Abstract
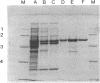
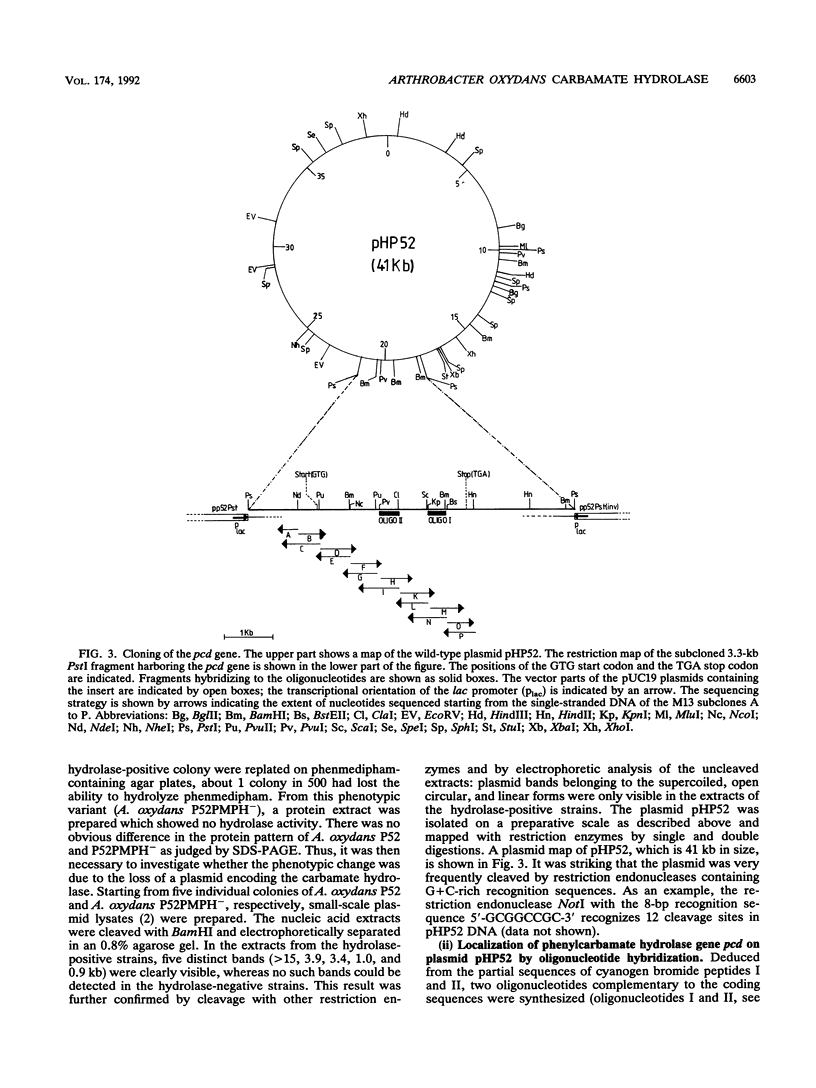
Arthrobacter oxydans P52 isolated from soil samples was found to degrade the phenylcarbamate herbicides phenmedipham and desmedipham cometabolically by hydrolyzing their central carbamate linkages. The phenylcarbamate hydrolase (phenmedipham hydrolase) responsible for the degradative reaction was purified to homogeneity. The enzyme was shown to be a monomer with a molecular weight of 55,000. A 41-kb wild-type plasmid (pHP52) was identified in A. oxydans P52, but not in a derivative of this strain that had spontaneously lost the ability to hydrolyze phenylcarbamates, indicating that the gene for phenylcarbamate degradation (pcd) is plasmid encoded. Determination of two partial amino acid sequences allowed the localization of the coding sequence of the pcd gene on a 3.3-kb PstI restriction fragment within pHP52 DNA by hybridization with synthetic oligonucleotides. The phenylcarbamate hydrolase was functionally expressed in Escherichia coli under control of the lacZ promoter after the 3.3-kb PstI fragment was subcloned into the vector pUC19. A stretch of 1,864 bases within the cloned Pst fragment was sequenced. Sequence analysis revealed an open reading frame of 1,479 bases containing the amino acid partial sequences determined for the purified enzyme. Sequence comparisons revealed significant homology between the pcd gene product and the amino acid sequences of esterases of eukaryotic origin. Subsequently, it was demonstrated that the esterase substrate p-nitrophenylbutyrate is hydrolyzed by phenmedipham hydrolase.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block M. D., Botterman J., Vandewiele M., Dockx J., Thoen C., Gosselé V., Movva N. R., Thompson C., Montagu M. V., Leemans J. Engineering herbicide resistance in plants by expression of a detoxifying enzyme. EMBO J. 1987 Sep;6(9):2513–2518. doi: 10.1002/j.1460-2075.1987.tb02537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brandsch R., Decker K. Isolation and partial characterization of plasmid DNA from Arthrobacter oxidans. Arch Microbiol. 1984 May;138(1):15–17. doi: 10.1007/BF00425400. [DOI] [PubMed] [Google Scholar]
- Brandsch R., Faller W., Schneider K. Plasmid pAO1 of Arthrobacter oxidans encodes 6-hydroxy-D-nicotine oxidase: cloning and expression of the gene in Escherichia coli. Mol Gen Genet. 1986 Jan;202(1):96–101. doi: 10.1007/BF00330523. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel W., Kopperschläger G., Hofmann E. An improved procedure for protein staining in polyacrylamide gels with a new type of Coomassie Brilliant Blue. Anal Biochem. 1972 Aug;48(2):617–620. doi: 10.1016/0003-2697(72)90117-0. [DOI] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Spierer P. The Ace locus of Drosophila melanogaster: structural gene for acetylcholinesterase with an unusual 5' leader. EMBO J. 1986 Nov;5(11):2949–2954. doi: 10.1002/j.1460-2075.1986.tb04591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. H., Stratowa C., Rutter W. J. Isolation of full-length putative rat lysophospholipase cDNA using improved methods for mRNA isolation and cDNA cloning. Biochemistry. 1987 Mar 24;26(6):1617–1625. doi: 10.1021/bi00380a020. [DOI] [PubMed] [Google Scholar]
- Hanzlik T. N., Abdel-Aal Y. A., Harshman L. G., Hammock B. D. Isolation and sequencing of cDNA clones coding for juvenile hormone esterase from Heliothis virescens. Evidence for a catalytic mechanism for the serine carboxylesterases different from that of the serine proteases. J Biol Chem. 1989 Jul 25;264(21):12419–12425. [PubMed] [Google Scholar]
- Johnson L. M., Talbot H. W., Jr Detoxification of pesticides by microbial enzymes. Experientia. 1983 Nov 15;39(11):1236–1246. doi: 10.1007/BF01990361. [DOI] [PubMed] [Google Scholar]
- Kearney P. C., Kaufman D. D. Enzyme from Soil Bacterium Hydrolyzes Phenylcarbamate Herbicides. Science. 1965 Feb 12;147(3659):740–741. doi: 10.1126/science.147.3659.740. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lockridge O., Bartels C. F., Vaughan T. A., Wong C. K., Norton S. E., Johnson L. L. Complete amino acid sequence of human serum cholinesterase. J Biol Chem. 1987 Jan 15;262(2):549–557. [PubMed] [Google Scholar]
- Long R. M., Satoh H., Martin B. M., Kimura S., Gonzalez F. J., Pohl L. R. Rat liver carboxylesterase: cDNA cloning, sequencing, and evidence for a multigene family. Biochem Biophys Res Commun. 1988 Oct 31;156(2):866–873. doi: 10.1016/s0006-291x(88)80924-0. [DOI] [PubMed] [Google Scholar]
- MacPhee-Quigley K., Taylor P., Taylor S. Primary structures of the catalytic subunits from two molecular forms of acetylcholinesterase. A comparison of NH2-terminal and active center sequences. J Biol Chem. 1985 Oct 5;260(22):12185–12189. [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Oakeshott J. G., Collet C., Phillis R. W., Nielsen K. M., Russell R. J., Chambers G. K., Ross V., Richmond R. C. Molecular cloning and characterization of esterase-6, a serine hydrolase of Drosophila. Proc Natl Acad Sci U S A. 1987 May;84(10):3359–3363. doi: 10.1073/pnas.84.10.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A. N., Barnett L., Brenner S. Transformation of Arthrobacter and studies on the transcription of the Arthrobacter ermA gene in Streptomyces lividans and Escherichia coli. Biochem J. 1987 Apr 15;243(2):431–436. doi: 10.1042/bj2430431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M., Camp S., Maulet Y., Newton M., MacPhee-Quigley K., Taylor S. S., Friedmann T., Taylor P. Primary structure of Torpedo californica acetylcholinesterase deduced from its cDNA sequence. 1986 Jan 30-Feb 5Nature. 319(6052):407–409. doi: 10.1038/319407a0. [DOI] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Shimada Y., Sugihara A., Tominaga Y., Iizumi T., Tsunasawa S. cDNA molecular cloning of Geotrichum candidum lipase. J Biochem. 1989 Sep;106(3):383–388. doi: 10.1093/oxfordjournals.jbchem.a122862. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorav J. L., Krejci E., Massoulié J. cDNA sequences of Torpedo marmorata acetylcholinesterase: primary structure of the precursor of a catalytic subunit; existence of multiple 5'-untranslated regions. EMBO J. 1987 Jul;6(7):1865–1873. doi: 10.1002/j.1460-2075.1987.tb02445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small D. H. Non-cholinergic actions of acetylcholinesterases: proteases regulating cell growth and development? Trends Biochem Sci. 1990 Jun;15(6):213–216. doi: 10.1016/0968-0004(90)90027-9. [DOI] [PubMed] [Google Scholar]
- Stalker D. M., McBride K. E., Malyj L. D. Herbicide resistance in transgenic plants expressing a bacterial detoxification gene. Science. 1988 Oct 21;242(4877):419–423. doi: 10.1126/science.242.4877.419. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek P. H., Karns J. S. Cloning of a carbofuran hydrolase gene from Achromobacter sp. strain WM111 and its expression in gram-negative bacteria. J Bacteriol. 1989 Jul;171(7):4038–4044. doi: 10.1128/jb.171.7.4038-4044.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]