemia and Lymphoma Society. This work was funded by NIH grant GM059447 awarded to P.R.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell. Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 2.Alfa, C., P. Fantes, J. Hyams, M. McLeod, and E. Warbrick. 1993. Experiments with fission yeast. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 3.al-Khodairy, F., and A. M. Carr. 1992. DNA repair mutants defining G2 checkpoint pathways in Schizosaccharomyces pombe. EMBO J. 11:1343-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boddy, M. N., B. Furnari, O. Mondesert, and P. Russell. 1998. Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280:909-912. [DOI] [PubMed] [Google Scholar]
- 5.Boddy, M. N., P. H. Gaillard, W. H. McDonald, P. Shanahan, J. R. Yates, 3rd, and P. Russell. 2001. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell 107:537-548. [DOI] [PubMed] [Google Scholar]
- 6.Boddy, M. N., A. Lopez-Girona, P. Shanahan, H. Interthal, W. D. Heyer, and P. Russell. 2000. Damage tolerance protein Mus81 associates with the FHA1 domain of checkpoint kinase Cds1. Mol. Cell. Biol. 20:8758-8766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boddy, M. N., and P. Russell. 2001. DNA replication checkpoint. Curr. Biol. 11:R953-R956. [DOI] [PubMed] [Google Scholar]
- 8.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51:463-471. [DOI] [PubMed] [Google Scholar]
- 9.Brondello, J. M., M. N. Boddy, B. Furnari, and P. Russell. 1999. Basis for the checkpoint signal specificity that regulates Chk1 and Cds1 protein kinases. Mol. Cell. Biol. 19:4262-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cha, R. S., and N. Kleckner. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602-606. [DOI] [PubMed] [Google Scholar]
- 11.Chen, X. B., R. Melchionna, C. M. Denis, P. H. Gaillard, A. Blasina, I. Van de Weyer, M. N. Boddy, P. Russell, J. Vialard, and C. H. McGowan. 2001. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell 8:1117-1127. [DOI] [PubMed] [Google Scholar]
- 12.Dalgaard, J. Z., and A. J. Klar. 2001. A DNA replication-arrest site RTS1 regulates imprinting by determining the direction of replication at mat1 in S. pombe. Genes Dev. 15:2060-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalgaard, J. Z., and A. J. Klar. 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102:745-751. [DOI] [PubMed] [Google Scholar]
- 14.Doe, C. L., J. S. Ahn, J. Dixon, and M. C. Whitby. 2002. Mus81-Eme1 and Rqh1 involvement in processing stalled and collapsed replication forks. J. Biol. Chem. 277:32753-32759. [DOI] [PubMed] [Google Scholar]
- 15.Du, L.-L., T. Nakamura, B. A. Moser, and P. Russell. 2003. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 23:6150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foss, E. J. 2001. Tof1p regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157:567-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis. ASM Press, Washington, D.C.
- 18.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 19.Gotter, A. L., T. Manganaro, D. R. Weaver, L. F. Kolakowski, Jr., B. Possidente, S. Sriram, D. T. MacLaughlin, and S. M. Reppert. 2000. A time-less function for mouse timeless. Nat. Neurosci. 3:755-756. [DOI] [PubMed] [Google Scholar]
- 20.Grewal, S. I., and A. J. Klar. 1997. A recombinationally repressed region between mat2 and mat3 loci shares homology to centromeric repeats and regulates directionality of mating-type switching in fission yeast. Genetics 146:1221-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirao, A., A. Cheung, G. Duncan, P. M. Girard, A. J. Elia, A. Wakeham, H. Okada, T. Sarkissian, J. A. Wong, T. Sakai, E. De Stanchina, R. G. Bristow, T. Suda, S. W. Lowe, P. A. Jeggo, S. J. Elledge, and T. W. Mak. 2002. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 22:6521-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 23.Hsiang, Y. H., M. G. Lihou, and L. F. Liu. 1989. Arrest of replication forks by drug-stabilized topoisomerase I-DNA cleavable complexes as a mechanism of cell killing by camptothecin. Cancer Res. 49:5077-5082. [PubMed] [Google Scholar]
- 24.Kalejta, R. F., and J. L. Hamlin. 1996. Composite patterns in neutral/neutral two-dimensional gels demonstrate inefficient replication origin usage. Mol. Cell. Biol. 16:4915-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kearsey, S. E., S. Montgomery, K. Labib, and K. Lindner. 2000. Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J. 19:1681-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, S. M., and J. A. Huberman. 2001. Regulation of replication timing in fission yeast. EMBO J. 20:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, W. J., S. Lee, M. S. Park, Y. K. Jang, J. B. Kim, and S. D. Park. 2000. Rad22 protein, a rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J. Biol. Chem. 275:35607-35611. [DOI] [PubMed] [Google Scholar]
- 28.Kolodner, R. D., C. D. Putnam, and K. Myung. 2002. Maintenance of genome stability in Saccharomyces cerevisiae. Science 297:552-557. [DOI] [PubMed] [Google Scholar]
- 29.Lindsay, H. D., D. J. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray, F. Osman, N. Walworth, and A. M. Carr. 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12:382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Parras, L., P. Hernandez, M. L. Martinez-Robles, and J. B. Schvartzman. 1992. Initiation of DNA replication in ColE1 plasmids containing multiple potential origins of replication. J. Biol. Chem. 267:22496-22505. [PubMed] [Google Scholar]
- 32.McGlynn, P., and R. G. Lloyd. 2002. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 3:859-870. [DOI] [PubMed] [Google Scholar]
- 33.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 34.Murakami, H., and H. Okayama. 1995. A kinase from fission yeast responsible for blocking mitosis in S phase. Nature 374:817-819. [DOI] [PubMed] [Google Scholar]
- 35.Myers, M. P., K. Wager-Smith, C. S. Wesley, M. W. Young, and A. Sehgal. 1995. Positional cloning and sequence analysis of the Drosophila clock gene, timeless. Science 270:805-808. [DOI] [PubMed] [Google Scholar]
- 36.Osborn, A. J., S. J. Elledge, and L. Zou. 2002. Checking on the fork: the DNA-replication stress-response pathway. Trends Cell Biol. 12:509-516. [DOI] [PubMed] [Google Scholar]
- 37.Ostermann, K., A. Lorentz, and H. Schmidt. 1993. The fission yeast rad22 gene, having a function in mating-type switching and repair of DNA damages, encodes a protein homolog to Rad52 of Saccharomyces cerevisiae. Nucleic Acids Res. 21:5940-5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paques, F., and J. Haber. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63:349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porter, S. E., and J. J. Champoux. 1989. The basis for camptothecin enhancement of DNA breakage by eukaryotic topoisomerase I. Nucleic Acids Res. 17:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saka, Y., F. Esashi, T. Matsusaka, S. Mochida, and M. Yanagida. 1997. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 11:3387-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sanchez, J. A., S. M. Kim, and J. A. Huberman. 1998. Ribosomal DNA replication in the fission yeast, Schizosaccharomyces pombe. Exp. Cell Res. 238:220-230. [DOI] [PubMed] [Google Scholar]
- 42.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 43.Segurado, M., M. Gomez, and F. Antequera. 2002. Increased recombination intermediates and homologous integration hot spots at DNA replication origins. Mol. Cell 10:907-916. [DOI] [PubMed] [Google Scholar]
- 44.Sogo, J. M., M. Lopes, and M. Foiani. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599-602. [DOI] [PubMed] [Google Scholar]
- 45.Takai, H., K. Naka, Y. Okada, M. Watanabe, N. Harada, S. Saito, C. W. Anderson, E. Appella, M. Nakanishi, H. Suzuki, K. Nagashima, H. Sawa, K. Ikeda, and N. Motoyama. 2002. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 21:5195-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell. Biol. 3:966-972. [DOI] [PubMed] [Google Scholar]
- 47.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 48.Yanow, S. K., Z. Lygerou, and P. Nurse. 2001. Expression of Cdc18/Cdc6 and Cdt1 during G2 phase induces initiation of DNA replication. EMBO J. 20:4648-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yonemasu, R., S. J. McCready, J. M. Murray, F. Osman, M. Takao, K. Yamamoto, A. R. Lehmann, and A. Yasui. 1997. Characterization of the alternative excision repair pathway of UV-damaged DNA in Schizosaccharomyces pombe. Nucleic Acids Res. 25:1553-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]