Abstract
MK5 (mitogen-activated protein kinase [MAPK]-activated protein kinase 5), also designated PRAK (p38-regulated and -activated kinase), was deleted from mice by homologous recombination. Although no MK5 full-length protein and kinase activity was detected in the MK5 knockout mice, the animals were viable and fertile and did not display abnormalities in tissue morphology or behavior. In addition, these mice did not show increased resistance to endotoxic shock or decreased lipopolysaccharide-induced cytokine production. Hence, MK5 deletion resulted in a phenotype very different from the complex inflammation-impaired phenotype of mice deficient in MK2, although MK2 and MK5 exhibit evolutional, structural, and apparent extensive functional similarities. To explain this discrepancy, we used wild-type cells and embryonic fibroblasts from both MK2 and MK5 knockout mice as controls to reexamine the mechanism of activation, the interaction with endogenous p38 MAPK, and the substrate specificity of both enzymes. In contrast to MK2, which shows interaction with and chaperoning properties for p38 MAPK and which is activated by extracellular stresses such as arsenite or sorbitol treatment, endogenous MK5 did not show these properties. Furthermore, endogenous MK5 is not able to phosphorylate Hsp27 in vitro and in vivo. We conclude that the differences between the phenotypes of MK5- and MK2-deficient mice result from clearly different functional properties of both enzymes.
In mouse and human genomes, a group of three mitogen-activated protein kinase (MAPK)-activated protein (MAPKAP) kinases (MKs), MK2, MK3/3pK, and MK5/PRAK (p38-regulated and -activated kinase), has been identified and the three kinases have been assigned neighboring positions in the phylogenetic tree (14). These three kinases also show apparent functional similarities: they carry a nuclear localization signal, show nuclear localization in resting cells (3, 17), and are exported from the nucleus upon activation (1, 3, 23, 26). Furthermore, the three kinases seem to have similar positions in the stress-activated p38 MAPK signaling pathway, i.e., all three seem to bind to p38 MAPK-α/SAPK2a (15, 26) and are phosphorylated and activated by p38 MAPK-α and -β/SAPK-2a and -2b (2, 15, 19, 21). The substrate specificities of these kinases also seem very similar. The small heat shock protein Hsp25/27 has been described as an efficient substrate for all three kinases in vitro (15, 19, 25) and also for MK2 in vivo (10).
Taking into account these similarities and the ubiquitous expression of these enzymes in the different cells and tissues analyzed (4, 19, 24), it was rather surprising that mice lacking only one of the three kinases, MK2, showed a clear phenotype: deletion of the gene for MK2 led to a defect in lipopolysaccharide (LPS)-induced biosynthesis of cytokines such as tumor necrosis factor (TNF), interleukin-6 (IL-6), and gamma interferon (IFN-γ) (10) and also to changes in cell migration (7, 11). Although the lack in catalytic activity of MK2 was responsible for the defects in cytokine biosynthesis, the lack of MK2 protein also resulted in decreased levels of p38 MAPK in several mouse tissues, probably due to MK2's transport or chaperone properties for p38 MAPK (11). This was unexpected, since both MK3 and MK5, which are coexpressed in most of the tissues analyzed, apparently also bind p38 MAPK and are also translocated from nucleus to cytoplasm. Hence, these kinases should be able to compensate for the loss of MK2 to at least a certain degree.
The reduction of biosynthesis of cytokines in the absence of MK2 is caused by a decrease of mRNA stability and translatability and depends on AU-rich elements in the 3′ nontranslated region of the cytokine mRNAs (16, 29). Interestingly, destabilization of the AU-rich element-containing mRNA of urokinase plasminogen activators in cells in which p38 MAPK signaling is inhibited could be reverted by overexpression of constitutively active MK2 but not by that of the constitutively active mutant of MK5 (6). Similarly, MK2 but not MK5 stimulates phosphorylation-dependent interaction between tuberin and 14-3-3 protein in transfected HEK293 cells (13). These data indicate that although MK2, MK3, and MK5 are similar in structure and assumed position in signaling, there could be significant differences in their functions in vivo. To analyze this possibility further, we deleted the MK5 gene from mice and compared the phenotypes of the resulting MK5 knockout mice and the MK2-deficient mice.
MATERIALS AND METHODS
Isolation of the mouse gene for MK5.
Two pairs of oligonucleotide PCR primers derived from MK5 cDNA (AF039840) were used to screen a 129SvJ P1 genomic library (Genome Systems, St. Louis, Mo.) from which one positive clone was obtained. The clone contained a part (about 13 kb) of the MK5 gene encoding exons 3 (homologous to genome locations 8162573 to 8162670 of the minus strand of the C57BL/6J strain) to 11 (genome locations 8149536 to 8149666 of the minus strand of C57BL/6J strain; NCBI Mouse Genome Resources) of the MK5 gene.
Construction of the targeting vector and transfection of ES cells.
SacI fragments of the P1 clone were subcloned into pBluescript II KS(+). The neomycin cassette was inserted between a 2.0-kb SacI fragment containing exon 5 and a 9.0-kb SpeI/SacI fragment containing exons 7 to 11 (Fig. 1), replacing parts of introns 5 and 6 and the entirety of exon 6. The targeting vector was linearized by digestion with ClaI. E14-1 embryonic stem (ES) cells which were derived from the 129/Ola substrain were a generous gift from Thomas Müller (MDC, Berlin, Germany). Trypsinized ES cells in suspension with the linearized targeting vector were electroporated using a Bio-Rad Gene Pulser II apparatus at 500 μF and 240 V/cm and subsequently seeded on neomycin-resistant mouse embryonic fibroblast (MEF) feeder cells. Selection of transfected ES cells in cell medium containing 385 μg of G-418/ml proceeded for 7 days. Subsequently, clones were picked with 50-μl pipette tips and placed into individual wells of a 96-well plate. After 5 days of cultivation in selection medium, half of the cells were frozen and the other half were allowed to grow to confluence on gelatin-coated 48-well plates for 5 to 7 days before Southern blot analysis was performed.
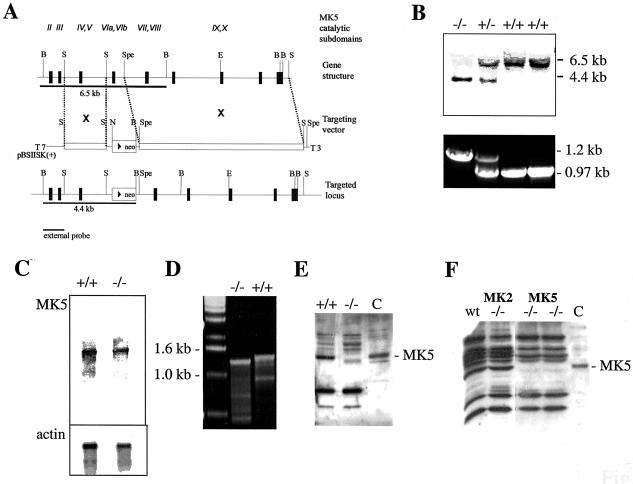
FIG. 1.
Generation of MK5-deficient mice by homologous recombination. (A) Schematic structure of the MK5 gene, the targeting vector, and the targeted locus. Restriction enzymes are indicated as follows: B, BamHI; E, EcoRV; N, NotI; S, SacI; Spe, SpeI. The neomycin cassette (neo) was cloned between SacI and SpeI sites, deleting exon 6 of MK5, which codes for part of kinase subdomains VIa and VIb. (B) Southern blot and PCR analysis of the F1 generation from the chimeric mice generated. For Southern hybridization after BamHI digestion, the external probe indicated in panel A was used. The targeted allele is represented by a 4.4-kb fragment, while the wild-type allele is represented by a 6.5-kb fragment. PCR was carried out using primers at both sides flanking the neo-cassette insertion and the same DNA as for Southern analysis. Insertion was detected by amplification of a 1.2-kb fragment, while experiments using the wild-type allele resulted in amplification of a 0.97-kb fragment. (C) Detection of MK5 mRNA isolated from macrophages of wild-type (+/+) and MK5−/− mice. As an equal loading control, actin mRNA was detected. (D) RT-PCR using macrophage total RNA (as analyzed in panel C) as the template. The fragment amplified from wild-type (+/+) macrophage mRNA was about 1.4 kb, while the fragment from MK5-deficient mRNA was about 100 bp shorter. (E and F) Western blot detection (using a polyclonal antiserum against recombinant MK5 [catalog no. 06-960; Upstate Biotechnology]) of MK5 protein from macrophages (E) and immortalized MEFs (F) derived from MK5-deficient (MK5−/−) mice and, as controls, from wild-type (wt) and MK2-deficient (MK2−/−) animals. The specific immunoreactive band of MK5 migrated with an apparent molecular mass of about 54 kDa. C, positive control (A431 cell lysate) (catalog no. 12-301; Upstate Biotechnology).
Southern blot analysis.
Genomic DNA was prepared from ES cells or mouse tail, digested by BamHI, separated by agarose gel electrophoresis, blotted onto nitrocellulose, and hybridized using the external probe P1 (BamHI-SacI fragment; Fig. 1A). Homologous recombination was detected by the appearance of an additional fragment of about 4.4 kb (Fig. 1B).
Generation of chimeras.
Two ES cell clones (60 and 149) were used for injection into C57BL/6 blastocysts at the EMBL Transgenic Service, where all further steps for generation of chimeras were also carried out by Kristina Vintersten. Coat-color chimera mice with chimerism levels between 80 to 100% were mated to C57BL/6 mice.
Genotyping.
Mouse tail genomic DNA was used for genotyping by PCR using primers 5′-cgtaacactagccacagttgtaactga and 5′-catatacttgtaagcacagctctgagtt. A 970-bp fragment was characteristic for the wild-type allele, while a 1.2-kb fragment represented the targeted allele (Fig. 1B).
Northern hybridization.
Total RNA of 5 × 106 macrophages was isolated by using a peqGold RNA Pure kit (PEQLAB), separated electrophoretically in 1.25% agarose-formaldehyde gels, and transferred to nitrocellulose. MK5 mRNA was detected by hybridization to 32P-labeled MK5 cDNA fragments (bp 707 to 2128 of AF039840).
RT-PCR.
Reverse transcription (RT) was performed with a 20-μl reaction mixture consisting of 1× display THERMO-RT buffer (Qiagen), 0.5 mM each deoxynucleoside triphosphate, 0.8 μg of total RNA, 1 μM T25 primer, and 2 μl of display THERMO-RT terminator mix. The reaction was carried out at 42°C for 40 min and stopped by exposure to 65°C for 10 min. RT solution (1 μl) was used for PCR in a 25-μl reaction mixture containing 1× PCR buffer (Qiagen), 0.2 mM each deoxynucleoside triphosphate, 0.2 μM primers atgtcggaggacagcgacatggagaaag and ctactggggctcgtggggaagggtctgc, and 2 U of HotStarTag polymerase (Qiagen), with thermal cycling as follows: first, 96°C for 15 min; then, 35 cycles of 96°C for 1 min, 50°C for 1 min, and 72°C for 1 min; and a final elongation step at 72°C for 5 min. Products are analyzed using 1.5% agarose gel electrophoresis. PCR products were extracted from the gel, cloned into TOPO vector, and sequenced.
Mouse tissue preparation.
During mouse autopsy, all tissues were fixated in neutral-buffered formalin, embedded in paraffin through graded series of alcohol, sectioned at 5-μm intervals, and stained with hematoxylin and eosin.
MEFs.
To immortalize primary MEFs from MK5−/− mice, cells were cotransfected with pSV40Tag encoding simian virus 40 large T antigen and pREP8 plasmid (Invitrogen) in a 10:1 mixture; colonies were selected with 3 mM histidinol (Sigma). MK2−/− and wild-type MEFs were obtained as described previously (10).
Western blotting.
For detection of MK5, proteins were extracted from 2 × 105 macrophages or MEFs, separated in sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (12% acrylamide), transferred to nitrocellulose, and analyzed by Western blotting using anti-PRAK antibodies (catalog no. 06-960; Upstate Biotechnology) and a secondary rabbit anti-sheep horseradish peroxidase-conjugated immunoglobulin G (IgG) antibody which was detected by enhanced chemiluminescence. As a positive control, A431 cell lysate (catalog no. 12-301; Upstate Biotechnology) was used. For determination of p38 MAPK levels, 50 μg of tissue lysate was separated for each lane by SDS-polyacrylamide gel electrophoresis and blotted onto nitrocellulose. p38 MAPK was detected using an affinity-purified polyclonal rabbit pan-p38 MAPK antiserum (catalog no. 9212; Cell Signaling Technology) diluted 1:1,000 in Odyssey blocking buffer (Li Cor). As a secondary antibody, Alexa Fluor 680 goat anti-rabbit IgG (Molecular Probes) was used in a dilution of 1:2,000. Blots were scanned and quantified using channel 700 of an Odyssey Infrared Imager and Odyssey 1.0 software (Li Cor).
Endotoxic shock and cytokine enzyme-linked immunosorbent assays.
LPS from Escherichia coli O26:B6 (catalog no. L8274; Sigma) and d-galactosidase (d-Gal) (Sigma) were diluted in pyrogen-free saline and injected intraperitoneally in a combination of LPS at 50 μg/kg of body weight and d-Gal at 1 g/kg. Spleen cells (107 cells/ml) from zymosan (Sigma)-primed animals were stimulated in vitro with 5 μg of LPS/ml. Release of cytokines was measured in the culture supernatant by enzyme-linked immunosorbent assays, as described previously (10).
TAP.
The cDNAs for mouse MK2 and MK5 were cloned into pcDNA3.1/NT-GFP-TOPO-TAP (Cellzome, Heidelberg) coding for eukaryotic expression of green fluorescent protein (GFP)--calmodulin-binding protein (CBP)-tobacco etch virus-protease cleavage site-protein A fusions. 293 cells were transiently transfected with the constructs and lysed at 48 h after transfection, and GFP A-CBP fusion protein interacting partner complexes were purified by subsequent binding to rabbit IgG beads (Sigma), TEV protease (Qiagen) cleavage, and binding to calmodulin affinity resin (Stratagene). Endogenous p38 MAPK bound to fusion protein was detected by Western blotting using pan-p38 MAPK antibodies (Cell Signaling Technology). Expression of equal amounts of endogenous p38 MAPK and tandem affinity purification (TAP) fusion protein in each cell lysate was demonstrated by Western blotting using the p38 MAPK antibody and the specific binding of the secondary antibody to the protein A domain of the fusion protein, respectively.
MK5 and MK2 kinase assay.
MEFs (2 × 106) were washed with ice-cold phosphate-buffered saline (PBS), lysed in 500 μl of lysis buffer (3), vortexed for 10 s, and put on ice for 5 min. After centrifugation at 10,000 × g at 4°C for 10 min, the supernatant was transferred to a new tube and the protein concentrations were determined using a Bio-Rad protein determination kit. Where indicated, kinase was immunoprecipitated from the lysate containing 1 mg of total protein by using 2 μg of PRAK antibody (catalog no. 06-960; Upstate Biotechnology) or 5 μl of a MK2-specific antiserum (3); otherwise, lysate (1 μl) was directly used in the kinase assay. For immunoprecipitations (IP), after 1 h of mixing at 4°C, 30 μl of protein G-agarose (Pharmacia) (a 50% slurry preequilibrated in lysis buffer) was added and the combination was mixed for another hour. Agarose was washed three times with lysis buffer containing 0.5 M NaCl and two times with 50 mM Tris-HCl (pH 7.5). Then, 40 μl of substrate buffer (0.5 mM EGTA, 0.5 mg of bovine serum albumin/ml, 30 μM PRAK substrate peptide, 50 mM Tris-HCl, pH 7.5) was added. After preheating at 30°C for 3 min, 10 μl of hot buffer (75 mM MgCl2, 0.5 mM ATP, 2 μl of [γ-33P]ATP) was added and a sample was incubated at 30°C for 20 min. A 20-μl aliquot was spotted on Whatman p81 paper and washed extensively in 1% phosphoric acid before radioactivity was measured. Detection of kinase activity in cell lysates was carried out using Hsp25 as substrate as described previously (5).
In vivo phosphate labeling, two-dimensional (2D) electrophoresis, phosphorimaging, and detection of phospho-Hsp25.
Wild-type MEF, MK2−/−, and MK5−/− cells were grown to 90% confluence on 10-cm-diameter dishes in 90% Dulbecco's minimal Eagle's medium supplemented with 10% fetal calf serum and antibiotic-antimycotic in a humidified atmosphere of 95% air and 5% CO2 at 37°C. The medium was changed to phosphate-free Dulbecco's minimal Eagle's medium (Sigma), and cells were incubated with 500 μCi of HCl-free [32P]orthophosphate (DuPont) for 2 h at 37°C. Cells were stimulated with 100 μM sodium arsenite for 45 min, washed with PBS, and lysed in 350 μl of lysis buffer containing 7 M urea, 2 M thiourea, 4% (wt/vol) CHAPS, 15 mM dithiothreitol (electrophoresis grade), 0.5% carrier ampholytes (pH 3 to 10), protease inhibitors (Roche), and 10 nM calyculin A (Calbiochem). The homogenate (about 350 μg) was solubilized by sonication on ice for 15 min followed by 20 min of centrifugation at 14,000 × g. Isoelectric focusing for 2D gel electrophoresis was performed using a Protean isoelectric focusing cell from Bio-Rad according to the instructions of the manufacturer. Supernatant was loaded on a 17-cm-long immobilized pH gradient strip (pH 3 to 10) and reswollen overnight at 50 V. Focusing was carried out for 1 h at 250 V, 1 h at 500 V, and 15 h at 7,000 V.
After equilibration in 50 mM Tris (pH 8.9), 6 M urea, 30% (wt/vol) glycerol, and 2% (wt/vol) SDS, gels were immediately applied to a vertical 12% (wt/vol) SDS gel without a stacking gel. Electrophoresis was carried out at 8°C with a constant current of 40 mA per gel. The gels of radioactively labeled MEF proteins were fixed in 30% ethanol-10% acetic acid and exposed. Radioactive spots visualized by autoradiography were excised. Gel pieces were washed sequentially for 10 min in tryptic digestion buffer (10 mM NH4HCO3) and digestion buffer-acetonitrile (1:1). These steps were repeated three times and led to a shrinking of the gel. It was reswollen with 2 μl of protease solution (Promega) (trypsin at 0.05 μg/μl) in digestion buffer and incubated overnight at 37°C.
Analysis of the tryptic peptides was carried out using an electrospray ion-trap mass spectrometer (MS) (LCQ Classic; Thermo Finnigan) directly coupled to a Nano-high-pressure liquid chromatography (HPLC) system (LC Packings; Dionex). The peptides were automatically transferred from the autosampler (Famos; Dionex) to the preconcentration column (C18 PepMap Nano Precolumn [0.3-mm i.d. by 1 mm long]). After preconcentration and washing for 10 min (40 μl/min in 0.1% trifluoroacetic acid), the peptides were automatically injected into a C18 PepMap Nano-HPLC column (LC Packings; Dionex) (75-μm i.d. by 250 mm long; 300-Å pore size; 5-μm-diameter particle size) using the Switchos system (LC Packings; Dionex). The inert HPLC pump (Ultimate; Dionex) was driven with a flow rate of 160 nl/min. The gradient (solution A, 0.1% formic acid-84% acetonitrile; solution B, 0.1% formic acid-84% acetonitrile) started at 5% of solution B and rose to 50% of solution B in 90 min. A dual-channel UV detector (LC-Packings; Dionex) with a 3-nl flow cell (LC Packings; Dionex) was used at wavelengths of 215 nm (peptide bond) and 295 nm (tryptophan side chains). Separated peptides were directly transferred to the MS via a heated capillary and a metal-backed coated glass needle (PicoTip [catalog no. FS360-20-10]; New Objective Incorporated). The following electrospray ion-trap parameters were used: spray voltage, 1.8 to 2.15 kV; capillary temperature, 200°C; capillary voltage, 42 V; tube lens offset, 30 V; and electron multiplier, −950 V. The collision time was set automatically, depending on the mass of the parent ion. The trapping time was set to 200 ms, and the automatic gain control was set to 107. The data were collected in centroid mode, with one full-MS experiment followed by three experiments investigating the MS-MS spectra of the three most intensive ions (intensity, at least 3 × 105). Dynamic exclusion was used for the data collection, with an exclusion duration of 5 min and an exclusion mass of ± 1.5 Da.
Hsp27 in-gel kinase assay.
For in-gel kinase assays 107 cells were treated with arsenite (150 μM for 45 min) or phorbol myristate acetate (100 nM for 45 min) in the presence of freshly prepared pervanadate solution (final concentration, 40 mM), washed with PBS, and lysed, and MK2 or MK5 was immunoprecipitated using immunoaffinity-purified non-cross-reacting MK5-specific antibodies (catalog no. 06-960; Upstate Biotechnology) (8), MK5 phosphorylation-specific antibodies (kind gift of Sir P. Cohen, Dundee, Scotland), anti-PRAK-antibodies (kind gift from J. Han, La Jolla, Calif.), and anti-MK2 antibodies (3) as described above. IPs were separated by electrophoresis in SDS-polyacrylamide gels containing 0.5 mg of recombinant Hsp27/ml or 0.5 mg of bovine serum albumin/ml as a control. After an in-gel kinase assay (27) was performed, phosphorylated proteins were visualized with a Fuji 1500 BAS phosphorimager and the signal was quantified by the use of TINA 2.09 software.
RESULTS
Deletion of MK5.
By homologous recombination in mice, we deleted MK5 exon 6, which codes for parts of the protein kinase catalytic core subdomains VIa and VIb (amino acids [aa] 131 to 161) required for kinase activity (Fig. 1). The MK5 exon 6-deficient allele was detected by Southern hybridization and PCR (Fig. 1B) and segregated nearly completely with Mendelian genetics. MK5−/− mice are viable and fertile and do not show behavioral abnormalities.
Total RNA was isolated from MK5 exon 6-deficient (MK5−/−) and wild-type (+/+) macrophages and analyzed by Northern blotting. In the analyses of both the wild-type and MK5−/− macrophages, an MK5-specific mRNA of about 2.4 kb was detected (Fig. 1C). RT-PCR of MK5 mRNA demonstrated that the mRNA of MK5−/− cells was lacking about 100 bp (Fig. 1D). Sequencing of the PCR fragment revealed that the fragment lacked the coding region corresponding to exon 6 (bases 1100 to 1189 of the MK5 cDNA [AF039840]; data not shown). The sequence of the neomycin cassette was removed from the transcript, obviously by alternative splicing. The deletion of parts of the strongly conserved protein kinase subdomains VIa and VIb (aa 132 to 161, containing the motif DLKPEN) led not only to a catalytically inactive enzyme but also to an instable protein probably caused by misfolding. In some experiments, there was a weaker and slightly faster-migrating band cross-reacting with the MK5 antiserum (catalog no. 06960; Upstate Biotechnology) which seemed specific for MK5-deficient cells (Fig. 1E and F). The possibility could be not completely excluded that this band represents some remaining truncated MK5 protein. However, from the results of transfection experiments performed using the deletion mutant (aa 132 to 161), we know that this truncated protein lacking aa 132 to 161 does not have any catalytic activity (data not shown).
Analysis of tissues of MK5−/− mice.
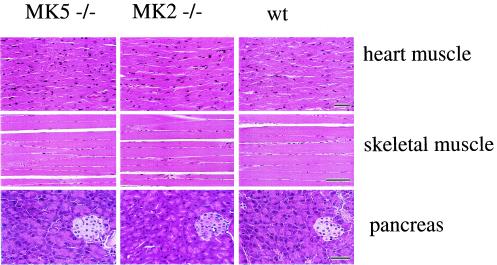
For a first analysis of the phenotype of the MK5−/− mice, all tissues of the mice were examined by a European College of Veterinary Pathologists Board-certified pathologist (A.D.G.) and compared to those of wild-type mice. In addition, we also decided to compare MK5−/− mice to MK2−/− mice (10) since MK2 and MK5 show 45% sequence identity (21) and because of the apparent functional similarities described in the introduction. However, no changes were found in the morphology of the tissues analyzed for either MK5-deficient or MK2-deficient animals. Comparisons of heart and skeletal as well as pancreas tissues, three tissues in which MK5 is highly expressed in wild-type mice (19, 21), are shown in Fig. 2.
FIG. 2.
Comparison of morphologies of selected tissues (heart muscle, skeletal muscle, and pancreas tissue [including an island]) from MK5-deficient (MK5−/−), MK2-deficient (MK2−/−), and wild-type (wt) mice. Tissues were stained with hematoxylin and eosin. Bars, 100 μm.
Susceptibility to endotoxic shock and LPS-induced inflammation.
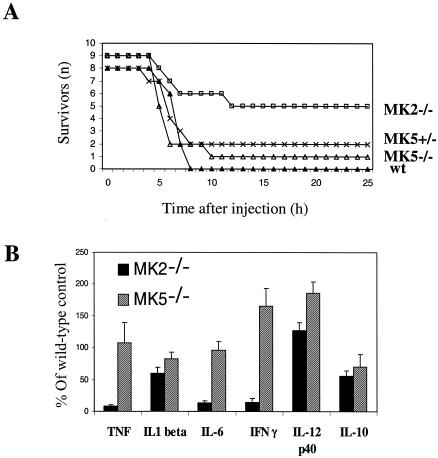
A characteristic phenotype of MK2-deficient mice is their resistance to endotoxic shock due to a decreased biosynthesis of certain inflammatory cytokines, such as TNF and IFN-γ. We analyzed whether MK5-deficient mice show similarities in this regard. When injected with a combination of LPS (50 μg per kg of body weight) and d-Gal (1 g per kg), MK5-deficient mice showed the same susceptibility to endotoxin-mediated liver failure as wild-type mice (Fig. 3A). Accordingly, there was also no significant reduction of biosynthesis of TNF, IL-6, or IFN-γ in LPS-stimulated spleen cells from MK5-deficient mice such as has been known to occur with MK2-deficient mice (Fig. 3B). Only IL-10 production was slightly decreased in both MK5−/− and in MK2−/− mice. There was even a slight increase in the production of IFN-γ and IL-12 p40 in the MK5-deficient mice. Since the mice were still in a mixed genetic background, these slight changes should be interpreted with caution.
FIG. 3.
Comparison of the phenotype of MK5 deficiency with the MK2-deficient inflammatory phenotype. (A) Survival of LPS-galactosamine-induced endotoxic shock. wt, wild type. (B) LPS-induced cytokine (TNF, IL, and IFN) production of spleen cell cultures from MK5- and MK2-deficient mice plotted as percentages of cytokine production of wild-type spleen cells. Results are shown as means ± standard errors of the means (n = 24 in each group).
MK5 did not stabilize p38 MAPK in vivo.
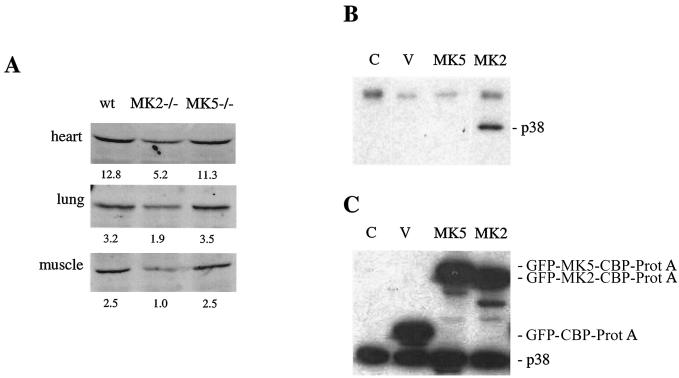
The lack of MK2 in mice leads to a significant reduction of p38 MAPK protein levels, which is caused by lack of chaperoning and/or nucleocytoplasmatic cotransport of p38 MAPK by MK2. Since p38 MAPK is supposed to be the activator for MK5 also and, hence, should also form a transient complex with endogenous MK5 (19, 23), we analyzed p38 MAPK protein levels in the MK5-deficient tissues. In contrast to MK2-deficient mice, in which Kotlyarov et al. detected the characteristic two- to threefold reduction of p38 MAPK protein levels in heart, muscle, and lung tissues (11), in the same tissues from MK5-deficient mice no significant reductions of p38 MAPK protein levels were observed (Fig. 4A). This indicates that MK5 does not fulfill the same chaperoning or transport function for p38 MAPK as MK2.
FIG. 4.
Analysis of interaction of MK5 with endogenous p38 MAPK. (A) Western blot detection of p38 MAPK levels in MK5-deficient mouse tissues and, as controls, in MK2-deficient and wild-type (wt) mouse tissues. Numbers below the bands represent the results of band quantification using channel 700 of an Odyssey Infrared Imager and Odyssey 1.0 software (Li Cor). (B) Western blot detection of endogenous p38 MAPK after TAP of proteins interacting with MK5 or MK2 from 293 cells. V, vector control without kinase fusion; C, control without transfection of TAP construct. (C) Expression control for TAP fusion proteins and p38 MAPK in 293 cell lysates before affinity purification. Lanes correspond to those shown in panel B.
MK5 does not specifically bind to endogenous p38 MAPK in vivo.
The failure of MK5 to stabilize p38 MAPK let us ask whether MK5 specifically binds to endogenous p38 MAPK. To test this, we used tandem-affinity purification of MK5- and (as a control) MK2-interacting proteins from 293 cells (22). GFP-CBP-protein A fusion proteins were expressed in transiently transfected 293 cells. After the two purification steps, endogenous p38 MAPK bound to GFP-MK2-CBP-protein A or GFP-MK5-CBP-protein A was detected using Western blotting (Fig. 4B). As an expression control, the hybrid bait proteins were detected by the IgG-binding properties of the protein A part and the endogenous p38 MAPK was detected by anti-pan-p38 MAPK Western blotting of the lysate proteins (Fig. 4C). Although expressed to a comparable level, only the MK2 fusion protein was able to bind detectable amounts of endogenous p38 MAPK of transfected 293 cells (Fig. 4B).
MK5 activity is not significantly increased by stimulation of the p38 MAPK cascade.
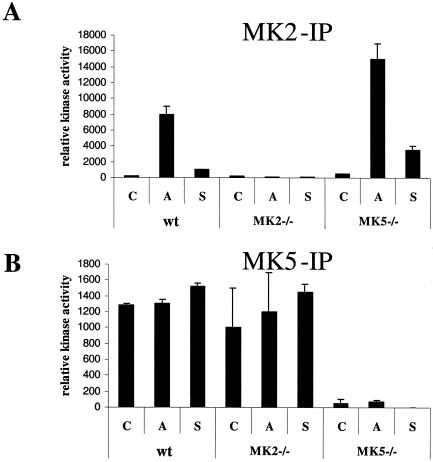
Since our data did not confirm a specific interaction of MK5 with p38 MAPK, we decided to analyze p38 MAPK-dependent activation of endogenous MK5. For that we used immortalized MEFs from the MK5-deficient mice and, as a positive control, MEFs derived from MK2-deficient and wild-type animals. Using a commercially available substrate for MK5, the “PRAKtide” KKLRRTLSVA (which was derived from glycogen synthase), and an IP kinase assay, MK5 activity was detected in nonstimulated wild-type and MK2-deficient cells but not in MK5-deficient cells. Interestingly, both treatment with arsenite and treatment with sorbitol (both of which are well known to stimulate the p38 MAPK) did not induce increased MK5 activity in these cells (Fig. 5A). As a control for p38 MAPK stimulation and MEF integrity, we also used MK2 IP and kinase assays to analyze MK2 reactivity with the peptide KKLRRTLSVA in these cells. As expected, no MK2 activity was detected in MK2-deficient cells. However, in wild-type and MK5-deficient cells, MK2 activity was greatly stimulated by the stress treatments used (Fig. 5B).
FIG. 5.
Analysis of stress-activated stimulation of MK5 and MK2 in MEFs by combined IP-kinase assays. Wild-type (wt) and MK2- and MK5-deficient MEFs were stimulated for 60 min with 250 μM arsenite (lanes A) or 300 mM sorbitol (lanes S) or left untreated (lanes C). Cells were lysed, and kinases were immunoprecipitated by MK2 (A)- and MK5 (B)-specific antibodies. Kinase activity in the IP was determined using [γ-33P]ATP and the peptide KKLRRTLSVA as the substrate. Incorporation of radioactive phosphate into the peptide was measured after binding to phosphocellulose filters was performed.
MK5 is not a major Hsp27 kinase.
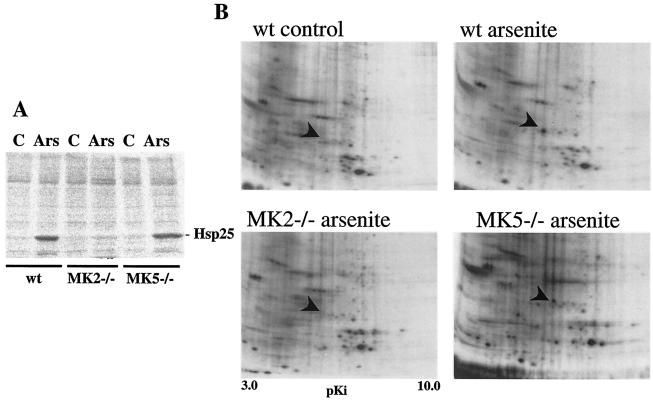
Catalytic reactivity of MK5 with Hsp27 (19), with a peptide derived from myosin light chain II (21), and with the PRAKtide KKLNRTLSVA or KKLRRTLSVA (derived from glycogen synthase) (8) has been described previously. In our experiments, the peptide KKLRRTLSVA was used to detect MK5 basal activity (see above). Although recombinant GST-MK5 was activated by p38 MAPK in vitro and then phosphorylated Hsp27, we were not able to phosphorylate Hsp27 with immunoprecipitated endogenous MK5 in vitro (data not shown). We then analyzed Hsp27 kinase activity in lysates from serum-starved MEFs lacking MK2 or MK5 (or serum-starved wild-type MEFs as a control) left unstimulated or stimulated by arsenite treatment. An approximately 3- to 10-fold stimulation of Hsp27 phosphorylating activity in wild-type and MK5-deficient cells was observed (Fig. 6A). Comparable results for other p38 MAPK-activating stimuli such as UV radiation were obtained with these cells (data not shown). Interestingly, a complete reduction of phosphorylation of exogenous Hsp27 after arsenite treatment was detected only in lysates from MK2-deficient cells (Fig. 6A).
FIG. 6.
Hsp25 phosphorylation in vitro and in vivo. (A) Cell lysates from wild-type (wt) and MK2- and MK5-deficient MEFs which were left nonstimulated (lanes C) or were stimulated by arsenite treatment (250 μM for 45 min) (lanes Ars) were incubated in vitro with recombinant Hsp25 and [γ-32P]ATP, and Hsp25 phosphorylating activity was detected by phosphorimaging. (B) 2D phosphoproteomics of wild-type and MK2- and MK5-deficient cells before and after stimulation by arsenite. The spot for phospho-Hsp25 was identified by mass spectroscopy and is indicated by an arrowhead. The autoradiograms shown are representative of the results of four separate experiments.
In a second approach, wild-type, MK2−/−, and MK5−/− MEFs were incubated with [32P]orthophosphate and stimulated by arsenite and proteins of the cell lysates were separated by 2D gel electrophoresis. The results determined using differential phosphoproteomes (Fig. 6B) demonstrated the phosphorylation of a protein identified by mass spectrometry as Hsp25, the mouse homologue of human Hsp27, in wild-type and MK5-deficient cells, indicating that in vivo MK2 was the only Hsp25 kinase stimulated by arsenite in these cells.
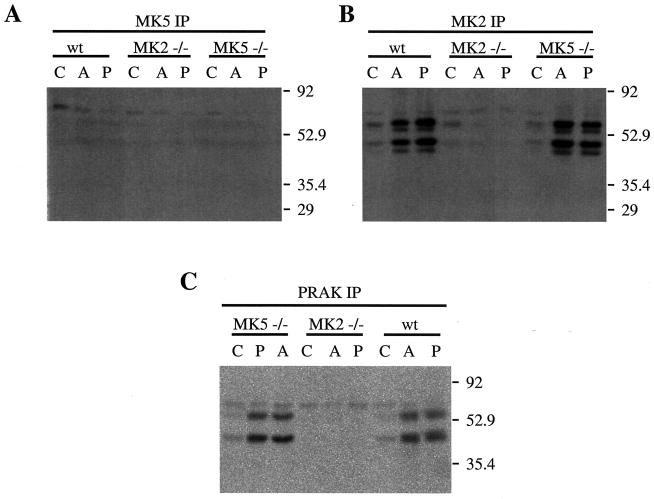
Since Hsp27 phosphorylation by MK5 has been shown using an in-gel assay, a remaining possibility was that the denaturation-renaturation procedure makes Hsp27 a better substrate. We immunoprecipitated MK5 with different antibody preparations and (as a control) MK2 from wild-type MEFs and MK2- and MK5-deficient cells and analyzed kinase activity in the IP with an in-gel assay using recombinant Hsp27 as the substrate (Fig. 7). In the anti-MK2 IP with wild-type and MK5-deficient but not with MK2-deficient cells, we could detect the two major bands (about 45 and 54 kDa) of Hsp27 phosphorylating activity known for MK2 (10) (Fig. 7B). In contrast, in the anti-MK5 IP using polyclonal affinity-purified sheep antibodies (8) (Fig. 7A) and a phosphorylation-specific antibody (kind gift of Sir P. Cohen; Dundee, Scotland; data not shown), no Hsp27 kinase activity could be detected. Interestingly, the antibody preparation against PRAK used for the first description of PRAK activity (19) (kind gift of J. Han, La Jolla, Calif.) was able to IP “MK5/PRAK” activity from MK5-deficient cells, indicating a cross-reactivity of this reagent with other sHsp kinases (Fig. 7C). Since these antibodies were not able to precipitate stress-induced Hsp27 kinase activity from MK2-deficient cells (Fig. 7C), it is questionable whether MK5 is precipitated by this reagent or whether MK5 shows any stress-inducible kinase reactivity with Hsp27.
FIG. 7.
IP-in-gel kinase assay (using different antibodies for IP of protein kinases) of reactivity with Hsp27. Wild-type (wt), MK2- and MK5-deficient MEFs were stimulated for 45 min with 250 μM arsenite (lanes A) or 100 nM phorbol myristate acetate (lanes P) or left untreated (lanes C). Cells were lysed, kinases were immunoprecipitated by (A) MK5-specific antibodies (8), (B) MK2-specific antibodies (3), or (C) anti-PRAK antibodies, and Hsp27 kinase activity was monitored by an in-gel kinase assay (10, 27).
DISCUSSION
MK5-deficient mice are viable and fertile and do not show changes in tissue morphology and behavior. They exhibit the same susceptibility to LPS-induced endotoxic shock as wild-type animals and do not show the defects in LPS-induced biosynthesis of inflammatory cytokines known to occur with MK2-deficient animals (10). Hence, despite the structural similarities (21) and evolutionary relatedness (14) between MK5 and MK2, analysis of MK5-deficient animals and cells clearly demonstrated the functional nonrelatedness of these enzymes. On the one hand, this was unexpected since besides the structural similarities, potential functional similarities such as activation by p38 MAPK, substrate specificity for the sHsps (19), nucleocytoplasmatic translocation in response to stress (23), and similar tissue expression patterns (4, 20, 21) have also been described previously. On the other hand, this finding explains the dramatic effects of deletion of MK2 (7, 10, 12, 16, 28), since MK5 is obviously not able to compensate for these effects, making MK2 a unique target for anti-inflammatory therapy.
How can the discrepancy between potential functional similarities for MK5 and MK2 kinases and the different phenotypes of the kinase-deficient mice (and the inability of MK5 to compensate for MK2 loss) be explained? We have provided experimental evidence that the functional similarities assumed so far do not represent the situation in mice and their cells in vivo. It is well established that phosphorylation and activation of MK5 can be obtained in vitro by the use of recombinant p42 ERK2 and recombinant p38 MAPK-α and -β but not by the use of c-Jun N-terminal kinase (19, 21). However, all data obtained so far demonstrating in vivo activation of endogenous MK5 (also designated PRAK) by p38 MAPK have been determined on the basis of the use of an antiserum for IP-kinase assays (19), which is demonstrated here to be unable to immunoprecipitate significant sHsp-phosphorylating activity from MK2-deficient cells. Since this antiserum is not able to precipitate sHsp-kinase activity from MK2-deficient cells but precipitates such activity from MK5-deficient cells, a cross-reaction of this antibody preparation with other sHsp-phosphorylating kinases such as MK2 or MK3 is likely. This means that the in vivo properties described for MK5/PRAK using this antibody preparation (19) result from IP of cross-reacting kinases.
Recently, thrombin-induced activation of PRAK in human platelets was analyzed by immunoaffinity-purified non-cross-reacting specific antibodies (8). In this case, relatively high basal activity but no significant stimulation (1.2-fold stimulation after 150 min) of PRAK was measured whereas significant stimulation of p38 MAPK and MNK1 was detected after 25 min. This result also makes activation of endogenous MK5/PRAK by p38 MAPK unlikely.
No significant stimulation of endogenous MK5 in living cells has been detected using the specific IP kinase assay so far. This might mean that the action of basal activity is regulated by protein-protein interaction and subcellular localization or simply that the right physiological stimuli or situations where MK5 is activated have not been identified so far. The recently available antiserum specific for phosphorylation of the regulatory site in the activation loop of the kinase (kind gift of P. Cohen) should help to answer this question in the future.
We further analyzed protein-protein interactions between p38 MAPK and MK5 and provided additional evidence that endogenous p38 MAPK is probably not acting the same way for activation of MK5 as for activation of MK2. In contrast to MK2-deficient tissues, in which p38 MAPK levels are reduced due to the missing chaperone and/or transporter function of MK2 (9), in full-length MK5-deficient tissues, for which significantly (at least) reduced MK5 truncated protein levels have been detected, no reduction of p38 MAPK levels has been observed. This indicates the existence of specific p38 MAPK-stabilizing properties of MK2. Expression levels of MK2 higher than those of MK5 could theoretically explain this observation also, but in the tissues analyzed a significant expression of MK5 mRNA (19, 21) comparable to the expression of MK2 mRNA (4) was detected. Another analysis of interaction of MK5 with endogenous p38 MAPK was carried out using TAP. We showed that endogenous p38 MAPK from 293 cells copurifies with MK2 but not with MK5. This strengthens the notion that there is no specific interaction between endogenous p38 MAPK and MK5.
Interaction between MK5 and p38 MAPK in vitro and for cells overexpressing tagged versions of both proteins has been described previously (18, 23). Furthermore, overexpression of both active and inactive p38 MAPK changes subcellular localization of MK5 (18, 23). Since artificial overexpression of signaling components can titrate out and quench other relevant endogenous signaling molecules, these data do not necessarily reflect the in vivo situation. It is interesting that the stress-induced kinetics of translocation of GFP-tagged MK5 is much slower than translocation of GFP-tagged MK2 and does not correlate with kinetics of p38 MAPK activation (18, 23). This also indicates different mechanisms of regulation of these processes by different upstream kinases.
In the evolutionarily conserved group of MAPKAP kinases, there is a third enzyme present in mice and humans that has been designated MK3 or 3pK (15, 24) (14). Since no mouse knockout is available for this kinase, the physiological role for this enzyme has not been defined so far. Since in MK2/MK5 double-knockout animals LPS-induced production of TNF is (in similarity to results with the MK2 knockout alone) reduced to about 10% of that of the wild type and is not further impaired (A. Kotlyarov and M. Gaestel, unpublished data), the question remains open whether MK3 activity is responsible for the remaining 10% of LPS-induced TNF biosynthesis. We would speculate that in regard to its functional properties MK3 seems more clearly related to MK2, since its primary structure includes the second regulatory phosphorylation site outside the catalytic domain (15) and the kinetics of its nuclear export (Kotlyarov and Gaestel, unpublished) is clearly more similar to that of MK2 than to that of MK5.
Acknowledgments
We thank Kristina Vintersten for her excellent work at the EMBL-Heidelberg Transgenic Service, Ole Morten Seternes and Ugo Moens for help with the MK5 kinase assay, Sir Philip Cohen (Dundee, Scotland) and Jiahuai Han (La Jolla, Calif.) for MK5 and PRAK antibodies, Michael Kracht for support in the TAP experiments, and Dorothea Krone and Stefanie Feldhege for excellent technical help.
The work was supported by grants Ga 453/7, Bu740/5, SFB621 TP-A3, and SFB 421 TP-B2 from the DFG.
REFERENCES
- 1.Ben-Levy, R., S. Hooper, R. Wilson, H. F. Paterson, and C. J. Marshall. 1998. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 8:1049-1057. [DOI] [PubMed] [Google Scholar]
- 2.Clifton, A. D., P. R. Young, and P. Cohen. 1996. A comparison of the substrate specificity of MAPKAP kinase-2 and MAPKAP kinase-3 and their activation by cytokines and cellular stress. FEBS Lett. 392:209-214. [DOI] [PubMed] [Google Scholar]
- 3.Engel, K., A. Kotlyarov, and M. Gaestel. 1998. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 17:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engel, K., K. Plath, and M. Gaestel. 1993. The MAP kinase-activated protein kinase 2 contains a proline-rich SH3-binding domain. FEBS Lett. 336:143-147. [DOI] [PubMed] [Google Scholar]
- 5.Engel, K., H. Schultz, F. Martin, A. Kotlyarov, K. Plath, M. Hahn, U. Heinemann, and M. Gaestel. 1995. Constitutive activation of mitogen-activated protein kinase-activated protein kinase 2 by mutation of phosphorylation sites and an A-helix motif. J. Biol. Chem. 270:27213-27221. [DOI] [PubMed] [Google Scholar]
- 6.Han, Q., J. Leng, D. Bian, C. Mahanivong, K. A. Carpenter, Z. K. Pan, J. Han, and S. Huang. 2002. Rac1-MKK3-p38-MAPKAPK2 pathway promotes urokinase plasminogen activator mRNA stability in invasive breast cancer cells. J. Biol. Chem. 277:48379-48385. [DOI] [PubMed] [Google Scholar]
- 7.Hannigan, M. O., L. Zhan, Y. Ai, A. Kotlyarov, M. Gaestel, and C. K. Huang. 2001. Abnormal migration phenotype of mitogen-activated protein kinase-activated protein kinase 2−/− neutrophils in zigmond chambers containing formyl-methionyl-leucyl-phenylalanine gradients. J. Immunol. 167:3953-3961. [DOI] [PubMed] [Google Scholar]
- 8.Hefner, Y., A. G. Borsch-Haubold, M. Murakami, J. I. Wilde, S. Pasquet, D. Schieltz, F. Ghomashchi, J. R. Yates III, C. G. Armstrong, A. Paterson, P. Cohen, R. Fukunaga, T. Hunter, I. Kudo, S. P. Watson, and M. H. Gelb. 2000. Serine 727 phosphorylation and activation of cytosolic phospholipase A2 by MNK1-related protein kinases. J. Biol. Chem. 275:37542-37551. [DOI] [PubMed] [Google Scholar]
- 9.Kotlyarov, A., and M. Gaestel. 2002. Is MK2 (mitogen-activated protein kinase-activated protein kinase 2) the key for understanding post-transcriptional regulation of gene expression? Biochem. Soc. Trans. 30:959-963. [DOI] [PubMed] [Google Scholar]
- 10.Kotlyarov, A., A. Neininger, C. Schubert, R. Eckert, C. Birchmeier, H. D. Volk, and M. Gaestel. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1:94-97. [DOI] [PubMed] [Google Scholar]
- 11.Kotlyarov, A., Y. Yannoni, S. Fritz, K. Laaβ, J.-B. Telliez, D. Pitman, L.-L. Lin, and M. Gaestel. 2002. Distinct cellular functions of MK2. Mol. Cell. Biol. 22:4827-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehner, M. D., F. Schwoebel, A. Kotlyarov, M. Leist, M. Gaestel, and T. Hartung. 2002. Mitogen-activated protein kinase-activated protein kinase 2-deficient mice show increased susceptibility to Listeria monocytogenes infection. J. Immunol. 168:4667-4673. [DOI] [PubMed] [Google Scholar]
- 13.Li, Y., K. Inoki, P. Vacratsis, and K. L. Guan. 2003. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J. Biol. Chem. 278:13663-13671. [DOI] [PubMed] [Google Scholar]
- 14.Manning, G., D. B. Whyte, R. Martinez, T. Hunter, and S. Sudarsanam. 2002. The protein kinase complement of the human genome. Science 298:1912-1934. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin, M. M., S. Kumar, P. C. McDonnell, S. Van Horn, J. C. Lee, G. P. Livi, and P. R. Young. 1996. Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J. Biol. Chem. 271:8488-8492. [DOI] [PubMed] [Google Scholar]
- 16.Neininger, A., D. Kontoyiannis, A. Kotlyarov, R. Winzen, R. Eckert, H. D. Volk, H. Holtmann, G. Kollias, and M. Gaestel. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277:3065-3068. [DOI] [PubMed] [Google Scholar]
- 17.Neufeld, B., A. Grosse-Wilde, A. Hoffmeyer, B. W. Jordan, P. Chen, D. Dinev, S. Ludwig, and U. R. Rapp. 2000. Serine/threonine kinases 3pK and MAPK-activated protein kinase 2 interact with the basic helix-loop-helix transcription factor E47 and repress its transcriptional activity. J. Biol. Chem. 275:20239-20242. [DOI] [PubMed] [Google Scholar]
- 18.New, L., Y. Jiang, and J. Han. 2003. Regulation of PRAK subcellular location by p38 MAP kinases. Mol. Biol. Cell 14:2603-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.New, L., Y. Jiang, M. Zhao, K. Liu, W. Zhu, L. J. Flood, Y. Kato, G. C. Parry, and J. Han. 1998. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 17:3372-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.New, L., M. Zhao, Y. Li, W. W. Bassett, Y. Feng, S. Ludwig, F. D. Padova, H. Gram, and J. Han. 1999. Cloning and characterization of RLPK, a novel RSK-related protein kinase. J. Biol. Chem. 274:1026-1032. [DOI] [PubMed] [Google Scholar]
- 21.Ni, H., X. S. Wang, K. Diener, and Z. Yao. 1998. MAPKAPK5, a novel mitogen-activated protein kinase (MAPK)-activated protein kinase, is a substrate of the extracellular-regulated kinase (ERK) and p38 kinase. Biochem. Biophys. Res. Commun. 243:492-496. [DOI] [PubMed] [Google Scholar]
- 22.Rigaut, G., A. Shevchenko, B. Rutz, M. Wilm, M. Mann, and B. Seraphin. 1999. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17:1030-1032. [DOI] [PubMed] [Google Scholar]
- 23.Seternes, O. M., B. Johansen, B. Hegge, M. Johannessen, S. M. Keyse, and U. Moens. 2002. Both binding and activation of p38 mitogen-activated protein kinase (MAPK) play essential roles in regulation of the nucleocytoplasmic distribution of MAPK-activated protein kinase 5 by cellular stress. Mol. Cell. Biol. 22:6931-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sithanandam, G., F. Latif, F. M. Duh, R. Bernal, U. Smola, H. Li, I. Kuzmin, V. Wixler, L. Geil, and S. Shrestha. 1996. 3pK, a new mitogen-activated protein kinase-activated protein kinase located in the small cell lung cancer tumor suppressor gene region. Mol. Cell. Biol. 16:868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stokoe, D., K. Engel, D. G. Campbell, P. Cohen, and M. Gaestel. 1992. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 313:307-313. [DOI] [PubMed] [Google Scholar]
- 26.Tanoue, T., R. Maeda, M. Adachi, and E. Nishida. 2001. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20:466-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Dam, H., D. Wilhelm, I. Herr, A. Steffen, P. Herrlich, and P. Angel. 1995. ATF-2 is preferentially activated by stress-activated protein kinases to mediate c-jun induction in response to genotoxic agents. EMBO J. 14:1798-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, X., L. Xu, H. Wang, P. R. Young, M. Gaestel, and G. Z. Feuerstein. 2002. Mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 deficiency protects brain from ischemic injury in mice. J. Biol. Chem. 277:43968-43972. [DOI] [PubMed] [Google Scholar]
- 29.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C. Y. Chen, A. B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18:4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]