Abstract
Tumor suppressor p53 controls cell cycle progression and apoptosis following DNA damage, thus minimizing carcinogenesis. Mutations in the human DDB2 gene generate the E subgroup of xeroderma pigmentosum (XP-E). We report here that XP-E strains are defective in UV irradiation-induced apoptosis due to severely reduced basal and UV-induced p53 levels. These defects are restored by infection with a p53 cDNA expression construct or with a DDB2 expression construct if and only if it contains intron 4, which includes a nonmutated p53 consensus-binding site. We propose that both before and after UV irradiation, DDB2 directly regulates p53 levels, while DDB2 expression is itself regulated by p53.
Xeroderma pigmentosum (XP) is a rare hereditary disease with a high incidence of UV light-induced skin cancers (7). Of eight known XP groups, XP-E has unique clinical and cellular phenotypes. For example, XP-E patients suffer from skin cancers without typical xerosis, and cell strains from XP-E patients are not abnormally sensitive to UV and have normal levels of DNA repair synthesis in vivo (26, 27). XP-E is caused by mutations in the DDB2 gene (26, 27, 37), which encodes the small subunit, p48DDB2, of a heterodimeric damage-specific DNA binding protein (DDB) that also contains p127DDB1 (10).
Expression of the DDB2 gene is perturbed by UV irradiation. In a human normal diploid fibroblast strain, IMR-90, after an initial reduction in DDB activity, there is a two- to fourfold increase in DDB2 mRNA levels 36 h after UV irradiation followed by a peaking of p48DDB2 protein and DDB activity 48 h after irradiation (27, 37). Since nucleotide excision repair (NER) is all but complete 48 h after UV irradiation (13), and since NER reconstitution studies have shown that DDB is not required for NER in vitro (1, 35), the direct function, if any, of DDB in DNA repair is unclear. In XP-E cell strains, p48DDB2 protein and DDB activity are undetectable after UV irradiation (except in strain XP82TO), yet DDB2 mRNA levels are abnormally high both before (2- to 4-fold) and after (11- to 37-fold) UV irradiation (27). However, the regulation of DDB2 induction after UV damage is poorly understood.
The tumor suppressor protein p53 is important in cellular responses to DNA damage and mutations in the TP53 gene are frequently identified in human cancers (12, 41, 45). For example, 40 to 60% of skin cancers have mutations in TP53, suggesting that abnormal p53 function is crucial in perhaps a majority of human skin cancers (8, 9, 16). Activation of p53 in response to DNA damage occurs as a result of a rapid increase in its levels through stabilization of the protein and a number of posttranslational modifications. These changes allow p53 to mediate transcriptional activation of a number of genes (3, 30, 40). The CDKN1A (WAF1) and BAX genes, whose products trigger cell cycle arrest and apoptosis, respectively, are regulated in this manner (20, 41).
While the degradation of p53 is known to be regulated by MDM2-mediated ubiquitination and consequent targeting for proteolysis (19, 21, 29), little is known about the regulation of basal p53 levels or of the initial enhancement of p53 levels and its stability after DNA damage. We report here that human primary XP-E fibroblast strains are defective in UV-induced apoptosis and have abnormally low or undetectable levels of p53 and its downstream-regulated proteins both before and after UV irradiation. These defects are restored functionally by endogenously regulated p53 cDNA or by a DDB2 construct harboring intron 4 of the DDB2 gene, if (and only if) this intron retains regulatory elements including a p53 consensus binding sequence (CBS). Furthermore, the intron is also required for DDB2 expression after UV irradiation. Thus, DDB2 is a critical regulator of p53, while DDB2 expression is itself regulated through functional interaction with p53.
MATERIALS AND METHODS
Cells and culture conditions.
The XP-E primary skin fibroblast strains were obtained as follows. XP2RO (GM02415) and GM01389 were purchased from the Coriell Institute Cell Repository (Camden, N.J.). XP3RO was a generous gift from J. H. J. Hoeijmakers (Erasmus University, Rotterdam, The Netherlands). XP82TO was obtained from S. Kondo (Tokyo Medical and Dental University, Tokyo, Japan). Ops1 was established in Kumamoto, Japan (24). IMR-90 (CRL-1262), a human normal primary lung fibroblast strain; Hs-27 (CRL-1634), CCD-32sk (CRL-1489), and CCD-33sk (CRL-1493), human normal primary skin fibroblast strains; and Bing (CRL-11554), an amphotropic envelope-expression packaging cell line, were obtained from the American Type Culture Collection (Rockville, Md.). All cells were cultured in Dulbecco's modified Eagle's minimum essential medium (Invitrogen) supplemented with 20% (vol/vol) fetal bovine serum (HyClone Laboratories, Inc.) at 37°C in a humidified 5% CO2 incubator.
UV survival assays.
Viable cell number was determined by dye exclusion as described previously (25). A total of 5 × 104 cells were plated in 60-mm-diameter dishes, and at the indicated times after UV-C (254 nm) irradiation at a fluence rate of 0.8 to 1.2 J m−2 s−1, cells were washed with 1× phosphate-buffered saline (PBS), detached with trypsin, and then collected. Viable cells were scored with 0.4% trypan blue stain (Invitrogen).
Caspase 3 assay.
Caspase 3 activity was measured with a caspase 3 activity assay kit (Roche Applied Sciences). Cells (0.2 × 106) were seeded onto 100-mm-diameter dishes and cultured into exponential growth. A total of 2.0 × 106 cell equivalents of extract was analyzed on microtiter plates. Units of caspase 3 activity were calculated with a standard curve of 7-amino-4-trifluoromethyl-coumarin and defined according to the manufacturer's instructions.
TP53 sequencing.
Genomic DNA was extracted from five XP-E cell strains (GM02415, XP3RO, XP82TO, GM01389, and Ops1) with the GenomicPrep Cells and Tissue DNA Isolation kit (Amersham Biosciences). PCR was performed with primer sets specific to exons 5 through 8 of TP53. PCR products were purified and directly sequenced.
Immunoblotting and immunoblot antibodies.
Cells (0.5 × 106) were seeded onto 100-mm-diameter dishes and cultured to exponential growth, washed once with PBS, and, when indicated, irradiated with 12 J of UV-C m−2. Culture medium was returned, and the cells were incubated and harvested by centrifugation at the indicated times.
To prepare whole-cell extracts, cell pellets were resuspended in hypotonic buffer (10 mM Tris-HCl [pH 8.0], 5 mM MgCl2, and protease inhibitors; Complete; Roche Applied Science) and lysed with a Dounce homogenizer. Two volumes of potassium glutamate buffer (250 mM Tris-HCl [pH 8.0]), 10 mM MgCl2, 0.3 M potassium glutamate [pH 7.8], 5 mM dithiothreitol [DTT], 50% glycerol, protease inhibitors) was added, and the extracts were incubated for 30 min at 4°C. The extracts were clarified by centrifugation for 1 h at 106,000 × g in a Beckman 100.3 rotor at 4°C and then dialyzed against a mixture containing 50 mM Tris-HCl (pH 8.0), 1 mM DTT, and 10% glycerol. Fifty micrograms of extract was resolved by 10 or 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10 or 12% polyacrylamide), and then polypeptides were transferred to a nitrocellulose filter, and the membrane was cut into appropriate sections and probed for p53, MDM2, p127DDB1, or p21CDKN1A as indicated. The same membrane was stripped and then probed for p48DDB2, actin, or p21BAX when indicated. Gel images were quantitatively analyzed with ImageQuaNT 1.2 (Molecular Dynamics). To normalize each protein level among different strains, samples without UV irradiation (0 h) were transferred to the same membrane and immunoblotted. The signals of each protein were quantitated with ImageQuaNT 1.2 and then normalized to their respective actin densities.
A rabbit polyclonal p48DDB2 antibody (27) was used for immunoblotting. Antibodies for p53 (Pab1801, DO-1, and FL-393), p21CDKN1A (F-5 and C-19), p21BAX (B-9), MDM2 (SMP-14), and actin (I-19) were purchased from Santa Cruz Biotechnology, Inc. Antibodies for p127DDB1, MDM2 (2A10), and p53 (Pab421) were purchased from Zymed Laboratories or Oncogene Research Products.
Search of putative transcription factor-binding elements.
The p53 CBS and other regulatory elements in intron 4 were identified by using genomic DNA sequences of the human DDB2 gene (24) with the TRANSFAC database (version 5.0, transfac.gbf.de/TRANSFAC) (32).
Retroviral vectors and infection.
To construct the cDNA under the control of the 5′-long terminal repeat (LTR) of Moloney leukemia virus, DDB2 cDNA with a BamHI adaptor was cloned into the pLXSN vector (BD Sciences Clontech) to produce pLXSN-DDB2 cDNA. To clone the 5′ untranslated region (5′UTR) containing the promoter region, 800 bp (nucleotides −1 to −800 from the ATG start site [+1]) of the 5′UTR of the human DDB2 gene were amplified by PCR from templates of phage clones harboring the human DDB2 gene using primers S1 (nucleotides −800 to −781) and AS1 (nucleotides −20 to −1). The purified PCR product was subcloned into the XhoI site of pBluescript (Stratagene) (pBS-5′UTR), sequenced, and then resubcloned into the pLXSN vector to yield pLXSN-5′UTR.
To construct the 5′UTR linked to the DDB2 cDNA (see Fig. 5A), PCR was performed with primers S1 and AS2 (nucleotides +1347 to +1366 from the ATG start site [+1] of the DDB2 cDNA) and templates of pBS-5′UTR and pLXSN-DDB2 cDNA. The 5′UTR and DDB2 cDNA were subcloned into the pLXSN vector to yield pLXSN-5′UTR+DDB2 cDNA.
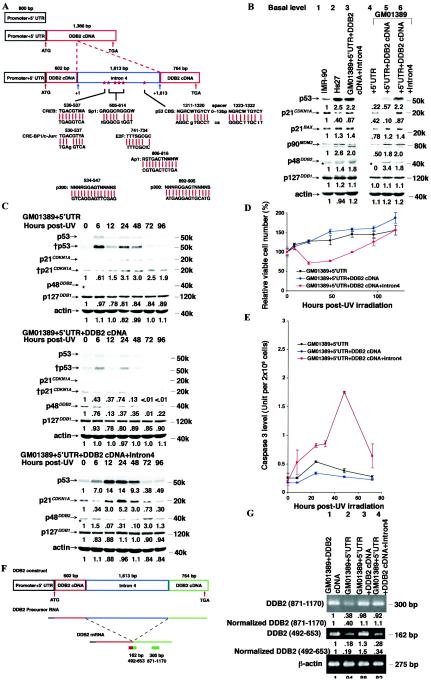
FIG.5.
Intron 4 of the DDB2 gene is required to restore basal and UV-induced levels of p53 and apoptosis to an XP-E strain and controls basal DDB2 mRNA levels. (A) Graphic representation of the DDB2 constructs. Intron 4 was correctly placed into its splice site of the DDB2 gene. Each of these constructs was subcloned into the pLXSN retroviral vector, and after virus infection, cells were selected for G418 resistance as described in Materials and Methods. (B) Basal protein levels in the XP-E strain GM01389 containing a retrovirus harboring the upper three constructs represented in panel A compared to levels in normal strains IMR-90 and Hs27. Each protein level of the IMR-90 was designated as 1.0, and the respective levels of these proteins in other strains with or without the expression constructs were normalized accordingly. Lanes 3 and 6 contained the same extract. The immunofluorescence exposure times were 30 s (p53), 1 min (p21CDKN1A), 5 s (p21BAX), 10 s (p90MDM2, p127DDB1, and actin), or 20 min (p48DDB2). The band marked with an asterisk lying just below p48DDB2 is nonspecific. (C) Protein levels before and after 12 J of UV irradiation m−2 in the XP-E strain GM01389 before and after infection of the constructs as shown in panel A. The exposure times were 30 s (p53), 2 min (†p53), 15 s (p21CDKN1A), 1 min (†p21CDKN1A), 20 min (p48DDB2), and 10 s (p127DDB1 and actin). Bands without numbers contained signals that were too low to quantitate. Each basal protein level (0 h) was designated as 1.0 to compare protein levels during the time course. The band marked with an asterisk lying just below p48DDB2 is nonspecific. (D) Cell viability of XP-E strains harboring retroviral constructs used in panel C after 12 J of UV irradiation m−2. Viable cell number was determined by dye exclusion. Within an experiment, each point was determined in triplicate, and each curve represents three such independent experiments, except that for the construct 5′UTR+DDB2 cDNA+Intron4 construct, the experiment was performed five times. (E) Caspase 3 activity of XP-E strains harboring retroviral constructs used in panel C after 12 J of UV irradiation m−2. Within an experiment, each point was determined in duplicate, and each curve represents two such independent experiments, except that for 5′UTR+DDB2 cDNA+Intron4, the experiment was performed four times. (F) Graphic representation of RNA processing of the DDB2 construct including intron 4. For detection of total DDB2 RNA including unspliced RNA, S5 and AS6 primers were used. For detection of spliced DDB2 RNA, S2 and AS3 primers were used. (G) Semiquantitative RT-PCR of DDB2 and β-actin. Semiquantitative RT-PCR was performed to determine the basal DDB2 expression levels in the XP-E strain infected with the DDB2 constructs as shown in panel A.
To make the DDB2 gene construct 5′UTR+DDB2 cDNA+intron4 (Fig. 5A), first intron 4 of the DDB2 gene was amplified with primers S2 (nucleotides +492 to +511) and AS3 (nucleotides +634 to +653) and templates of phage clones harboring the human DDB2 genomic DNA (24). Then two DDB2 cDNAs containing nucleotides −32 to +600 or +601 to +1345 were amplified with primers S3 (nucleotides −32 to −13) and AS4 (nucleotides +581 to +600) or S4 (nucleotides +601 to +620) and AS5 (nucleotides +1326 to +1345), respectively, and a template of pLXSN-DDB2 cDNA. To provide the final construct, the three PCR products were purified, mixed, and used as templates with primers S1 and AS2. This final PCR product was verified by sequencing and subcloned into pLXSN. The junction sequence of exon 4 and intron 4 or intron 4 and exon 5 were verified by DNA sequencing. For the constuct carrying the 955-bp deletion of intron 4 (see Fig. 6A), the construct with intron 4 was digested by HindIII (Roche applied science), and then the appropriate purified fragments were ligated.
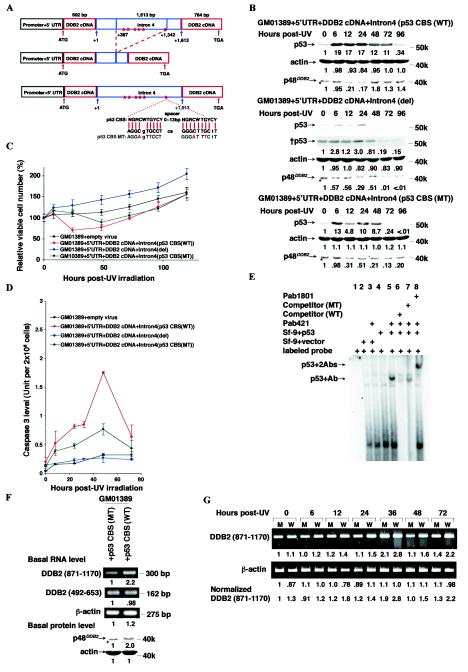
FIG.6.
Intron 4, including the p53 CBS of the DDB2 gene, is important for controlling levels of DDB2 RNA, p48DDB2, and p53, and apoptosis. (A) Graphic representation of mutant DDB2 constructs. Both constructs were made from the DDB2 construct including the wild-type intron 4. For the deletion mutant, a 955-bp fragment (+387 to + 1342) of intron 4 (+1 is a first nucleotide of intron 4) was removed. For the p53 CBS mutant, 4 nucleotides were changed to alter both elements of the p53 CBSs. (B) Protein levels before and after 12 J of UV irradiation m−2 in the XP-E strain GM01389 with the mutant DDB2 constructs. The construct with the wild-type p53 CBS was used as a control. The exposure times were 30 s (p53), 2 min (†p53), 15 s (p21CDKN1A), 1 min (†p21CDKN1A), 20 min (p48DDB2), 40 min (†p48DDB2), or 10 s (actin). The band marked with an asterisk lying just below p48DDB2 is nonspecific. Each basal protein level (0 h) was designated as 1.0 to compare protein levels during the time course. (C) Cell viability of XP-E strains harboring retroviral constructs used in panel B after 12 J of UV irradiation m−2. Viable cell number was determined by dye exclusion. Within an experiment, each point was determined in triplicate and each curve represents at least two such independent experiments, except that for 5′UTR+DDB2 cDNA+Intron 4 [p53 CBS (MT)] the experiment was performed once. (D) Caspase 3 activity of XP-E strains harboring retroviral constructs used in panel B after 12 J of UV irradiation m−2. Within an experiment, each point was determined in duplicate, and each curve represents at least two such independent experiments, except that for 5′UTR+DDB2 cDNA+Intron4 [p53 CBS (MT)], the experiment was performed once. (E) Mobility shift assays with p53 and a probe harboring the p53 CBS found in intron 4 of the DDB2 gene. 32P-labeled 34-bp oligonucleotides containing the wild-type or mutated p53 CBS as shown in panel A were incubated with extracts from Sf-9 cells infected with empty or p53 cDNA-containing viruses, anti-p53 antibodies, and unlabeled competitor oligonucleotides as indicated. Unbound labeled oligonucleotides were run off of the gel. p53+Ab, a band observed only when p53 and Pab421 are present; p53 + 2Abs, a band observed only when p53, Pab421, and Pab1801 are present. (F) Basal DDB2 and β-actin RNA and p48DDB2 and β-actin protein levels in the XP-E GM01389 strain with the DDB2 constructs with the wild-type (WT) or mutant (MT) p53 CBS. Each of RNA or protein levels was determined by semiquantitative RT-PCR or immunoblotting, respectively. The exposure times for immunoblotting were 20 min (p48DDB2) and 10 s (actin). The band marked with an asterisk lying just below p48DDB2 is nonspecific. Each protein level from cells infected with p53 CBS (MT) was designated as 1.0, and the respective levels of these proteins in p53 CBS (WT) were normalized accordingly. (G) Semiquantitative RT-PCR of DDB2 and β-actin in the XP-E GM01389 strain with the DDB2 constructs with the wild-type or mutant p53 CBS before and after 12 J of UV irradiation m−2. M, DDB2 construct with the mutant p53 CBS; W, DDB2 construct with the wild-type p53 CBS.
For the construct harboring the mutant p53 CBS embedded in the construct 5′UTR+DDB2 cDNA+intron4, the QuikChange mutagenesis kit (Stratagene) was used according to the manufacturer's instructions. PCR was performed with primers S-MT (5′-GGTGTGGGGGGCAGGAGTTCCTCAGGGATTTCTTTATGGTATAAGC) and AS-MT (5′-GCTTATACCATAAAGAAATCCCTGAGGAACTCCTGCCCCCCAGACC). The mutated sequence was verified by DNA sequencing.
For the p53 cDNA construct, human wild-type p53 cDNA was cloned from HeLa RNA by reverse transcriptase PCR (RT-PCR) and then cloned into the pLXSN vector. The sequence was verified.
For viral infections, 106 Bing cells were plated in 60-mm-diameter dishes, incubated for 24 h, and then transfected with Lipofectamine 2000 (Invitrogen) with 8 μg of retroviral plasmid. After 24 h, the medium was removed and saved, and fresh medium was added. After 24 h, this medium was removed. The two virus-containing medium samples were filtered (0.45-μm-pore-diameter filter; Doc Frugal Scientific) and supplemented with 4 μg of Polybrene per ml (Sigma) (first and second supernatants). Target cell strains were plated at 105 cells per 60-mm-diameter dish and incubated overnight. For infections, the culture medium was replaced with the first supernatant and then incubated for 24 h. The infection was repeated with the second supernatant. After 48 h, cells were replated in 100-mm-diameter dishes and incubated overnight. The infected cells were selected with 150 μg of G418 per ml (Invitrogen) for 1 week before starting the experiments. Within each experiment, cells proliferated from the same seeding culture were utilized throughout.
EMSA.
Baculovirus containing a p53 expression construct (14) was a kind gift from M. Botchan (University of California, Berkeley). As a control, baculovirus without an insert was used. Cells were lysed in a mixture containing 50 mM Tris (pH 8.0), 150 mM sodium chloride, and 1% Nonidet P-40 and sonicated with a microtip. After incubation for 2 h on ice, the material was centrifuged at 12,000 × g for 30 min at 4°C and the supernatant was dialyzed against 50 mM Tris (pH 8.0), 10% glycerol, and 1 mM DTT. For electrophoretic mobility shift assays (EMSAs), a DNA probe (34 bp) was made by annealing oligonucleotides 5′-GGGGGCAGGCGTGCCTCAGGGCTTGCTTTATGGT to 5′-ACCATAAGCAAGCCCTGAGGCACGCCTGCCCCC (The p53 consensus binding sites are underlined). Extracts (5 μg) prepared as described above were preincubated with 500 ng of p53 antibodies Pab421 and Pab1801 as indicated in a mixture containing 25 mM Tris-HCl (pH 8.0), 5% glycerol, 5 mM MgCl2, 60 mM KCl, 0.1 mg of bovine serum albumin ml−1, and 0.5 μg of sonicated salmon sperm DNA for 30 min at 4°C, followed by addition of 88 fmol of 32P-labeled double-stranded oligonucleotide with or without 40 pmol of competitor as indicated (unlabeled 34-bp double-stranded oligonucleotide or p53 CBS MT oligonucleotide made by annealing oligonucleotides 5′-GGGGGCAGGAGTTCCTCAGGGATTTCTTTATGGT to 5′-ACCATAAAGAAATCCCTGAGGAACTCCTGCCCCC). The incubation was continued for an additional 15 min at room temperature, and then samples were resolved by PAGE (4% polyacrylamide) in 0.5× Tris-borate-EDTA buffer. The gels were dried and exposed to X-ray film at −80°C for 24 h.
Semiquantitative RT-PCR.
RT-PCR was performed as described previously (27). The following DDB2 primers were used for semiquantitative RT-PCR: S5 (nucleotides +871 to +890) and AS6 (nucleotides +1151 to +1170) and S2 and AS3.
RESULTS
XP-E strains are defective in UV-induced apoptosis.
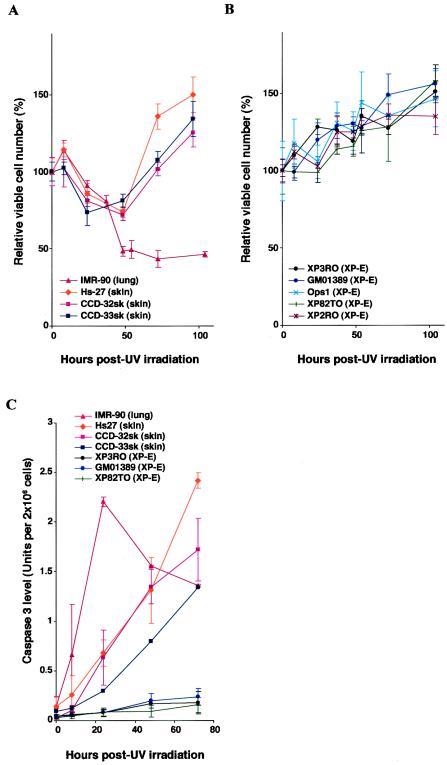
XP-E diploid fibroblast strains are resistant to UV irradiation as assessed by colony-forming ability when compared to strains in other XP groups (24, 26). To precisely assess this resistance of XP-E strains compared to normal diploid fibroblasts, we measured cell survival as judged by dye exclusion (Fig. 1A and B). After exposure to 12 J of UV irradiation m−2, approximately 30% of normal skin strains or 50% of a normal lung strain were killed, whereas all XP-E strains showed no apparent killing. This result suggests that XP-E strains could be defective in UV-induced apoptosis.
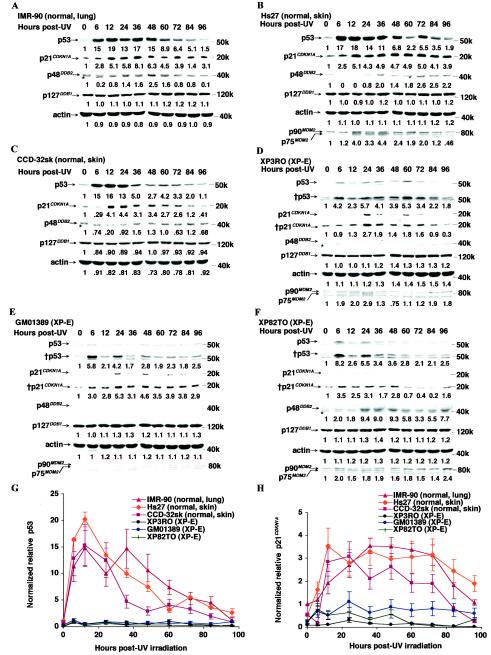
FIG. 1.
XP-E strains are defective in apoptosis after UV irradiation. (A and B) Cell viability of normal (IMR-90, Hs27, CCD-32sk, and CCD-33sk) (A) and XP-E (XP3RO, GM01389, Ops1, XP82TO, and XP2RO) (B) diploid fibroblast strains after UV irradiation. Viable cell number was determined by dye exclusion at times indicated after UV irradiation at a dose of 12 J m−2. Within an experiment, each point was determined in triplicate, and each curve represents three such independent experiments. (C) Caspase 3 activity of normal (IMR-90, Hs27, CCD-32sk, and CCD-33sk) and XP-E (XP3RO, GM01389, and XP82TO) strains before and after 12 J m−2 of UV irradiation. Caspase 3 activity was determined as described in Materials and Methods. Within an experiment, each point was determined in duplicate, and each curve represents three (Hs27 and GM01389), two (IMR-90, CCD-32sk, XP3RO, and XP82TO), or one (CCD-33sk) such experiment.
Apoptosis is a program leading to cell death in response to DNA damage, and defects in apoptosis lead to expansion of a population of neoplastic cells and cancer (2, 17, 23). To confirm whether the observed resistance of XP-E strains to UV irradiation was caused by a defect of apoptosis, we measured the induction of caspase 3, a downstream effector of apoptosis. XP-E strains clearly showed impaired induction of caspase 3 activity compared to that of normal strains (Fig. 1C), implying that p48DDB2 is required for UV-induced apoptosis. Interestingly, both normal lung and skin strains were killed by the radiation, but the time courses varied (Fig. 1A), and this difference was reflected in different expression patterns of caspase 3 (Fig. 1C).
XP-E strains have reduced basal and UV-induced p53.
Since cancers often contain mutations in TP53 that preclude apoptosis and presumably lead to carcinogenesis, we examined sequences of genomic DNAs extracted from each of five XP-E strains of exons 5 through 8 of TP53, which are known to be hot spots of human cancers. No mutations were found in any of the XP-E strains (data not shown).
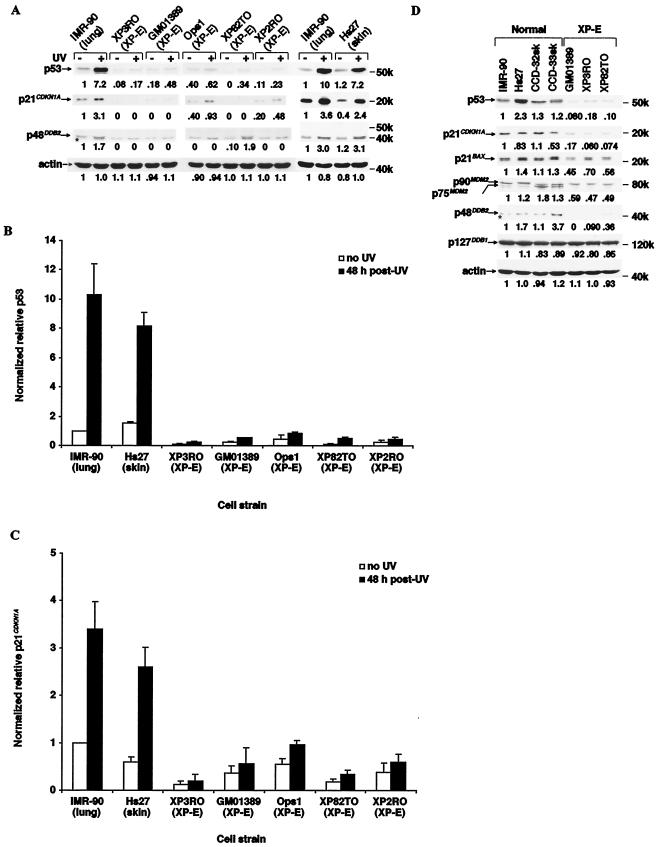
To test whether the apoptotic defect was related to functional abnormality of p53 we examined the basal and UV-induced levels of the protein in normal and XP-E strains (Fig. 2). Levels of p48DDB2 and p21CDKN1A—an inhibitor of cyclin-cyclin-dependent kinase complexes (18) and a growth-inhibitory factor upregulated by p53 (11), respectively—were simultaneously measured. Both p53 and p21CDKN1A levels were 7- to 13 -fold and 2- to 4-fold upregulated by UV irradiation in normal lung and skin strains, respectively (Fig. 2). In contrast, in the XP-E strains, both p53 and p21CDKN1A levels were severely reduced both prior to and 48 h after irradiation. In agreement with previous observations (27, 37), p48DDB2 protein was undetectable in four of the XP-E strains despite the fact that DDB2 RNA was overexpressed and was increased two- to threefold by UV irradiation of normal strains. (In the fifth strain, XP82TO, with a mild clinical manifestation, p48DDB2 protein was expressed as previously reported (27).
FIG. 2.
Reduced levels of p53 and several of its target gene products in XP-E strains. (A) Levels of p53, p21CDKN1A, and p48DDB2 proteins and actin before and 48 h after 12 J of UV irradiation m−2. Samples were prepared, analyzed by Western blotting, and quantitated as described inMaterials and Methods. Each level in the normal IMR-90 strain was designated as 1.0. The chemiluminescence exposure times were 20 s (p53), 30 s (p21CDKN1A), 20 min (p48DDB2), or 10 s (actin). The band marked with an asterisk lying just below p48DDB2 is nonspecific. (B and C) Quantitation of p53 (B) and p21CDKN1A (C) levels before and 48 h after 12 J of UV irradiation m−2. p53 and p21CDKN1A protein levels were normalized to those of actin, and then the level in the unirradiated IMR-90 strain was designated as 1.0. Each value was determined by 6 (IMR-90), 4 (XP3RO), or 3 (Hs27, GM01389, Ops1, and XP2RO) experiments for p53 levels; 10 (Hs27), 8 (IMR-90), 5 (XP2RO), 4 (XP3RO and GM01389), or 3 (Ops1) experiments for p21CDKN1A levels. (D) The basal protein levels of p53, p21CDKN1A, p21BAX, MDM2, p48DDB2, p127DDB1, and actin. The chemiluminescence exposure times were 20 s (p53), 30 s (p21CDKN1A), 5 s (p21BAX), 10 s (MDM2, p127DDB1, and actin), or 20 min (p48DDB2). The band marked with an asterisk lying just below p48DDB2 is nonspecific.
Basal levels of several other proteins in three XP-E strains were also compared to those in four normal strains (Fig. 2D). MDM2 mediates proteolytic degradation of p53, so it was important to determine if it were overproduced and hence responsible for the abnormally low basal levels of p53. However, MDM2 was instead found to be reduced by 60% in the DDB2 mutant strains. BAX is a member of the Bcl-2 protein family that promotes apoptosis by inhibiting Bcl-2 (38), and it is normally upregulated by p53 (34). It was also reduced about 50% in the XP-E strains. In contrast, levels of actin and p127DDB1 (the large subunit of the DDB heterodimer) were normal, while p21CDKN1A and p48DDB2 again were severely reduced in the experiment shown in Fig. 2A.
In diploid human skin fibroblast strains, p53 is stabilized as an early event 9 to 12 h after UV irradiation (46), whereas p48DDB2 is upregulated as a late event 48 h after UV irradiation (27, 37). To determine detailed time courses of induction of each of the proteins in the XP-E strains, we compared the levels of p53, p21CDKN1A, MDM2, p127DDB1, and p48DDB2 in normal and XP-E strains up to 96 h after UV irradiation (Fig. 3). As expected for the normal strains, p53 levels increased to a maximum 12 h after irradiation (Fig. 3A to C and G) followed shortly by increased p21CDKN1A (Fig. 3A to C and H). The p53 and p21CDKN1A levels had increased 11- to 22-fold and 2- to 4-fold, respectively, although the time courses differed somewhat between lung and skin fibroblasts. For XP-E strains, in contrast, both p53 and p21CDKN1A levels were increased by the radiation, but remained far below the levels in normal cells (Fig. 3D to H). Likewise, MDM2 protein levels, while increased up to twofold by irradiation of the XP-E strains, still remained below normal, so that the reduced level of p53 protein present after irradiation was not attributable to an enhancement of MDM2 (compare Fig. 3B to Fig. 3D to F).
FIG.3.
p53 levels after UV irradiation are severely reduced in XP-E strains. (A to F) Protein levels of p53, p21CDKN1A, p48DDB2, p127DDB1, and actin after 12 J of UV irradiation m−2 in normal (A to C) and XP-E (D to F) strains. The chemiluminescence exposure times were 20 s (p53), 2 min (†p53), 15 s (p21CDKN1A), 3 min (†p21CDKN1A), 20 min (p48DDB2), or 10 s (MDM2, p127DDB1, and actin). The band marked with an asterisk lying just below p48DDB2 is nonspecific. Each basal protein level (0 h) was designated as 1.0 to compare protein levels during the time course. (G and H) Quantitation of p53 (G) and p21CDKN1A (H). All protein levels were normalized of the protein levels in the IMR-90 cells at 0 h after being normalized for the amount of actin. Each value was determined by six (XP3RO), four (CCD-32sk and GM01389), three (IMR-90 and Hs27), or two (XP82TO) experiments for p53 levels; six (Hs 27), three (IMR-90, CCD-32sk, GM01389, and XP82TO), or two (XP3RO) experiments for p21CDKN1A levels.
In sum, the naturally occurring mutations in DDB2 found in XP-E strains result in substantially reduced levels of p53 and its downstream-regulated proteins both before and after UV irradiation.
A p53 cDNA, but not a DDB2 cDNA expression construct, rescues the XP-E phenotype.
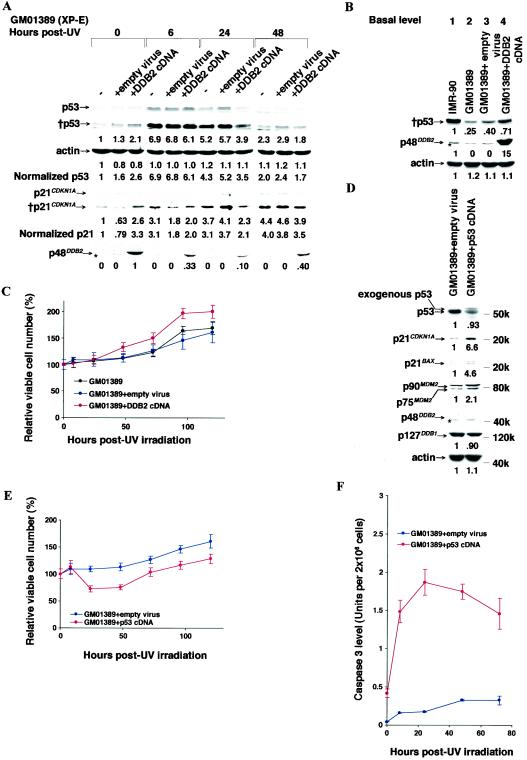
We attempted to complement the XP-E phenotype by overexpressing wild-type p48DDB2 in the XP-E GM01389 strain by introducing the DDB2 cDNA in a Moloney murine leukemia virus with the 5′-LTR as a promoter. The GM01389 strain itself carries compound heterozygous mutations (delN349 and L350P) in the DDB2 gene (27, 37). G418 resistance was carried by the virus and resistance to the drug ensured infection and integration among the surviving cells. The basal p48DDB2 protein levels increased roughly 10-fold in the infected XP-E strain compared to that in uninfected normal IMR-90 cells (Fig. 4B, lane 4 compared to lane 1). The basal levels of both p53 and p21CDKN1A were both increased approximately threefold, so that while p21CDKN1A in the XP-E strain was restored to the level of normal cells without the virus, that of p53 remained about 60% of normal (Fig. 4A, 0 h, and 4B, lane 4).
FIG. 4.
Effect of overexpression of DDB2 or p53 cDNAs upon p53 responses and cell viability after UV irradiation. (A and B) Levels of p53, p21CDKN1A, and p48DDB2 proteins and actin before and after 12 J of UV irradiation m−2 in the XP-E strain GM01389 stably infected with a Moloney murine leukemia virus that carried the DDB2 cDNA, as indicated. The stably infected cells were selected with G418 as described in Materials and Methods. −, no infection; +empty virus, infection with the empty viral vector and G418 selection; +DDB2 cDNA, infection with a DDB2 cDNA construct and G418 selection. The chemiluminescence exposure times were 20 s (p53), 3 min (†p53), 15 s (p21CDKN1A), 3 min (†p21CDKN1A), and 20 min (p48DDB2). The band marked with an asterisk lying just below p48DDB2 is nonspecific. (A) Levels of p53 and p21CDKN1A proteins were normalized to the basal protein level (0 h) of GM01389, which was designated as 1.0, and then the levels were normalized for the relative band intensities of actin. In the case of p48DDB2, the basal level (0 h) of GM01389 with the DDB2 cDNA was designated as 1.0 because endogenous p48DDB2 was undetectable. (B) Each protein level of the IMR-90 was designated as 1.0, and then the other protein levels werenormalized accordingly. (C) Effect of overexpression of DDB2 cDNA in the XP-E strain GM01389 upon cell viability as determined by dye exclusion after 12 J of UV irradiation m−2. The numbers of viable cells were determined by the dye exclusion assay before and after irradiation as described in Materials and Methods. Within an experiment, each point was determined in triplicate; each curve represents three (no infection and DDB2 cDNA infection) or four (empty virus infection) independent experiments. (D) Effect of exogenous p53 cDNA expression in the XP-E strain GM01389 upon basal protein levels of p53, p21CDKN1A, p21BAX, MDM2, p48DDB2, p127DDB1, and actin. (E and F) Cell viability as determined by dye exclusion (E) or caspase 3 activity (F) before and after 12 J of UV irradiation m−2. GM01389 cells were infected with virus containing the p53 cDNA, and stably infected cells were selected with G418. (D) The chemiluminescence exposure times were 3 min (p53 and p21CDKN1A), 5 s (p21BAX), 30 s (MDM2), 20 min (p48DDB2), and 10 s (p127DDB1 and actin). Each protein level of the GM01389+empty virus construct was designated as 1.0, and the respective levels of these proteins in the cells infected with p53 cDNA-containing virus were normalized to these values. The band marked with an asterisk is nonspecific. Note that in the XP-E strain infected with p53 cDNA, the bands are smeared. (E) The curve for p53 cDNA infection was determined by two independent experiments. (Within an experiment, each point was determined in triplicate.) (F) Caspase 3 activity was determined as described in Materials and Methods. Within an experiment, each point was determined in duplicate, and each curve represents two independent experiments.
When these comparisons were made after UV irradiation, p53 levels in the infected XP-E strain increased about sevenfold above its low basal level, therefore still remaining much lower than the induced level in the normal strain (Fig. 4A). Similarly p21CDKN1A was also increased, but still remained at roughly 50% normal in the XP-E strain. Most surprisingly, when killing by UV irradiation was measured by dye exclusion, DDB2 overexpression did not restore apoptotic killing, but actually enhanced survival (Fig. 4C). A similar enhancement was obtained even in the normal IMR-90 strain by infection of DDB2 cDNA (data not shown). In sum, overexpression itself of DDB2 cDNA under the viral promoter did not rescue the XP-E phenotype.
To clarify whether abnormally low p53 levels was the direct cause of XP-E strains being resistant to UV irradiation, p53 cDNA was introduced into the XP-E GM01389 strain by using the same viral vector. Exogenous p53 expression, which yields a protein showing a migration pattern upon SDS gels different from that of endogenous p53 (39), rescued expression levels of its downstream genes, CDKN1A, BAX, and MDM2 (Fig. 4D). However, in these cells, which had undergone the G418 selection and outgrowth, the total p53 expression levels were slightly reduced (Fig. 4D) presumably because high p53 expression itself would have killed the cells during the G418 selection, so that viability was limited to cells with somewhat lower-than-normal p53 expression. Importantly, apoptotic cell killing and caspase 3 induction by UV irradiation were clearly restored to the XP-E strain by the p53 cDNA expression (Fig. 4E and F), although the basal caspase 3 level was higher and its induction was much earlier than in normal strains (compare Fig. 4F to Fig. 1C). Thus, p53 expression functionally rescued the XP-E phenotype and substituted for DDB2 function at least in part.
A DDB2 expression construct containing intron 4 is required to restore the XP-E phenotype.
To attempt to restore normal regulation of p48DDB2 and p53 and UV sensitivity to XP-E strains, we explored the inclusion of genomic regulatory elements of the DDB2 gene in the exogenous DDB2 cDNA construct. In examining the human DDB2 gene sequence to this end, we found several interesting regulatory elements in intron 4 including CREB, CRE-BP1/c-Jun, p300, Sp1, E2F, Ap1, and a p53 consensus binding site that is the only highly conserved p53 CBS in the DDB2 gene (Fig. 5A). We therefore introduced three other DDB2 constructs into the XP-E strain via retroviral infection with G418 selection (Fig. 5A): the DDB2 promoter and 5′UTR alone, the DDB2 promoter and 5′UTR linked to the DDB2 cDNA, and the DDB2 promoter and 5′UTR linked to the cDNA containing intron 4. (Note that a partial match to the p53 CBS was reported to be in the proximal promoter of the DDB2 gene [43], and this region was included in the promoter construct.)
The effect of the construct containing the promoter and 5′UTR was essentially the same as that of the vector alone in the GM01389 strain (Fig. 5B, lane 4, compared to Fig. 4B, lane 3, for the basal protein level; Fig. 5C, GM01389 + 5′UTR, compared to Fig. 4A, +empty virus, for the UV response). As for basal protein levels, neither the promoter and 5′UTR construct nor the promoter and 5′UTR linked to the cDNA restored the basal levels of p53 or p21CDKN1A to normal, even though the basal p48DDB2 level with the latter construct was roughly threefold higher than that of normal strains (Fig. 5B, lane 4 or 5, compared to lanes 1 and 2). In contrast, the basal p53 and p21CDKN1A levels were both restored to the levels of normal strains by the construct containing the promoter, 5′UTR, and DDB2 cDNA containing intron 4 (Fig. 5B, lanes 3 and 6, compared to lanes 1 and 2). It is noteworthy that the basal p48DDB2 level was reduced to about half by the addition of intron 4 to the cDNA (Fig. 5B, lanes 3 and 6, compared to lane 5).
Turning to p53 and p21CDKN1A expression after UV irradiation, again the induction levels in the XP-E strain were restored only by the construct that contained the promoter, 5′UTR, and cDNA with intron 4 (Fig. 5C, GM01389+5′UTR+DDB2 cDNA+Intron4). Moreover, apoptotic cell killing of the XP-E strain by the radiation was similarly restored only when the intron was present (Fig. 5D). This response was paralleled by caspase 3 induction (Fig. 5E)—i.e., caspase 3 induction was restored only when the intron was included. Interestingly, the p53 induction and other responses to the radiation were even more abnormal with the construct including the promoter and 5′UTR linked to the DDB2 cDNA without the intron (Fig. 5C to E) and were similar to those with the construct of the DDB2 cDNA under the viral promoter (Fig. 4A and C).
The DNA sequences of the p53 CBS in intron 4 were normal in each of five XP-E strains (data not shown). To study whether transcriptional and/or posttranscriptional regulation mediated by intron 4 might cause the differences in basal p48DDB2 levels among the DDB2 constructs, total and spliced DDB2 RNA (mRNA) levels were determined by semiquantitative RT-PCR (Fig. 5F). The total DDB2 RNA levels were almost equal after infection with the various constructs (Fig. 5G, lanes 1 to 4), but the level of spliced DDB2 mRNA was only 30% when the intron was present (Fig. 5G, lane 4 compared to lane 1 or 3). Moreover, the p48DDB2 levels reflect these relative mRNA levels among the constructs (compare Fig. 5B and G). Thus, reduced mRNA levels would contribute to keeping basal p48DDB2 levels low (normal), suggesting that intron 4 of the DDB2 gene controls p48DDB2 protein levels through posttranscriptional mechanisms that might include splicing and/or other mechanisms, such as RNA interference, for which RNA might be coded for in intron 4. Intron 4 contains no open reading frames, but does contain several candidate sequences for interfering RNAs.
The p53 CBS is crucial for p53 and p48DDB2 regulation.
To further investigate the function of intron 4 and its p53 CBS, we utilized two other constructs: a 955-bp deletion from intron 4 that removed all of the putative regulatory elements shown in Fig. 5A and a construct with base pair substitutions in the p53 CBS (Fig. 6A). GM01389 XP-E cells infected with the construct with the deletion, while maintaining the outer regions of intron 4, had the low p53 levels before and after irradiation characteristic of XP-E strains (Fig. 6B). The construct harboring the mutant p53 CBS in intron 4 only somewhat restored the p53 levels either before or after UV irradiation, while in the same experiment, the construct harboring the wild-type p53 CBS completely restored the p53 levels to normal (Fig. 6B). In addition, apoptosis and caspase 3 activity after UV irradiation were not restored or were slightly restored by the construct harboring the deletion or the mutant p53 CBS, respectively (Fig. 6C and D). These results show that intron 4 with the wild-type p53 CBS is indispensable for normal p53 levels and the p53-mediated responses to UV irradiation.
To verify that p53 actually binds to the putative p53 CBS in intron 4, we made oligonucleotides containing the wild-type or mutant p53 CBS as shown in Fig. 6A and then conducted EMSAs using p53 expressed in insect cells (Fig. 6E). (Since p53 alone is generally insufficient for binding to a p53 CBS in in vitro experiments, whereas a mixture of p53 and anti-p53 antibody does show specific binding [15, 44], we included the anti-p53 Pab421 antibody.) Indeed, retardation of mobility occurred only with the wild-type p53 CBS oligonucleotide in the presence of both p53 and the antibody (Fig. 6E). In addition, this band was competed out by unlabeled wild-type p53 CBS oligonucleotide, but not by unlabeled mutant p53 CBS oligonucleotide (Fig. 6E, lanes 6 and 7). Finally, the retarded band was supershifted by the addition of a second anti-p53 antibody, Pab1801 (Fig. 6E, lane 8). These results indicate that p53 does selectively bind to the p53 CBS in intron 4 of the DDB2 gene.
Although intron 4 of the DDB2 gene controlled basal DDB2 RNA levels (Fig. 5G), the mechanisms for regulating basal DDB2 RNA and/or p48DDB2 levels by the p53 CBS in intron 4 are unclear. The total DDB2 RNA level mediated by the presence of the DDB2 construct harboring the mutant p53 CBS (Fig. 6A) was about half that mediated by the wild-type p53 CBS (Fig. 6F), suggesting that the wild-type p53 CBS in intron 4 of the DDB2 gene regulates total DDB2 RNA levels. However, the spliced DDB2 mRNA levels were essentially equal for the two constructs. Turning to basal p48DDB2 protein levels, the level mediated by the mutant p53 CBS was about 50% that mediated by the wild-type p53 CBS (Fig. 6F), suggesting that the basal p48DDB2 level is also regulated by translation and/or posttranslational modifications. In sum, the p53 CBS in intron 4 of the DDB2 gene is important for regulation of both DDB2 RNA and p48DDB2 protein levels.
A final question is whether the p53 CBS in intron 4 is required for transcriptional activation of the DDB2 gene after UV irradiation. To this end, DDB2 RNA levels were determined by semiquantitative RT-PCR before and after UV irradiation using the XP-E GM01389 strain infected with each of the four DDB2 constructs. Neither the construct with DDB2 cDNA alone nor that with the 5′UTR linked to DDB2 cDNA induced DDB2 RNA levels up to 72 h after UV irradiation (data not shown), suggesting that intron 4 of the DDB2 gene is important for UV-induced transcriptional activation of DDB2. In contrast, the constructs with intron 4 containing the wild-type or the mutant p53 CBS increased DDB2 RNA levels after UV irradiation (Fig. 6G). Moreover, in these cases, the RNA level was maximal 36 h after UV irradiation, as is the case with the wild-type strain, IMR-90 (27, 37). It is noteworthy that the normal transcriptional activation by the wild-type CBS is clearly more efficient than that by the mutant CBS before (basal) and after UV irradiation (Fig. 6F and G). This suggests that the p53 CBS in the intron 4 is crucial for the transcription activation of the DDB2 gene through coordination with other regulatory elements of intron 4.
Taken all together the normal basal level of p48DDB2 protein as well as p53 binding to its CBS in intron 4 of the DDB2 gene are required for normal levels of both basal and UV-induced p53 as well as for normal p53-mediated functions including apoptosis. In addition, these are required for normal DDB2 induction after UV damages have been removed.
DISCUSSION
Signal transduction pathways involving p53 are generally studied utilizing overexpression of cDNAs in immortalized or cancer cell lines. However, such systems are often not good models for normal cells, because the regulation of the cell cycle and/or apoptosis are already defective in such cell lines. Furthermore, overexpression of the cDNAs themselves perturbs their normal regulation and that of their downstream target genes. In this study utilizing only normal diploid human fibroblasts, the overexpression of the DDB2 cDNA by itself did not complement, but rather exaggerated the abnormality of the p53 response in an XP-E strain. While human primary cell strains suffer from senescence and limited life span, we feel that they are generally better for evaluating regulatory pathways than are immortal cell lines.
As referenced above and confirmed in this study, after UV irradiation, p48DDB2 protein is maximal 48 h or later after irradiation of normal strains, whereas p53 levels are maximal 9 to 12 h after UV irradiation, and the peaks of p21CDKN1A and MDM2 follow shortly thereafter. From these time courses, activated p53 is required for the p48DDB2 induction, which would be consistent with previous observations that led to a proposed involvement of p53 in p48DDB2 induction (22, 28). Moreover, the p53 CBS in intron 4 of the DDB2 gene is required for normal levels of p48DDB2 both before and after UV irradiation.
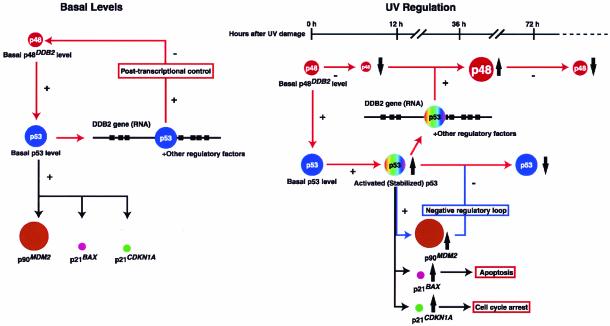
Regulatory interactions between p48DDB2 and p53 suggested by this study are schematized in Fig. 7. XP-E strains, which have mutations in the coding regions of the DDB2 gene and undetectable p48DDB2 protein, showed low basal levels of p53 and its downstream proteins p21CDKN1A, p21BAX, and p90MDM2. These basal levels were restored to normal only by the wild-type DDB2 construct that included intron 4, suggesting that the normal basal level of wild-type p48DDB2 protein is required for normal basal levels of p53 and its downstream proteins. Likewise, it would appear that the basal p48DDB2 level is controlled by the p53 level through its interaction with the p53 CBS in intron 4 of the DDB2 gene, because the presence of the CBS regulated basal DDB2 RNA and p48DDB2 protein levels (Fig. 6F). This regulation is complex, however, because the wild-type p53 CBS mediated transcriptional activation of the DDB2 gene (i.e., positive regulation), but p48DDB2 protein levels were negatively regulated by the wild-type p53 CBS in intron 4. However, overexpression of DDB2 cDNA appears to be toxic, presumably at least in part due to the disturbance of normal p53 regulation.
FIG. 7.
Regulatory interactions between p48DDB2 and p53. Details of the schemes are described in the text. Not shown are stimulation of E2F1-activated transcription by simultaneous overexpression of DDB1 and DDB2 cDNAs (42), stimulation of p53 through the activation of transcription of p14CDKN2A (ARF) by E2F1 (4), and degradation of p48DDB2 by Cullin 4A (36).
Turning to regulation after UV irradiation, XP-E strains showed low levels of UV-induced p53 and its downstream proteins and were defective in UV-induced apoptosis. These defects were also complemented only by a DDB2 construct that included intron 4 with the wild-type p53 CBS. The p53 CBS was also required for the transcriptional activation of the DDB2 gene after irradiation. The entire regulatory regime of p48DDB2 protein after UV irradiation is not completely clear, however, because the protein level is reduced very early after UV irradiation and then is increased through transcriptional activation by p53 and other regulatory factors 48 to 72 h after UV irradiation. With the degradation of activated p53 by MDM2, the p48DDB2 level finally returns to the normal basal level.
Since the reduction of p48DDB2 levels after UV irradiation did not strictly correlate with its RNA levels, the protein levels must be controlled by posttranscriptional mechanisms. Since p53 selectively binds to cdk4 RNA and inhibits its translation (6, 33), p53 might bind not only DDB2 DNA in intron 4 but also DDB2 RNA and control splicing and/or translation of p48DDB2. Another possibility might be posttranslational modification of p48DDB2 by Cullin 4A (36). Cullin 4A, an E3 ubiquitin ligase, promotes p48DDB2 degradation. Other factors are likely to be involved in these regulatory interactions. To fully understand apoptosis and anticancer responses after exposure to DNA-damaging agents, these factors and interactions must be clearly defined.
As a final note, XP-E patients have low, but detectable levels of normal p53, while Li-Fraumeni syndrome (LFS) patients have mutant forms of the protein. LFS patients have a broad spectrum of malignancies, with cancer risks of 50% by age 40 and up to 90% by age 60 (5, 31). XP-E patients have mild photoreactions, but cancers also appear at middle age or later. Interestingly, one of five XP-E patients (Ops1 cells), who has a homozygous nonsense mutation in the DDB2 gene, suffered from multiple melanomas (24), one of the most common malignancies of LFS patients, but not of other XP groups. Given the XP-E phenotype, it may be that other XP-E individuals have weak sun sensitivity and malignancies that are not characteristic of XP, and therefore these individuals may not have been identified as XP-E.
Acknowledgments
We thank J. E. Cleaver for arranging the TP53 sequencing for the XP-E strains, K. Yoshida and D. E. Koshland, Jr., for help with the retroviral experiments, M. Botchan for baculovirus containing the p53 construct, R. Tjian for critical reading of the manuscript, J. Thorner, and Q. Zhou for valuable suggestions, J. H. J. Hoeijmakers and S. Kondo for cell strains, A. Fischer for providing cell culture, and Y. Itoh for encouragement.
This work was supported in part by the Naito Foundation; the Kao Foundation for Arts and Sciences; the Nakatomi Foundation; and grants-in-aid from the Ministry of Education, Science, Sports and Culture, Japan to T.I.; and USPHS grants P30ES08196 and GM59424 to S.L. and 2RO1ES/CA08061 to J.E.C.
REFERENCES
- 1.Aboussekhra, A., M. Biggerstaff, M. K. K. Shivji, J. A. Vilpo, V. Monocollin, V. N. Podust, M. Protic, U. Hubscher, J.-M. Egly, and R. D. Wood. 1995. Mammalian DNA excision repair reconstituted with purified protein components. Cell 80:859-868. [DOI] [PubMed] [Google Scholar]
- 2.Adams, J. M., and S. Cory. 2001. Life-or-death decisions by the Bcl-2 protein family. Trends Biochem. Sci. 26:61-66. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft, M., and K. Vousden. 1999. Regulation of p53 stability. Oncogene 18:7637-7643. [DOI] [PubMed] [Google Scholar]
- 4.Bates, S., A. C. Phillips, P. A. Clark, F. Scott, G. Peters, R. L. Luwig, and K. H. Vousden. 1998. p14ARF links the tumour suppressors RB and p53. Nature 395:124-125. [DOI] [PubMed] [Google Scholar]
- 5.Birch, J. M., R. D. Alston, R. J. McNally, D. G. Evans, A. M. Kelsey, M. Harris, O. B. Eden, and J. M. Varley. 2001. Relative frequency and morphology of cancers in carriers of germline TP53 mutations. Oncogene 20:4621-4628. [DOI] [PubMed] [Google Scholar]
- 6.Cassiday, L. A., and L. J. Maher III. 2002. Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 30:4118-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleaver, J. E., and K. H. Kraemer. 1995. Xeroderma pigmentosum and Cockayne syndrome, p. 4393-4419. In C. R. Scriver, A. L. Beaudet, W. S. Sly, and D. Valle (ed.), The metabolic and molecular basis of inherited diseases. McGraw-Hill, New York, N.Y.
- 8.Cleaver, J. E., and E. Crowley. 2002. UV damage, DNA repair and skin carcinogenesis. Front. Biosci. 7:1024-1043. [DOI] [PubMed] [Google Scholar]
- 9.de Gruijl, F. R., H. J. van Kranen, and L. H. F. Mullenders. 2001. UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J. Photochem. Photobiol. 63:19-27. [DOI] [PubMed] [Google Scholar]
- 10.Dualan, R., T. Brody, S. Keeney, A. F. Nichols, A. Admon, and S. Linn. 1995. Chromosomal localization and cDNA cloning of the genes (DDB1 and DDB2) for the p127 and p48 subunits of a human damage-specific DNA binding protein. Genomics 29:62-69. [DOI] [PubMed] [Google Scholar]
- 11.El-Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 12.Evan, G. I., and K. H. Vousden. 2001. Proliferation, cell cycle and apoptosis in cancer. Nature 411:342-348. [DOI] [PubMed] [Google Scholar]
- 13.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis, p. 283-316. American Society for Microbiology, Washington, D.C.
- 14.Friedman, P. N., S. E. Kern, B. Vogelstein, and C. Prives. 1990. Wild-type, but not mutant, human p53 proteins inhibit the replication activities of simian virus 40 large tumor antigen. Proc. Natl. Acad. Sci. USA 87:9275-9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Funk, W. D., D. T. Pak, R. H. Karas, W. E. Wright, and J. W. Shay. 1992. A transcriptionally active DNA-binding site for human p53 protein complexes. Mol. Cell. Biol. 12:2866-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hainaut, P., T. Hernandez, A. Robinson, P. Rodriguez-Tome, T. Flores, M. Hollstein, C. C. Harris, and R. Montesano. 1998. IARC data base of p53 gene mutations in human tumors and cell lines: updated complication, revised formats and new visualization tolls. Nucleic Acids Res. 26:205-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 18.Harper, J. W., G. R. Adami, N. Wei, K. Keyomarsi, and S. J. Elledge. 1993. The p21 Cdk interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805-816. [DOI] [PubMed] [Google Scholar]
- 19.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 20.Hickman, E. S., M. C. Moroni, and K. Helin. 2002. The role of p53 and pRB in apoptosis and cancer. Curr. Opin. Genet. Dev. 12:60-66. [DOI] [PubMed] [Google Scholar]
- 21.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 22.Hwang, B. J., J. M. Ford, P. C. Hanawalt, and G. Chu. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA 96:424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igney, F. H., and P. H. Krammer. 2002. Death and anti-death: tumour resistance to apoptosis. Nat. Rev. Cancer 2:277-287. [DOI] [PubMed] [Google Scholar]
- 24.Itoh, T., T. Mori, H. Ohkubo, and M. Yamaizumi. 1999. A newly identified patient with clinical xeroderma pigmentosum phenotype has a nonsense mutation in the DDB2 gene and incomplete repair in (6-4) photoproducts. J. Investig. Dermatol. 113:251-257. [DOI] [PubMed] [Google Scholar]
- 25.Itoh, T., and M. Yamaizumi. 2000. UVs syndrome: establishment and characterization of fibroblastic cell lines transformed with simian virus 40 DNA. J. Investig. Dermatol. 114:101-106. [DOI] [PubMed] [Google Scholar]
- 26.Itoh, T., S. Linn, T. Ono, and M. Yamaizumi. 2000. Reinvestigation of the classification of five cell strains of xeroderma pigmentosum group E with reclassification of three of them. J. Investig. Dermatol. 114:1022-1029. [DOI] [PubMed] [Google Scholar]
- 27.Itoh, T., A. Nichols, and S. Linn. 2001. Abnormal regulation of DDB2 expression in xeroderma pigmentosum group E strains. Oncogene 20:7041-7050. [DOI] [PubMed] [Google Scholar]
- 28.Kannan, K., N. Amariglio, G. Rechavi, and D. Givol. 2000. Profile of gene expression regulated by induced by p53: connection to the TGF-β family. FEBS Lett. 470:77-82. [DOI] [PubMed] [Google Scholar]
- 29.Kubbutat, M. H. G., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by mdms. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 30.Lakin, N. D., and S. P. Jackson. 1999. Regulation of p53 in response to DNA damage. Oncogene 18:7644-7655. [DOI] [PubMed] [Google Scholar]
- 31.Lustbader, E. D., W. R. Williams, M. L. Bondy, S. Strom, and L. C. Strong. 1992. Segregation analysis of cancer in families of childhood soft-tissue-sarcoma patients. Am. J. Hum. Genet. 51:344-356. [PMC free article] [PubMed] [Google Scholar]
- 32.Matys V., E. Ficke, R. Geffers, E. Goβling, M. Haubrock, R. Hehl, K. Hornischer, D. Karas, A. E. Kel, O. V. Kel-Margoulis, D.-U. Kloos, S. Land, B. Lewicki-Potapov, H. Michael, R. Munch, I. Reuter, S. Rotert, H. Saxel, M. Scheer, S. Thiele, and E. Wingender. 2003. TRANSFAC: transcriptional regulation, form patterns to profiles. Nucleic Acids Res. 31:374-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, S. J., T. Suthiphongchai, G. P. Zambetti, and M. E. Ewen. 2000. p53 binds selectively to the 5′ untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor β- and p53-mediated translational inhibition of cdk4. Mol. Cell. Biol. 20:8420-8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miyashita, T., and J. C. Reed. 1995. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80:293-299. [DOI] [PubMed] [Google Scholar]
- 35.Mu, D., C. H. Park, T. Matsunaga, D. S. Hsu, J. T. Reardon, and A. Sancar. 1995. Reconstitution of human DNA repair excision nuclease in a highly defined system. J. Biol. Chem. 270:2415-2418. [DOI] [PubMed] [Google Scholar]
- 36.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of Cullin 4A in mammalian cells. Mol. Cell. Biol. 21:6738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichols, A. F., T. Itoh, J. A. Graham, W. Liu, M. Yamaizumi, and S. Linn. 2000. Human damage-specific DNA binding protein p48. Characterization of XPE mutations and regulation following UV irradiation. J. Biol. Chem. 272:21422-21428. [DOI] [PubMed] [Google Scholar]
- 38.Oltvai, A. N., C. L. Milliman, and S. J. Korsmeyer. 1993. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74:609-619. [DOI] [PubMed] [Google Scholar]
- 39.Ookawa, K., T. Kudo, S. Aizawa, H. Saito, and S. Tsuchida. 2002. Transcriptional activation of the MUC2 gene by p53. J. Biol. Chem. 277:48270-48275. [DOI] [PubMed] [Google Scholar]
- 40.Oren, M. 1999. Regulation of the p53 tumor suppressor protein. J. Biol. Chem. 274:36031-36034. [DOI] [PubMed] [Google Scholar]
- 41.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 42.Shiyanov, P., S. A. Hayes, M. Donepudi, A. F. Nichols, S. Linn, B. L. Slagle, and P. Raychaudhuri. 1999. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 19:4935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan, T., and G. Chu. 2002. p53 binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Mol. Cell. Biol. 22:3247-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka, H., H. Arakawa, T. Yamaguchi, K. Shiraishi, S. Fukuda, K. Matsui, Y. Takei, and Y. Nakamura. 2000. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature 404:42-49. [DOI] [PubMed] [Google Scholar]
- 45.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 46.Yamaizumi, M., and T. Sugano. 1994. UV-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene 9:2775-2784. [PubMed] [Google Scholar]