Abstract
In vivo, histone tails are involved in numerous interactions, including those with DNA, adjacent histones, and other, nonhistone proteins. The amino termini are also the substrates for a number of enzymes, including histone acetyltransferases (HATs), histone deacetylases, and histone methyltransferases. Traditional biochemical approaches defining the substrate specificity profiles of HATs have been performed using purified histone tails, recombinant histones, or purified mononucleosomes as substrates. It is clear that the in vivo presentation of the substrate cannot be accurately represented by using these in vitro approaches. Because of the difficulty in translating in vitro results into in vivo situations, we developed a novel single-cell HAT assay that provides quantitative measurements of endogenous HAT activity. The HAT assay is performed under in vivo conditions by using the native chromatin structure as the physiological substrate. The assay combines the spatial resolving power of laser scanning confocal microscopy with simple statistical analyses to characterize CREB binding protein (CBP)- and P300-induced changes in global histone acetylation levels at specific lysine residues. Here we show that CBP and P300 exhibit unique substrate specificity profiles, consistent with the developmental and functional differences between the two HATs.
Since the initial identification of specific enzymes with nuclear histone acetyltransferase (HAT) activity in 1996 (7, 42), an enormous research effort has centered upon defining the nuclear functions in which these enzymes participate. Two members of the nucleoplasmic HAT A family, CBP and its close homologue P300, are of particular interest because of their involvement in important cellular processes, including differentiation, development, and apoptosis. CBP and P300 exhibit a high degree of sequence similarity and have similar domain and motif structures (8, 27). Both have a nuclear receptor binding domain, three zinc fingers, a bromodomain, and a large internal HAT domain (11). More than 40 different proteins have been shown to interact with CBP and/or P300 in vitro, and they fall roughly into four different categories: nuclear receptors, transcription factors, signaling molecules, and viral proteins (5, 8, 11, 12, 25). CBP and P300 were first identified as transcriptional coactivators proposed to function by bridging the basal transcription machinery to upstream regulatory sequences (3, 9, 27). They have since been shown to positively and negatively regulate a number of gene activation pathways and to be integral players in a variety of developmental processes, including apoptosis, cell proliferation, and embryogenesis (reviewed in references 5, 11, 12, and 25).
Central to the functions ascribed to CBP and P300 is their ability to alter chromatin structure through histone acetylation. The nucleosome is the fundamental repeating unit of chromatin. It is composed of an octameric core particle consisting of two molecules each of histones H2A, H2B, H3, and H4, around which 146 bp of DNA is wrapped. The amino-terminal “unstructured” domains found on each of the four core histones undergo several types of posttranslational modifications (e.g., acetylation, phosphorylation, and methylation) that are believed to influence histone function, possibly through chromatin alterations (4, 45).
Histone acetylation, in particular, has been strongly correlated with the chromatin remodeling that is associated with transcriptional activation and characterized by DNase I sensitivity. The individual nucleosomes are connected by short intervening DNA sequences (termed linker DNA) forming the 10-nm chromatin fiber, or “beads on a string.” Although the 10-nm fiber conformation has been well characterized in vitro by electron microscopy, nucleosomal arrays do not adopt this fully extended conformation unless very low ionic strength buffers are utilized (10, 14, 19, 40). Hence, the chromatin substrate is rarely, if ever, presented to the nuclear HATs as 10-nm extended nucleosomal fibers. Rather, emerging evidence indicates that euchromatin and heterochromatin are found in structures that are folded beyond the 30-nm level to fibers of 80 to 200 nm in diameter (14, 16). In this environment, the tails are essential and are engaged in contacts within and between chromatin fibers. Therefore, the presentation of the histone N termini is not reconstituted in vitro by current approaches.
In vitro biophysical studies performed on isolated nucleosomes and model nucleosomal arrays have clearly demonstrated that the histone N termini function in a variety of interactions, including nucleosome-DNA, nucleosome-nucleosome, and higher-order fiber-fiber interactions (1, 2, 14, 16, 38, 39). One conclusion of these studies is that the interactions that engage the N termini in vitro are highly dependent upon the model system employed and the ionic composition of buffers. Consequently, the presentation of core histone tails in vitro (i.e., as histones, purified nucleosomes, or short nucleosomal arrays) as HAT substrates may differ significantly from their presentation in vivo within native chromatin. The most obvious difference is the presence of higher-order chromatin compaction states (i.e., those beyond the 30-nm fiber) that are highly dependent on the histone tails which regulate the fiber-fiber interactions (15). The addition of a broad spectrum of chromatin-associated proteins is also anticipated to add to the complexity of the system and impact histone tail presentation. For example, histone H1 has been shown to modulate histone acetylation activity in vivo and to contribute to the conformation of the endogenous chromatin substrate (13). Consequently, the current in vitro assays, which do not account for any structure beyond the nucleosome, may not accurately reflect the accessibility to the substrate and/or the specificity of the nuclear HAT in situ.
Histone acetylation is not a binary process whereby chromatin is either acetylated or not. Rather, there are basal steady-state acetylation levels for all acetylated histone lysine residues present throughout euchromatic and heterochromatic regions of the nucleus. These steady-state levels arise through the balance of continually acting HATs and histone deacetylases (HDACs), with the specific site utilization reflecting the net result of this balance. In 1990, Thorne et al. (35) demonstrated that basal histone acetylation levels in unsynchronized HeLa cells were relatively low. For example, only one of four (histone H4) or five (histone H3) possible acetylation sites was typically occupied, and the most frequently acetylated sites were K14 (for H3) and K16 (for H4). The existence of di-, tri-, tetra-, and penta-acetylated histones at steady state has been demonstrated; however, they are much less abundant, particularly tetra- and penta-acetylated forms (35). More recent determinations by Schiltz et al. (30) suggest that approximately 60% of K16 sites are acetylated in mononucleosomes isolated from HeLa cells, while other sites, such as K5 of histone H4, are found on a very small subset of the chromatin at steady state. These observations collectively argue that all potential acetylation sites will be significantly represented in chromatin. Therefore, we can assess differences in global acetylation at specific histone residues that are contingent upon minimal increases in CBP and P300 expression.
In this study, we examined CBP/P300 HAT specificities in vivo by using the native chromatin structure as a substrate. We demonstrate that both HATs induce increased acetylation on specific lysine residues in endogenous chromatin. By developing a single-cell quantitative HAT assay, we were able to investigate HAT specificities under conditions where the chromatin is compact and the histone tails are engaged in their native molecular interactions. Our studies demonstrate that CBP and P300 are able to acetylate histones in a concentration-dependent manner in vivo and that these proteins are capable of acting on large regions of the genome rather than being confined to a small number of promoter or enhancer sequences.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa (human epithelioid cervical carcinoma), COS-7 (African green monkey kidney), IM (male Indian Muntjac skin fibroblast), and 10T1/2 (C3H mouse embryo fibroblast) cells were cultured in Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS), RPMI 1640 plus 10% FBS, Ham's F-10 medium plus 20% FBS, and α-MEM plus 10% FBS, respectively, in a 37°C incubator under 5% CO2. Cells were plated onto sterilized glass coverslips so that they were 50 to 80% confluent the following day. Cells were transiently transfected with the appropriate construct by using Effectene (Qiagen Inc.) with slight modifications to the manufacturer's instructions. Briefly, 400 ng of plasmid DNA and 4 μl of Effectene were used per coverslip. Plasmids used in these experiments include a previously described full-length murine CBP construct fused to green fluorescent protein (GFP) (6), a full-length hemagglutinin (HA)-P300 fusion construct (21-179; Upstate), and a negative HAT control, alternative splicing factor (ASF) fused to GFP (31). Cells were cultured for 18 to 20 h to allow for transient protein expression while minimizing the coactivator function of CBP and P300. To eliminate the characterization of artifacts resulting from high levels of overexpression, only cells expressing minimal amounts of the proteins of interest were selected for imaging and subsequent analysis. All cells expressing large amounts of GFP-CBP, HA-P300, or ASF-GFP were manually eliminated.
Indirect immunofluorescence.
Transiently transfected cells were fixed 18 to 20 h posttransfection with 4.0% paraformaldehyde in phosphate-buffered saline (PBS) (pH 7.5) for 5 min at room temperature. Cells were permeabilized with PBS containing 0.5% Triton X-100 for 5 min. Next, cells were washed twice with PBS, inverted onto 30-μl aliquots of an appropriate primary antibody, and incubated at room temperature for 30 min. Coverslips were rinsed with PBS containing 0.1% Triton X-100 and washed twice with PBS prior to a 30-min incubation with an appropriate secondary antibody conjugated to a fluorophore. Cells were rinsed with PBS containing 0.1% Triton X-100 and washed twice with PBS. Coverslips were mounted onto slides containing approximately 10 μl of a 90% glycerol-PBS-based medium containing 1 mg of paraphenylenediamine/ml and 0.5 μg of 4′,6′-diamidino-2-phenylindole (DAPI)/ml. HA-P300 was visualized with a mouse anti-HA monoclonal antibody (HA-7; Sigma) and an anti-mouse Alexa-Fluor 488 secondary antibody (A-11001; Molecular Probes).
A panel of commercially available rabbit primary antibodies, directed against various acetylated histone tail lysine residues, was used to detect the specificity preferences of GFP-CBP and HA-P300. A second panel of antibodies specifically recognizing HDACs and HATs was used to quantify total protein expression levels and included HDAC-1 (1:100; Upstate), HDAC-2 (1:200; Santa Cruz Biotechnology), HDAC-3 (1:100; a gift from E. Verdin), HDAC-4 (1:50; a gift from E. Verdin), PCAF (1:200; a gift from X. Yang), TAFII250 (1:400; Santa Cruz Biotechnology), TIP60 (1:200; Upstate), and an anti-MYST (1:200; Upstate) family antibody recognizing ESA1, TIP60, MOZ, MOF, SAS2, and SAS3/YBF. Additionally, antibodies specifically recognizing either CBP (Ac238; Neomarkers) or P300 (Rockland) were used at 1:500 and 1:200, respectively. Secondary antibodies included both anti-mouse and anti-rabbit antibodies conjugated with either Alexa-Fluor 488 (Molecular Probes) or cyanin 3 (Cy3) (Jackson ImmunoReseach Laboratories, Inc.) and were used at 1:200.
Image acquisition and visualization.
To investigate the localization patterns and three-dimensional (3D) spatial relationships, 3D optical (z-) series were collected using a Zeiss Axioplan 2 digital imaging microscope equipped with a 100× (1.4 numerical aperture [NA]) plan-apochromat lens and a Coolsnap HQ cooled-CCD camera (Photometrics/Roper Scientific). Z-series extending above and below individual nuclei were collected at 200-nm intervals. Images were processed by maximum-likelihood-expectation deconvolution in softWoRx (Applied Precision) by using a constrained iterative algorithm and theoretical optical transfer files generated for wavelengths of 485 (DAPI), 535 (GFP or Alexa-488), and 610 (Cy3) nm. Deconvolved images were visualized in Imaris (Bitplane AG), and 3D projections and rotations were generated. The localization patterns of endogenous and transfected CBP and P300 were compared visually, as were the 3D spatial relationships between endogenous or transiently expressed CBP/P300 and either acetylation sites or chromatin. The analysis was performed by comparing the spatial relationships between endogenous or transiently expressed CBP/P300 with a highly represented steady-state acetylation site, K14, or a much less abundant steady-state acetylation site, K5. Endogenous P300 and CBP localization patterns were investigated in untransfected paraformaldehyde-fixed cells by using the anti-P300 and anti-CBP antibodies listed above. Composite montages of collected images were assembled in Adobe Photoshop, version 7.0.
Confocal microscopy was employed to investigate total-protein expression levels and to perform the HAT assay. Briefly, 12-bit images were collected using a Zeiss 510 laser-scanning microscope equipped with a 40× (1.3NA) lens. For each antibody trial, the detector gain was first optimized by sampling various regions of the coverslip and then fixed for each channel. Once set, the detector gain value was kept constant throughout the image acquisition process. As a result, qualitative measurements for signal intensities from identical channels of different images could be compiled to permit quantitative measurements and statistical comparisons. For each assay, a minimum of 50 transiently expressing interphase cells were imaged for each antibody treatment. Individual channels were separated with Zeiss LSM 510, version 3.0 (Carl Zeiss, Inc.), and subjected to quantitative analysis.
Qualitative and quantitative image analyses.
Quantitative image analysis was performed using Metamorph (version 4.6r9; Universal Imaging Corp.) on individual channels previously extracted from the collected series of confocal images. For the HAT assay, the DAPI channel was used to determine total DNA content and define the nuclear area, the Cy3 channel provided information about the acetylation status of a specific epitope, and the GFP or Alexa 488 channel identified those cells transiently expressing the construct of interest (i.e., GFP-CBP, HA-P300, or ASF-GFP). For the total-protein expression assays, the DAPI channel identified the nuclear shape, while the Cy3 and GFP or Alexa 488 channels provided information about total fluorescent signal intensities (or protein expression levels) and identified transfected cells, respectively. In both the HAT and protein expression assays, background noise was subtracted from each image prior to making any qualitative measurements. A binary DAPI (nuclear) mask was then generated through signal intensity thresholding of the DAPI channel. The nuclear mask was applied to all channels, and the total signal intensities for each individual channel were measured and exported as raw data into Microsoft Excel. For each assay and each antibody trial, signal intensities from identical channels were compiled. For the HAT assay only, the Cy3 data (acetylation [Ac] level) were normalized to the DAPI data (normalized acetylation [N-Ac]) to account for variation in the amounts of DNA and histones arising from variable cell cycle status (e.g., pre- or post-S phase). Next, the data from all channels were imported into Prism (GraphPad Software Inc.), where various statistical analyses were performed.
Statistical analyses.
To visually depict the heterogeneity that was observed in determining total-protein expression levels in interphase cells, box-and-whisker plots were generated in Prism. By representing the total distribution ranges of untransfected and GFP-CBP- or HA-P300-transfected populations as box-and-whisker plots, we were able to make simple comparisons between populations. For reference purposes, the entire distribution range is signified by the whiskers, while the 25th percentile, the mean, and the 75th percentile are identified by the bottom of the box, the middle horizontal line, and the top of the box, respectively. Next, specific differences in mean total intensity (TI) values between the untransfected and transfected populations were investigated by unpaired t tests (two tailed) using Prism. Finally, to quantify the mean fold increase in CBP or P300 expression in GFP-CBP- or HA-P300-expressing cells, respectively, ratios between the mean TIs from transfected and untransfected populations were calculated. The ratio represents the average fold increase in expression that is anticipated to occur throughout the HAT assay as a result of transient GFP-CBP or HA-P300 expression.
The HAT assay takes advantage of four different statistical approaches to define the substrate specificity preferences of CBP and P300. Recall that all acetylation intensity values were normalized to DAPI signal intensities to account for DNA and histone content variation occurring due to the location in the cell cycle. First, histograms were generated to assess the global increase in N-Ac intensities expected to occur in instances where substrate specificity preferences occur. By comparing the N-Ac distributions for untransfected controls with those for GFP-CBP-, HA-P300-, or ASF-GFP-expressing cells, clues as to specific substrates were gleaned. Next, scatter plots were generated by comparing the GFP or Alexa 488 TIs of only the transiently expressing cells to the N-Ac intensity for each antibody directed against specific acetylated histone tail lysine residues in the COS-7, HeLa, and IM cell lines. Best-fit lines were applied to reveal potential correlations between protein expression (i.e., GFP or Alexa-Fluor 488 TI) and N-Ac intensities. Linear regression analysis was performed in Prism, and R2 values were calculated to describe the dependency of specific increases in acetylation on specific increases in GFP-CBP or HA-P300 expression. Finally, unpaired t tests (two tailed) were performed to compare the mean N-Ac values calculated for the transiently expressing (GFP-CBP and HA-P300) and untransfected control populations for each antibody trial. The specific substrate preferences of CBP and P300 were ordered by comparing the results of all four statistical analyses obtained from COS-7, HeLa, and IM cell lines and ranking the increases in acetylation contingent upon GFP-CBP or HA-P300 expression. Composite montages of exported graphs were assembled in Adobe Photoshop 7.0.
RESULTS
Development of an in vivo HAT assay.
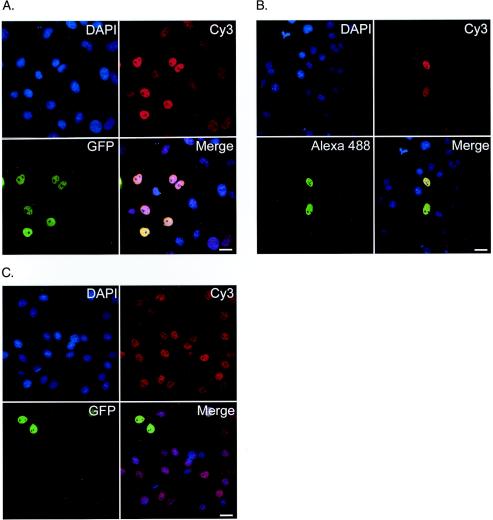
Differences in global steady-state acetylation in cells treated with HDAC inhibitors have been characterized by microscopy previously (17, 21). As a result, we reasoned that it should be possible to use fluorescence microscopy to detect and quantify differences in specific acetylation levels in cells transfected with constructs encoding HATs and to compare them with untransfected controls. Initial experiments were conducted to determine whether or not transiently expressing GFP-CBP or HA-P300 would alter the steady-state acetylation level of lysine 5 (K5) of histone H4. Figure 1 shows fluorescence images of K5 acetylation in cells transfected with GFP-CBP (Fig. 1A) or HA-P300 (Fig. 1B). Figure 1C shows a control transfection of a nuclear protein that does not contain endogenous HAT activity (ASF-GFP). Although increases in acetylation were observed in cells expressing the HAT constructs, a direct relationship between HAT expression and specific increases in acetylation could not be conclusively demonstrated until several key control experiments and selection criteria were performed (detailed below).
FIG. 1.
GFP-CBP and HA-P300 expression correlates with increased lysine 5 acetylation. Confocal micrographs depict the K5 (histone H4) acetylation status in COS-7 cells transiently expressing GFP-CBP (A), HA-P300 (B), or ASF-GFP (C). DNA is stained with DAPI (blue), acetylated K5 is identified by a secondary antibody conjugated to Cy3 that recognizes the acetylated K5 antibody (red), and GFP-CBP, HA-P300, and ASF-GFP expression is shown in green. The merge image is an overlay of the three channels. Bars, 25 μm.
GFP-CBP and HA-P300 recapitulate endogenous expression patterns.
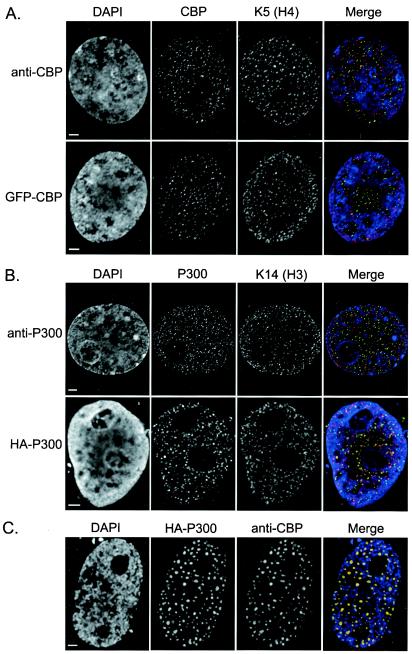
When cells were transfected with the HAT fusion proteins, a significant range of protein expression was observed at the time of fixation (18 to 20 h posttransfection). When cells were examined 48 to 72 h posttransfection, a significant amount of cell death had occurred and most viable cells expressed high levels of the transfected protein. By employing several key selection criteria, our image-based approach to determining in vivo HAT specificities has several distinct advantages over conventional biochemical approaches. For example, we are able to select only those cells which recapitulate the expression patterns of the endogenous proteins (Fig. 2A and B) and to eliminate those cells which are apoptotic, express high levels of the protein, or exhibit atypical subcellular protein distributions (Fig. 2C).
FIG.2.
(A and B) Subnuclear distribution of transiently expressed GFP-CBP or HA-P300 and endogenous CBP/P300 in relation to acetylation and chromatin. Representative images are shown to highlight the ability of transiently expressed GFP-CBP (A) or HA-P300 (B) to recapitulate the expression patterns of their endogenous counterparts. Cells were paraformaldehyde fixed, costained with either anti-K5 (H4) (A) or anti-acetylated K14 (H3) (B), and counterstained with DAPI to visualize bulk chromatin. Labels on left identify cells stained for endogenous CBP or P300 (anti-CBP or anti-P300, respectively) or transiently expressing either GFP-CBP or HA-P300. Merged images depict the 3D interrelationship between either CBP, GFP-CBP, P300, or HA-P300 (green) and acetylated K5 or K14 (red) or chromatin (blue) in a single plane isolated from deconvolved z-series collected using digital imaging microscopy. (C) Example of the type of cell excluded from subsequent analysis based on morphological abnormalities (i.e., large HA-P300 aggregates concomitant with the reorganization of CBP into similar regions). HA-P300 and endogenous CBP appear green and red, respectively, in the merged image, and DAPI staining appears blue. Bars, 2 μm.
In order to verify that the majority of transfected cells were suitable for analysis, we determined whether or not transfected cells organized CBP or P300 into the several hundred subnuclear foci that are characteristic of this class of nuclear protein (18). In addition, we confirmed that the endogenous and transiently expressed HATs had similar subcellular distributions relative to acetylated chromatin. The endogenous staining patterns of CBP and P300 were investigated in paraformaldehyde-fixed COS-7, HeLa, IM, and 10T1/2 cells by using anti-CBP (Ac238) or anti-P300 (Rockland). In agreement with previous descriptions (17, 25), CBP, P300, and specific histone acetylations localize to between 200 and 500 discrete foci scattered nonrandomly throughout the nucleoplasm (Fig. 2). These foci are massive on a molecular scale, with an estimated diameter of 100 nm each. If we assume that the acetylation foci and the nucleosomes are spherical, with radii of 50 and 5.5 nm, respectively, we can estimate the total number of nucleosomes per focus to be approximately 750 ( πr focus3 / πr nucleosome3). CBP, P300, and acetylation foci are noticeably absent in nucleoli and are depleted in chromatin-dense regions (identified by highly intense DAPI staining).
Transient expression of GFP-CBP or HA-P300 in COS-7, HeLa, IM, and 10T1/2 cells yielded two distinct localization patterns—a predominant pattern indistinguishable from that with the endogenous counterpart (Fig. 2A and B) and a much less abundant pattern that formed larger aggregates, typically numbering between 50 and 75 per nucleus (Fig. 2C). Cells with the latter pattern (i.e., large aggregates) exhibited striking increases in overall GFP-CBP or HA-P300 fluorescence compared to the predominant focal pattern. Since structures of this size were never observed with the endogenous proteins, these cells were excluded from all subsequent analyses.
To investigate the spatial relationship between transient GFP-CBP/HA-P300 expression and acetylation (K14 and K5 of histones H3 and H4, respectively), 3D digital imaging microscope (DIM) images were collected and compared to those obtained for endogenous CBP/P300 and acetylation. Endogenous CBP/P300, GFP-CBP/HA-P300, and acetylation (K14 and K5) were nonrandomly distributed throughout the nucleoplasm, absent in nucleoli, and significantly depleted in dense regions of chromatin. Although a focal staining pattern was observed in all cases, a general lack of colocalization was apparent between CBP/P300 or GFP-CBP/HA-P300 and acetylated K14 or K5. Figure 2 shows representative images depicting the spatial relationship between endogenous CBP/P300 or transiently expressed GFP-CBP/HA-P300 and acetylation. Despite the general lack of colocalization, a striking complementarity between the HAT (endogenous CBP/P300 and GFP-CBP/HA-P300) distribution and the acetylated chromatin (K14 and K5) was observed, as has been previously described (17, 18, 21). Briefly, the acetylated chromatin often localizes to the intervening spaces between the chromatin-dense regions (DAPI-intense regions) and the HAT foci.
Transient HAT expression results in minimal increases in total-protein expression.
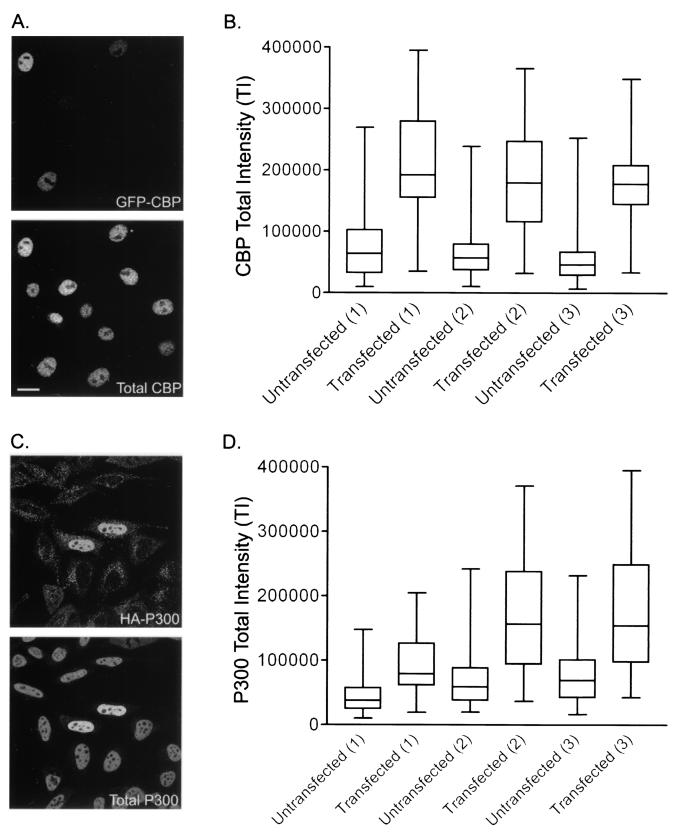
To address the possibility that high overexpression could artificially impact our results, we decided to quantify total CBP and P300 expression levels in transfected cells and compare them with the respective levels in untransfected controls. Total protein levels were determined by using a single-cell approach, similar to that of the HAT assay, which combines confocal microscopy, quantitative image analysis, and statistical analyses. Because the detector gain is held constant throughout the image acquisition process, the fluorescent signal intensities in nuclei of one image can be compared to those in another. In other words, total CBP or P300 signal intensities can be used to quantify their respective protein expression levels. This allows quantitative comparisons between untransfected cells and GFP-CBP- or HA-P300-expressing cells.
Total CBP and P300 expression levels were calculated in three separate experiments each for GFP-CBP- and HA-P300-expressing cells, respectively. As shown in Fig. 3, both transfected and untransfected cells showed significant variability in CBP (Fig. 3A and B) and P300 (Fig. 3C and D) expression levels. A general increase in total signal intensity over that for untransfected controls was observable for cells transiently expressing either GFP-CBP or HA-P300. Upon further inspection of the box-and-whisker plots, two additional trends were noted. First, the CBP/P300 TI distributions for the transfected populations are supershifted compared to their untransfected counterparts. Second, although all CBP/P300 TI distributions are supershifted in the transfected populations, a predominant proportion (approximately 75%) of this distribution still remains within the normal CBP/P300 TI distribution range calculated for the respective untransfected population.
FIG. 3.
Determination of total HAT expression in GFP-CBP- and HA-P300-expressing cells. Shown are total CBP (A and B) and P300 (C and D) expression levels in GFP-CBP-expressing HeLa cells and HA-P300-expressing 10T1/2 cells, respectively. (A and C) Representative images used in the calculation. (B and D) Box-and-whisker plots depict the distribution ranges for three separate experiments examining the TI distributions of both untransfected and transfected populations for GFP-CBP- and HA-P300-expressing cells, respectively. Experimental pairs of untransfected and transfected populations are identified by the number 1, 2, or 3 in parentheses. Whiskers delineate the entire TI distribution, while the lower and upper horizontal lines of each box indicate the 25th and 75th percentiles, respectively. The middle horizontal line represents the mean calculated for each population. Bars, 15 μm.
To determine the mean fold increase in CBP/P300 expression in GFP-CBP- or HA-P300-expressing cells, it was first necessary to discern if the differences in mean TI between untransfected and transfected populations were statistically significant. Unpaired t tests were performed to evaluate the differences between mean TI values for the untransfected and transfected populations (for both CBP and P300 expression) in each triplicate experiment (Table 1). In all cases, P values of <0.05 were calculated, demonstrating statistically significant differences between mean TI values of untransfected and transfected populations for both CBP and P300. Since total fluorescent signal intensities are proportional to protein expression levels, mean TI ratios for untransfected and transfected populations were calculated to quantify the mean fold overexpression. Cells transiently expressing GFP-CBP exhibited a mean fold increase of 2.89 to 3.50 in total CBP expression, while in HA-P300-expressing cells, the mean fold increase in P300 expression varied between 2.34 and 2.84 (Table 1). Although statistically significant increases in the mean TI were calculated, the majority of cells transiently expressing GFP-CBP or HA-P300 (approximately 75%) still exhibited total CBP and P300 expression levels within the normal distribution ranges of untransfected cells.
TABLE 1.
Total CBP and P300 expression levels in cells transiently expressing GFP-CBP and HA-P300
| Protein and expt | Populationa | No. of cells | Mean TIb (104) | SEM (103) | Pc | Ratiod |
|---|---|---|---|---|---|---|
| CBP | ||||||
| Trial 1 | Untransfected | 291 | 7.37 | 2.95 | <0.001 | 2.89 |
| Transfected | 78 | 21.3 | 10.0 | |||
| Trial 2 | Untransfected | 457 | 6.14 | 1.50 | <0.001 | 3.00 |
| Transfected | 118 | 18.4 | 7.20 | |||
| Trial 3 | Untransfected | 331 | 5.18 | 1.87 | <0.001 | 3.50 |
| Transfected | 97 | 18.1 | 5.86 | |||
| P300 | ||||||
| Trial 1 | Untransfected | 299 | 4.37 | 1.49 | <0.001 | 2.34 |
| Transfected | 47 | 10.2 | 7.78 | |||
| Trial 2 | Untransfected | 328 | 7.01 | 2.33 | <0.001 | 2.84 |
| Transfected | 42 | 19.9 | 15.0 | |||
| Trial 3 | Untransfected | 431 | 7.82 | 2.21 | <0.001 | 2.44 |
| 55 | 19.0 | 13.5 |
Populations comprised either untransfected control cells or cells transfected with GFP-CBP or HA-P300.
Calculated by averaging the total signal intensity for each nucleus in a population.
Unpaired t tests (two tailed) were performed to identify statistical differences between the mean TI values for the transfected versus untransfected populations. A P value of <0.05 is considered statistically significant.
The ratio between the mean TI for transfected cells and the mean TI for untransfected cells describes the mean fold overexpression in transfected cells.
Based on these findings, all subsequent analyses were restricted to cells exhibiting normal physiologic behavior. Cells exhibiting atypical protein distributions, abnormal cellular morphologies, or high levels of transiently expressed protein (GFP-CBP, HA-P300, or ASF-GFP) were manually eliminated from all subsequent analyses.
Transient HAT expression does not alter HDAC or other HAT expression levels.
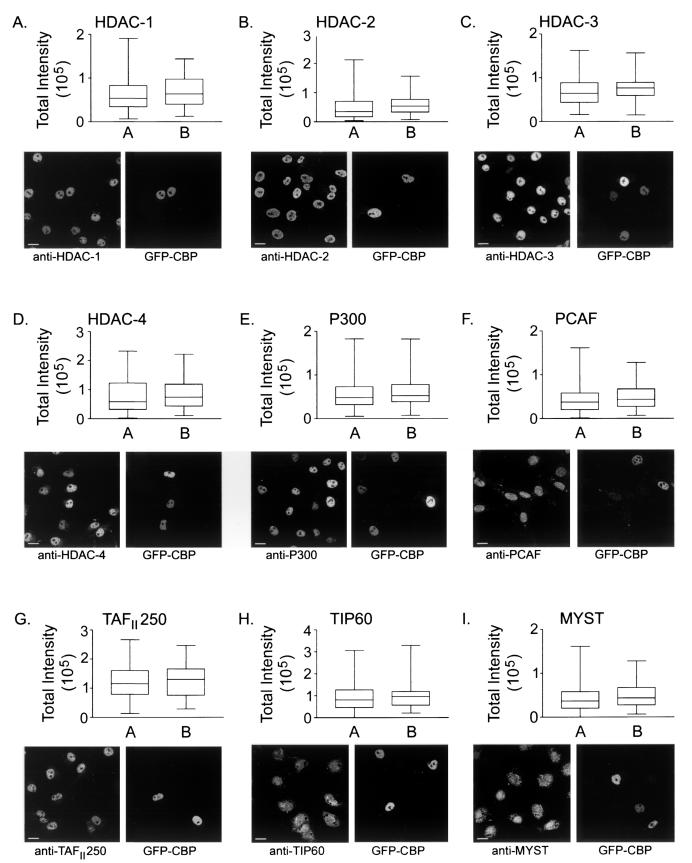
From the initial experiments, we were able to determine that minimal increases in CBP or P300 expression resulted in detectable increases in K5 acetylation. However, prior to examining subsequent acetylated epitopes, it was necessary to demonstrate that additional HAT and HDAC genes were not induced. Moreover, both P300 and CBP have been found in protein complexes that can include additional HATs such as PCAF. Conceivably, if p300 and/or CBP is required to form complexes with other nuclear proteins in order to function as a HAT, we would anticipate that those proteins would be upregulated in order to generate the increased cellular HAT activity we observed. Furthermore, CBP and p300 are also transcriptional coactivators that could indirectly alter steady-state acetylation levels by inducing endogenous HAT and/or HDAC expression. As a result, efforts were first made to minimize or optimize the time between transfection and fixation so as to minimize the coactivator potential of CBP/P300 while still maintaining appropriate protein expression levels for performing the HAT assay. Accordingly, we investigated the expression levels of several critical nuclear HATs and HDACs in cells transiently expressing GFP-CBP or HA-P300 and compared them to those in untransfected controls.
Using the same procedure that we applied to quantify increases in total HAT expression, we employed a panel of antibodies to quantify the expression levels of HDACs 1, 2, 3, and 4 and of the HATs CBP, P300, PCAF, TAFII250, and TIP60 and the MYST family of HATs including ESA1, TIP60, MOZ, MOF, SAS2, and SAS3/YBF. Figure 4 displays a representative image for each HDAC (Fig. 4A to D) or HAT (Fig. 4E to I) investigated in GFP-CBP-expressing cells and a box-and-whisker-plot describing the TI distributions for each HDAC or HAT for both untransfected and transfected populations. Particularly notable among all experiments is the expression level variability that is normally observed in all interphase nuclei. All HDACs and HATs investigated exhibited considerable heterogeneity in the amounts of total protein expressed. Importantly, the ranges characterized for GFP-CBP- and HA-P300-expressing cells generally fall within the larger range of the untransfected populations. Unpaired t tests were performed to identify potential differences in mean TI values between the transfected and untransfected populations. The results for GFP-CBP- and HA-P300-expressing cells are shown in Tables 2 and 3, respectively. No statistically significant differences (i.e., P values of <0.05) between the mean TI values of the untransfected and transfected populations were observed in cells expressing either GFP-CBP or HA-P300. Since the cross-reactivity of anti-CBP and anti-P300 antibodies is often a concern, it was necessary to demonstrate that neither cross-reacts with the opposite HAT. The specificity of each antibody is demonstrated by the fact that no statistically significant increases in expression levels were evident when GFP-CBP-expressing cells were stained with anti-P300 (P = 0.12 [Table 2]) or when HA-P300-expressing cells were stained with anti-CBP (P = 0.21 [Table 3]).
FIG. 4.
Determination of HDAC and HAT expression levels in GFP-CBP-expressing cells. Distribution ranges for cells transiently expressing GFP-CBP were determined for the HDACs and HATs given at the top of each panel. Each panel includes a box-and-whisker plot defining the TI distribution range (whiskers), the 25th and 75th percentiles (bottom and top lines of the box, respectively), and the mean TI (middle horizontal bar) for the untransfected (A) and transfected (B) populations. Representative images are included for reference purposes. Bars, 15 μm. Note that in panel F (PCAF), 10T1/2 cells were substituted for COS-7 cells because of the species specificity of the anti-PCAF antibody.
TABLE 2.
Calculation of total HDAC and HAT expression levels in GFP-CBP-expressing COS-7 cells
| Protein | Populationa | No. of cells | Mean TIb (104) | SEM (103) | Pc |
|---|---|---|---|---|---|
| HDAC-1 | Untransfected | 320 | 6.34 | 2.30 | 0.25 |
| Transfected | 42 | 7.10 | 5.69 | ||
| HDAC-2 | Untransfected | 320 | 4.87 | 2.31 | 0.13 |
| Transfected | 42 | 5.88 | 5.49 | ||
| HDAC-3 | Untransfected | 184 | 6.98 | 2.44 | 0.25 |
| Transfected | 48 | 7.59 | 4.46 | ||
| HDAC-4 | Untransfected | 180 | 7.61 | 4.20 | 0.32 |
| Transfected | 42 | 8.55 | 7.79 | ||
| P300 | Untransfected | 167 | 5.31 | 2.43 | 0.12 |
| Transfected | 55 | 6.05 | 4.03 | ||
| PCAFd | Untransfected | 204 | 4.31 | 2.01 | 0.19 |
| Transfected | 42 | 4.96 | 4.47 | ||
| TAFII250 | Untransfected | 100 | 5.94 | 2.68 | 0.30 |
| Transfected | 54 | 6.42 | 3.66 | ||
| MYSTe | Untransfected | 207 | 4.25 | 2.01 | 0.15 |
| Transfected | 42 | 4.96 | 4.47 | ||
| TIP60 | Untransfected | 124 | 9.00 | 5.26 | 0.29 |
| Transfected | 38 | 10.2 | 9.86 |
Either untransfected control cells or cells transfected with GFP-CBP.
Calculated by averaging the total signal intensity for each nucleus in a population.
Unpaired t tests (two tailed) were performed to identify statistical differences between the mean TI values for the transfected versus untransfected populations. A P value of <0.05 is considered statistically significant.
10T1/2 cells were substituted for COS-7 cells due to the specificity of the anti-PCAF antibody for murine CBP.
The MYST antibody collectively recognizes six members of the MYST HAT family including ESA1, TIP60, MOZ, MOF, SAS2, and SAS3/YBE.
TABLE 3.
Calculation of total HDAC and HAT expression levels in HA-P300-expressing HeLa cells
| Protein | Populationa | No. of cells | Mean TIb (106) | SEM (104) | P valuec |
|---|---|---|---|---|---|
| HDAC-1 | Untransfected | 255 | 2.50 | 9.13 | 0.15 |
| Transfected | 63 | 2.79 | 14.4 | ||
| HDAC-2 | Untransfected | 232 | 1.92 | 6.93 | 0.15 |
| Transfected | 40 | 2.18 | 15.8 | ||
| HDAC-3 | Untransfected | 202 | 2.43 | 8.44 | 0.18 |
| Transfected | 45 | 2.68 | 15.1 | ||
| HDAC-4 | Untransfected | 234 | 3.21 | 10.7 | 0.15 |
| Transfected | 64 | 3.53 | 18.4 | ||
| CBP | Untransfected | 180 | 2.20 | 13.8 | 0.21 |
| Transfected | 47 | 2.57 | 23.2 | ||
| PCAF | Untransfected | 328 | 2.03 | 6.35 | 0.15 |
| Transfected | 47 | 2.28 | 17.6 | ||
| TAFII250 | Untransfected | 239 | 2.11 | 7.97 | 0.44 |
| Transfected | 68 | 2.23 | 14.5 | ||
| MYSTd | Untransfected | 338 | 2.17 | 6.94 | 0.13 |
| Transfected | 52 | 2.46 | 14.8 |
Either untransfected control cells or cells transfected with HA-P300.
Calculated by averaging the total signal intensity for each nucleus in a population.
Unpaired t tests (two tailed) were performed to identify statistical differences between the mean TI values for the transfected versus untransfected populations. A P value of <0.05 is considered statistically significant.
The MYST antibody collectively recognizes six members of the MYST HAT family including ESA1, TIP60, MOZ, MOF, SAS2, and SAS3/YBF.
Transient and minimal increases in HAT expression correlate with acetylation increases.
By employing criteria that allow the exclusion of cells significantly overexpressing the transfected protein and/or cells that have altered cellular or nuclear distributions of the transfected protein, we were able to accurately assay for CBP and P300 HAT activities. To determine the in vivo substrate specificity preferences of CBP and P300, cells were transiently transfected with GFP-CBP, HA-P300, or ASF-GFP and immunofluorescently stained for specific acetylated histone epitopes. Images were collected, and fluorescent intensity values were extracted for the transfected protein and each specific acetylated epitope. A panel comprising nine different sequence-specific anti-acetylation antibodies was used and is listed in Table 4. ASF-GFP serves as a negative control, because it localizes to the nucleus (22). ASF-GFP does not have, nor does it recruit, HAT activity.
TABLE 4.
Anti-acetylation antibody panel
| Acetylated epitopea (antibody designation) | Histone | Antibody dilution | Company |
|---|---|---|---|
| K5 (K5) | H4 | 1:200 | Upstate |
| K8 (K8) | H4 | 1:8,000 | Upstate |
| K5, K8, K12, K16 (AcH4) | H4 | 1:15,000 | Upstate |
| K9 (K9) | H3 | 1:400 | Upstate |
| K14 (K14) | H3 | 1:200 | Upstate |
| K12 (K12) | H4 | 1:2,000 | Serotec |
| K16 (K16) | H4 | 1:100 | Serotec |
| K9, K14 (Ac3) | H3 | 1:500 | Serotec |
| K5, K12, K15, K20 (H2B) | H2B | 1:800 | Serotec |
Each number represents the acetylated lysine (K) residue contained within the N-terminal tail of the indicated histone that is recognized by the respective antibody. Antibody designations in parentheses indicate the nomenelature used in the text to describe the acetylated epitope.
Eighteen to 20 h posttransfection, approximately 5 to 60% of COS-7, HeLa, IM, or 10T1/2 cells expressed the protein of interest (data not shown). Figure 1 is a representative image used for the analysis and specifically depicts the relationship between GFP-CBP, HA-P300, and ASF-GFP expression and acetylation at K5 of histone H4. Note that the images for this data set were prepared to illustrate the range of staining for each trial (i.e., GFP-CBP, HA-P300, or ASF-GFP). The apparently higher steady-state acetylation in the ASF-GFP panels is actually equal in intensity to that in the weakly staining nontransfected cells of the GFP-CBP and HA-P300 panels. Figure 1 shows that increases in K5 acetylation were observed for cells expressing GFP-CBP or HA-P300, but not ASF-GFP, relative to untransfected controls. For all cell lines investigated, increases in acetylation signal intensities were most obvious for K14 and K5, and smaller apparent increases were observed for H2B, Ac3, and AcH4 (data not shown). Not surprisingly, no acetylation increases for any epitope were observed in ASF-GFP-expressing cells.
GFP-CBP and HA-P300 alter the steady-state levels of specific acetylated species of histones.
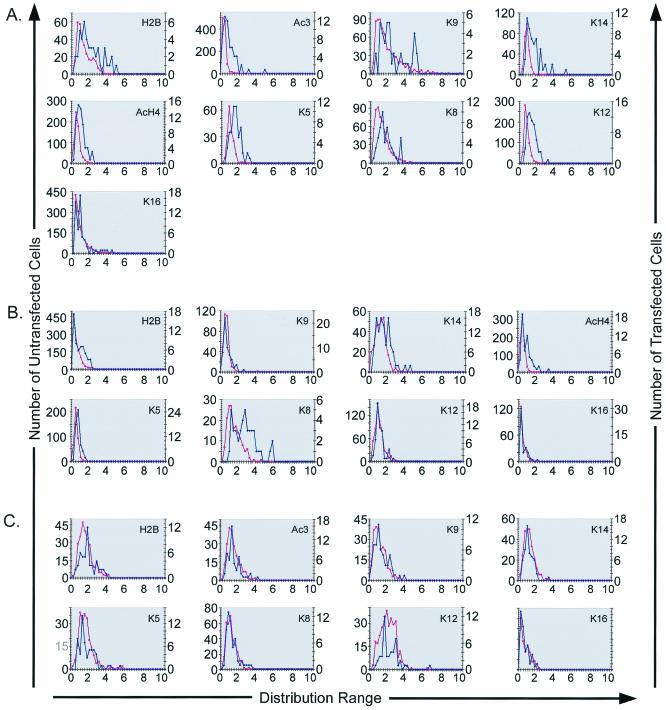
Histograms comparing N-Ac (see Materials and Methods) levels for three separate cell lines were generated to quantify the differences observed between transfected and untransfected cells. The results obtained with COS-7 cells are shown in Fig. 5 and are consistent with those obtained with the other cell lines (data not shown). Distribution increases for the transfected versus the untransfected population were expected to occur only if the acetylation levels are dependent on GFP-CBP or HA-P300 expression and HAT activity. Histone acetylation was found to be increased on several lysines when GFP-CBP- or HA-P300-transfected cells were compared to their untransfected counterparts (Fig. 5A and B). In contrast, no significant relationship was observed between ASF-GFP expression and specific histone acetylations (Fig. 5C). In cells transfected with GFP-CBP, increases in N-Ac distributions were found for H2B, Ac3 (H3), K14 (H3), AcH4 (H4), K5 (H4), and K12 (H4) but not for K9 (H3), K8 (H4), or K16 (H4) (Fig. 5A). In HA-P300-expressing cells, N-Ac distribution increases were found for H2B, K14, AcH4, K5, and K8 but not for K9, K12, or K16 (Fig. 5B). These results indicate that both CBP and P300 expression may induce specific changes in global nuclear histone acetylation levels.
FIG. 5.
Distribution of N-Ac intensities. Histograms comparing the N-Ac distributions for COS-7 cells transfected with GFP-CBP (A), HA-P300 (B), or ASF-GFP (C) with those for untransfected cells are shown. The histograms generated are representative of the results obtained for the IM and HeLa cell lines. The first y axis represents the number of untransfected cells (pink), the second y axis represents the number of transfected cells (blue), and the x axis represents the distribution range for the N-Ac intensities for the acetylated epitope indicated in the top right corner of each graph.
Transient HAT expression correlates with increased histone acetylation.
The results from the preceding sections demonstrate that CBP and P300 can induce specific acetylation increases that are independent of the upregulation of additional HATs or downregulation of common HDACs. However, it is not apparent whether the observed result is direct or indirect. To further examine the relationship between CBP or P300 expression and the amount of histone acetylation, scatter plots were generated comparing GFP-CBP or HA-P300 fluorescence with acetylated histone fluorescence (Fig. 6A and B, respectively).
FIG. 6.
Relationship between CBP and P300 expression and acetylation. Scatter plots were generated for cells expressing GFP-CBP (A), HA-P300 (B), or ASF-GFP (C) to identify potential relationships between protein expression and N-Ac intensities. Linear regression analysis was performed, and R2 values were calculated and are given for each epitope investigated. Larger R2 values identify instances where N-Ac intensities are dependent on protein expression levels.
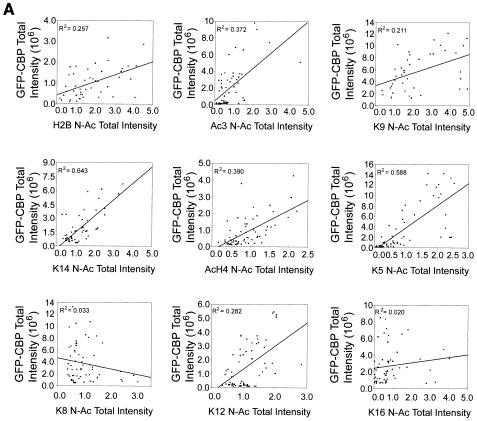
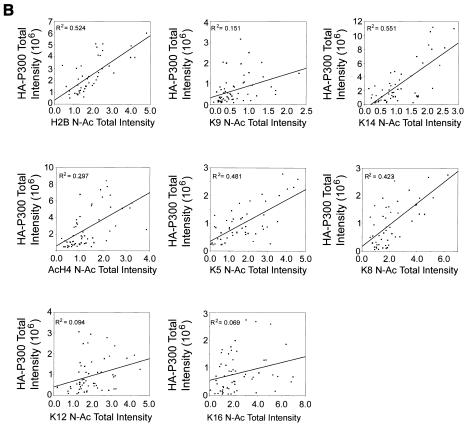
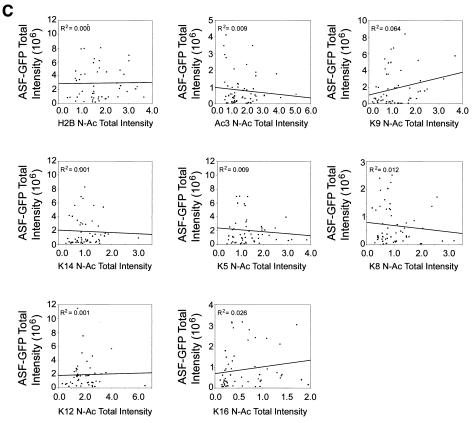
As expected, no significant relationship between ASF-GFP expression and N-Ac levels was observed (Fig. 6C). In contrast, positive linear relationships between GFP-CBP or HA-P300 expression and N-Ac levels were observed for several specific lysine residues. Figure 6 graphically depicts the results obtained from COS-7 cells. Similar results were obtained with the HeLa and IM cell lines (data not shown). Strong positive correlations were observed for H2B, Ac3, K9, K14, AcH4, K5, and K12 in GFP-CBP-expressing cells, while in HA-P300-expressing cells, strong positive correlations were observed for H2B, K9, K14, AcH4, K5, and K8. With the HeLa and IM cell lines, Ac3 was also observed to exhibit a strong positive relationship in cells expressing HA-P300 (data not shown). Therefore, under the conditions of our single-cell-based assay, the observed increases in global acetylation levels are dependent on the amount of CBP or P300 expressed.
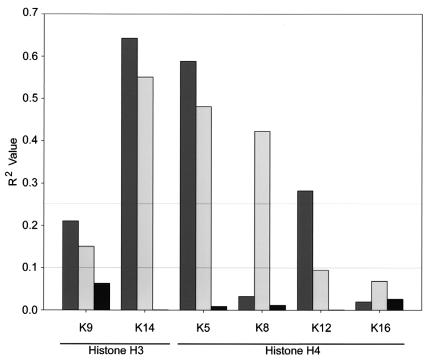
Because there was an apparent correlation between the CBP or P300 expression level and N-Ac values for several acetylated epitopes, linear regression analysis was performed to describe the correlation. An R value of 0 signifies no correlation between HAT expression and N-Ac values, whereas values of +1 or −1 identify perfect positive and negative correlations, respectively. By squaring the R value (R2), a better understanding of correlation is achieved. Linear regression analysis was performed for COS-7, HeLa, and IM cells, and the calculated R2 values for COS-7 cells are presented in each scatter plot in Fig. 6. Figure 7 graphically depicts the R2 values calculated for each acetylated epitope of histone H3 and H4. While both proteins were closely correlated with K14 acetylation of histone H3 and K5 acetylation of histone H4, CBP expression was closely correlated with K12 acetylation but not with K8 acetylation, whereas P300 exhibited the opposite relationship. Variability in R2 values was observed and can be roughly grouped into three distinct populations: (i) those for which R2 was less than 0.100, (ii) those for which R2 was between 0.100 and 0.250, and (iii) those for which R2 was greater than 0.250. For GFP-CBP, the R2 values for only two epitopes, K8 and K16, fell below 0.100, while for all the remaining epitopes except K9 (R2 = 0.211), the R2 values were greater than 0.250. For HA-P300-expressing cells, only the R2 values for K12 and K16 were less than 0.100, while those for H2B, K14, AcH4, K5, and K8 were greater than 0.250 and that for K9 was 0.151.
FIG. 7.
Relationships between HAT expression and acetylation of specific lysine residues. R2 values calculated for the correlation between GFP-CBP (dark shaded bars), HA-P300 (light shaded bars), or ASF-GFP (solid bars) and specific acetylated lysine residues (indicated on the x axis) contained in histones H3 and H4 are graphically represented. Horizontal lines mark R2 values of 0.100 and 0.250 for reference purposes. See the text for details.
Transient HAT expression induces acetylation at specific histone residues.
Since it was anticipated that increases in bulk acetylation would occur in cells where a specificity preference was demonstrated, unpaired t tests (two tailed) were used to compare mean N-Ac values for transfected and untransfected populations. Table 5 summarizes the results calculated for COS-7 cells transiently expressing GFP-CBP, HA-P300, or ASF-GFP. Additional experiments were conducted in the HeLa and IM cell lines with very similar results (data not shown). Recall that only cells minimally expressing the protein of interest were included in the analysis. GFP-CBP-expressing COS-7 cells exhibited statistical significance for a number of acetylated epitopes including H2B, Ac3, K14, AcH4, K5, and K12 but not for K9, K8, or K16. HA-P300-expressing cells exhibited statistical significance for H2B, K14, AcH4, K5, and K8 but did not exhibit significance for K9, K12, or K16. The P value calculated for the K9 epitope in the COS-7 HA-P300 trial fell just outside the cutoff value (P < 0.05) at 0.06. Subsequent t tests performed for HeLa and IM cells revealed results similar to those outlined above (data not shown): no statistical significance was calculated for K9, K8, or K16 in GFP-CBP-expressing cells or for K9, K12, or K16 in HA-P300-expressing cells. Together, these results strongly corroborate those detailed in the two preceding sections. By comparing the results generated from histograms, regression analysis, and t tests for all three cell lines, we ordered (from highest to lowest) the acetylation site specificity changes induced by GFP-CBP (K14, K5, K12, and H2B) and HA-P300 (K14, H2B, K5, and K8). Interestingly, although GFP-CBP and HA-P300 induce different changes in steady-state histone acetylation, three of the top four acetylated species (K14, K5, and H2B) are altered by both proteins, with the final substrate residues, K12 or K8, representing unique targets for GFP-CBP and HA-P300, respectively.
TABLE 5.
Calculations for the difference in mean N-Ac intensities for both transfected and untransfected GFP-CBP, HA-P300, and ASF-GFP populations
| Transfected protein | Antibody | Histone | Cells transfected | No. of cells | Mean N-Ac intensitya | SD | P |
|---|---|---|---|---|---|---|---|
| GFP-CBP | H2B | H2B | + | 56 | 0.7253 | 0.5518 | <0.0001 |
| − | 1,110 | 0.4779 | 0.4660 | ||||
| Ac3 | H3 | + | 58 | 0.8406 | 0.7604 | <0.0001 | |
| − | 1,111 | 0.3128 | 0.2452 | ||||
| K9 | H3 | + | 55 | 0.4212 | 0.3041 | 0.1159 | |
| − | 657 | 0.3504 | 0.3218 | ||||
| K14 | H3 | + | 54 | 1.3152 | 0.5448 | <0.0001 | |
| − | 174 | 0.7429 | 0.3138 | ||||
| AcH4 | H4 | + | 58 | 1.3168 | 0.8019 | <0.0001 | |
| − | 714 | 0.6266 | 0.4563 | ||||
| K5 | H4 | + | 64 | 0.8724 | 0.7799 | <0.0001 | |
| − | 680 | 0.2260 | 0.1762 | ||||
| K8 | H4 | + | 49 | 1.6284 | 1.1293 | 0.5633 | |
| − | 219 | 1.7391 | 1.2277 | ||||
| K12 | H4 | + | 52 | 1.4327 | 0.7891 | 0.0006 | |
| − | 548 | 1.0440 | 0.7728 | ||||
| K16 | H4 | + | 58 | 0.8548 | 0.9250 | 0.3053 | |
| − | 1,406 | 0.7436 | 0.8044 | ||||
| HA-P300 | H2B | H2B | + | 58 | 1.7423 | 1.4153 | 0.0032 |
| − | 211 | 1.2667 | 0.9650 | ||||
| K9 | H3 | + | 58 | 0.6341 | 0.4576 | 0.0612 | |
| − | 327 | 0.5390 | 0.3342 | ||||
| K14 | H3 | + | 56 | 1.4416 | 0.7895 | <0.0001 | |
| − | 302 | 1.0685 | 0.5528 | ||||
| AcH4 | H4 | + | 52 | 0.3951 | 0.3808 | 0.0271 | |
| − | 210 | 0.2937 | 0.2697 | ||||
| K5 | H4 | + | 49 | 1.7152 | 1.2147 | <0.0001 | |
| − | 273 | 0.9958 | 0.9285 | ||||
| K8 | H4 | + | 54 | 1.3439 | 0.8745 | <0.0001 | |
| − | 247 | 0.6052 | 0.2436 | ||||
| K12 | H4 | + | 49 | 2.1554 | 1.3265 | 0.4543 | |
| − | 221 | 2.3570 | 1.7754 | ||||
| K16 | H4 | + | 57 | 2.3785 | 1.7211 | 0.1913 | |
| − | 242 | 2.7992 | 2.2755 | ||||
| ASF-GFP | H2B | H2B | + | 51 | 1.7145 | 0.7998 | 0.0527 |
| − | 274 | 1.4940 | 0.7327 | ||||
| Ac3 | H3 | + | 55 | 1.3870 | 0.7336 | 0.2753 | |
| − | 222 | 1.2790 | 1.3870 | ||||
| K9 | H3 | + | 54 | 1.1687 | 0.7666 | 0.1943 | |
| − | 224 | 1.0369 | 0.6430 | ||||
| K14 | H3 | + | 55 | 1.0264 | 0.4807 | 0.5536 | |
| − | 259 | 1.0722 | 0.5293 | ||||
| K5 | H4 | + | 59 | 1.6588 | 0.9061 | 0.2021 | |
| − | 220 | 1.5051 | 0.7956 | ||||
| K8 | H4 | + | 55 | 1.0354 | 0.6548 | 0.1778 | |
| − | 323 | 0.9260 | 0.5370 | ||||
| K12 | H4 | + | 48 | 1.9610 | 1.0373 | 0.1747 | |
| − | 309 | 1.7756 | 0.8518 | ||||
| K16 | H4 | + | 57 | 0.5930 | 0.4817 | 0.6581 | |
| − | 311 | 0.6239 | 0.4845 |
Calculated for either the untransfected or the transfected cell population as the mean ratio of Cy3 intensity to DAPI intensity.
DISCUSSION
The HAT assay detailed here is conceptually similar in design to standard reporter gene assays. Transfection of cells with a construct encoding an enzymatic activity, such as luciferase, results in cellular production of an enzymatic activity that can be quantitatively correlated with a separate activity, such as DNA binding or transcription. An important distinction and improvement over traditional transfection assays is that the HAT assay employs additional selection criteria to maximize the representation of normal physiologic behavior. The major advantages of our approach are that (i) each cell used in the final measurement contributes equally and (ii) criteria can be employed to specifically eliminate the higher overexpressors from the analysis. In conventional biochemical approaches, large numbers of cells are pooled to make a single measurement. As a result, the contribution of a single cell within the total population is not equal, but rather directly proportional, to its relative expression level. By specifically eliminating the highest-expressing cells and those most likely to generate expression-related artifacts, this assay is likely to be a more reliable measure of physiologically relevant activities.
The histone tails are presented in a much different manner in vivo than they are in vitro. In vivo, histone N termini associate with DNA and adjacent nucleosomes contained within the same fiber or an adjacent fiber to facilitate further chromatin compaction states (14-16). The relative absence of 10- and 30-nm chromatin fibers in vivo (10, 19, 40) implies that the substrates typically utilized in vitro, namely, histone tails, purified histones, or reconstituted mononucleosomes, do not accurately recapitulate the endogenous counterparts that they are meant to recapitulate. In fact, the prevalence of higher-order chromatin (i.e., beyond the 30-nm fiber) implies that the HAT substrates may be much less accessible in vivo than in vitro. Consistent with the hypothesis that certain types of higher-order chromatin structure (i.e., at or beyond the 200-nm chromonema fiber level) may limit the accessibility of the histone N termini to HATs, nucleoplasmic regions exhibiting intense DAPI staining fail to incorporate any of the acetylations associated with transient CBP or P300 expression. The in vivo HAT assay, therefore, presents the substrate in a natural conformation and relies on statistical analyses to determine if there is a relationship between a specific HAT (i.e., CBP or P300) and a specific acetylated lysine residue contained within the histone N termini. Because we observe significant increases in global acetylation for several histone residues, we can conclude that either (i) CBP and P300 do not require additional activities (i.e., cofactors) to acetylate the endogenous chromatin or (ii) such cofactors are not limiting relative to the endogenous levels of CBP and P300.
In this study, we have developed a novel imaging approach to characterize CBP- and P300-dependent histone acetylation. Most importantly, the assay is performed under physiologically relevant conditions and uses native chromatin structure as the substrate. The complexity of the physiological in vivo environment inherent in our quantitative HAT assay does not allow us to conclusively state that GFP-CBP and HA-P300 HAT activities directly regulate the global acetylation levels of specific subsets of histone lysine residues. For example, although we demonstrated that GFP-CBP and HA-P300 expression did not alter the expression levels of many additional HATs and HDACs, we cannot exclude the possibility that their expression may alter the enzymatic activity of other HATs or HDACs. While this remains possible, the striking similarity between the utilization of specific lysines by P300 in vitro (27) and the changes in acetylation induced when P300 is transiently expressed in vivo and its distinction from other in vitro HAT activities (26, 32, 36) are more consistent with the notion that the HAT activity inherent to the transfected enzymes is responsible for the histone acetylations we observe. Furthermore, the short posttransfection period prior to fixation and analysis limits the potential for translation-dependent events from CBP- or P300-regulated genes to impact on the observed acetylation pattern. Finally, by employing viability, morphological, and expression criteria, we confine our analysis to cells that are expressing very close to physiological levels, further minimizing the limitations of conventional transfection-based assays. Therefore, although we cannot conclude with certainty that the HAT activity inherent in the expressed GFP-CBP or HA-P300 is directly responsible for the changes in histone acetylation that we observe, we can conclude that the changes in acetylation that we observe upon transfection are directly dependent on CBP and P300 expression levels. Hence, either CBP and P300 are directly responsible for these acetylations or they are responsible indirectly, by regulating the activity of HATs or HDACs without a requirement for increasing or decreasing their protein levels.
In the past, CBP and P300 have largely been considered functionally redundant proteins. The overlapping changes in histone acetylation, specifically at K5 and K14, may account for the observed functional redundancies. More recently, however, functional differences between CBP and P300 have been described (20, 23, 24, 37, 44). For example, genetic studies involving CBP or P300 hemizygous or knockout mice clearly demonstrate that one HAT cannot simply substitute for the other (23, 28, 29, 33, 34, 41, 43). The observation of distinct phenotypes associated with P300 and CBP knockout mice in embryogenesis and development further underscores the disparate roles for each. Studies involving antisense oligodeoxynucleotides and hammerhead ribozymes specific to either CBP or P300 mRNAs have also identified distinct roles for CBP and P300 in retinoic acid-induced differentiation, cell cycle exit, and apoptosis of embryonal carcinoma F9 cells (20, 37). Although it is clear that CBP and P300 have some overlapping and redundant functions, it is now equally apparent that they have distinct and separable functions. Currently, little or no evidence is available to adequately account for the underlying functional differences observed between them. Our characterization of novel differences in the acetylation induced by GFP-CBP (K12) and HA-P300 (K8) represents the first differences demonstrated between these two HATs which may account for the observed functional differences.
In conclusion, we have developed a novel in vivo HAT assay and used it to characterize the HAT specificity preferences of CBP and P300. This approach couples the spatial resolving power of a laser scanning confocal microscope and the quantitative analytical power of basic statistical approaches. This approach overcomes conventional biochemical limitations by enabling cells to be analyzed either on an individual basis or on a population basis. Because it is performed under normal physiologic conditions and on endogenous substrates, the conformational differences of histone tails that exist between different substrates in vitro (i.e., purified histone tails, mononucleosomes, or short polynucleosome arrays) are avoided. We believe that this methodology offers many advantages over conventional biochemical approaches and should be generally amenable to the determination of any substrate specificity with minimal modifications.
Acknowledgments
We thank Darin McDonald and Roseline Godbout for critical reading of the manuscript. We also thank E. Verdin (University of California, San Francisco) and X. Yang (McGill University) for the generous gifts of anti-HDACs 3 and 4 and anti-PCAF, respectively.
We thank the Canadian Institutes of Health Research (CIHR) and the Alberta Heritage Foundation for Medical Research (AHFMR) for operational funding support. During the course of this study K.J.M. was supported by studentships from the Natural Sciences and Engineering Research Council of Canada, CIHR, and AHFMR and by the Department of Oncology endowed PhD studentship. M.J.H. is supported by scholarships from the CIHR and AHFMR.
REFERENCES
- 1.Annunziato, A. T., and J. C. Hansen. 2000. Role of histone acetylation in the assembly and modulation of chromatin structures. Gene Expr. 9:37-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausio, J., D. W. Abbott, X. Wang, and S. C. Moore. 2001. Histone variants and histone modifications: a structural perspective. Biochem. Cell Biol. 79:693-708. [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 5.Blobel, G. A. 2000. CREB-binding protein and p300: molecular integrators of hematopoietic transcription. Blood 95:745-755. [PubMed] [Google Scholar]
- 6.Boisvert, F. M., M. J. Kruhlak, A. K. Box, M. J. Hendzel, and D. P. Bazett-Jones. 2001. The transcription coactivator CBP is a dynamic component of the promyelocytic leukemia nuclear body. J. Cell Biol. 152:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 8.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 9.Chrivia, J. C., R. P. Kwok, N. Lamb, M. Hagiwara, M. R. Montminy, and R. H. Goodman. 1993. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature 365:855-859. [DOI] [PubMed] [Google Scholar]
- 10.Georgel, P. T. 2002. Chromatin structure of eukaryotic promoters: a changing perspective. Biochem. Cell Biol. 80:295-300. [DOI] [PubMed] [Google Scholar]
- 11.Giles, R. H., D. J. Peters, and M. H. Breuning. 1998. Conjunction dysfunction: CBP/p300 in human disease. Trends Genet. 14:178-183. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 13.Gunjan, A., D. B. Sittman, and D. T. Brown. 2001. Core histone acetylation is regulated by linker histone stoichiometry in vivo. J. Biol. Chem. 276:3635-3640. [DOI] [PubMed] [Google Scholar]
- 14.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361-392. [DOI] [PubMed] [Google Scholar]
- 15.Hayes, J. J. 2002. Changing chromatin from the inside. Nat. Struct. Biol. 9:161-163. [DOI] [PubMed] [Google Scholar]
- 16.Hayes, J. J., and J. C. Hansen. 2001. Nucleosomes and the chromatin fiber. Curr. Opin. Genet. Dev. 11:124-129. [DOI] [PubMed] [Google Scholar]
- 17.Hendzel, M. J., M. J. Kruhlak, and D. P. Bazett-Jones. 1998. Organization of highly acetylated chromatin around sites of heterogeneous nuclear RNA accumulation. Mol. Biol. Cell 9:2491-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendzel, M. J., M. J. Kruhlak, N. A. MacLean, F. Boisvert, M. A. Lever, and D. P. Bazett-Jones. 2001. Compartmentalization of regulatory proteins in the cell nucleus. J. Steroid Biochem. Mol. Biol. 76:9-21. [DOI] [PubMed] [Google Scholar]
- 19.Horn, P. J., and C. L. Peterson. 2001. The bromodomain: a regulator of ATP-dependent chromatin remodeling? Front. Biosci. 6:D1019-D1023. [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki, H., R. Eckner, T. P. Yao, K. Taira, R. Chiu, D. M. Livingston, and K. K. Yokoyama. 1998. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393:284-289. [DOI] [PubMed] [Google Scholar]
- 21.Kruhlak, M. J., M. J. Hendzel, W. Fischle, N. R. Bertos, S. Hameed, X. J. Yang, E. Verdin, and D. P. Bazett-Jones. 2001. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J. Biol. Chem. 276:38307-38319. [DOI] [PubMed] [Google Scholar]
- 22.Kruhlak, M. J., M. A. Lever, W. Fischle, E. Verdin, D. P. Bazett-Jones, and M. J. Hendzel. 2000. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J. Cell Biol. 150:41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kung, A. L., V. I. Rebel, R. T. Bronson, L. E. Ch'ng, C. A. Sieff, D. M. Livingston, and T. P. Yao. 2000. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by CBP. Genes Dev. 14:272-277. [PMC free article] [PubMed] [Google Scholar]
- 24.Lipinski, K. S., P. Fax, B. Wilker, H. Hennemann, D. Brockmann, and H. Esche. 1999. Differences in the interactions of oncogenic adenovirus 12 early region 1A and nononcogenic adenovirus 2 early region 1A with the cellular coactivators p300 and CBP. Virology 255:94-105. [DOI] [PubMed] [Google Scholar]
- 25.McManus, K. J., and M. J. Hendzel. 2001. CBP, a transcriptional coactivator and acetyltransferase. Biochem. Cell Biol. 79:253-266. [PubMed] [Google Scholar]
- 26.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAFII250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 27.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 28.Oike, Y., A. Hata, T. Mamiya, T. Kaname, Y. Noda, M. Suzuki, H. Yasue, T. Nabeshima, K. Araki, and K. Yamamura. 1999. Truncated CBP protein leads to classical Rubinstein-Taybi syndrome phenotypes in mice: implications for a dominant-negative mechanism. Hum. Mol. Genet. 8:387-396. [DOI] [PubMed] [Google Scholar]
- 29.Oike, Y., N. Takakura, A. Hata, T. Kaname, M. Akizuki, Y. Yamaguchi, H. Yasue, K. Araki, K. Yamamura, and T. Suda. 1999. Mice homozygous for a truncated form of CREB-binding protein exhibit defects in hematopoiesis and vasculo-angiogenesis. Blood 93:2771-2779. [PubMed] [Google Scholar]
- 30.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189-1192. [DOI] [PubMed] [Google Scholar]
- 31.Sleeman, J., C. E. Lyon, M. Platani, J. P. Kreivi, and A. I. Lamond. 1998. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp. Cell Res. 243:290-304. [DOI] [PubMed] [Google Scholar]
- 32.Smith, E. R., A. Eisen, W. Gu, M. Sattah, A. Pannuti, J. Zhou, R. G. Cook, J. C. Lucchesi, and C. D. Allis. 1998. ESA1 is a histone acetyltransferase that is essential for growth in yeast. Proc. Natl. Acad. Sci. USA 95:3561-3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanaka, Y., I. Naruse, T. Hongo, M. Xu, T. Nakahata, T. Maekawa, and S. Ishii. 2000. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev. 95:133-145. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka, Y., I. Naruse, T. Maekawa, H. Masuya, T. Shiroishi, and S. Ishii. 1997. Abnormal skeletal patterning in embryos lacking a single Cbp allele: a partial similarity with Rubinstein-Taybi syndrome. Proc. Natl. Acad. Sci. USA 94:10215-10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thorne, A. W., D. Kmiciek, K. Mitchelson, P. Sautiere, and C. Crane-Robinson. 1990. Patterns of histone acetylation. Eur. J. Biochem. 193:701-713. [DOI] [PubMed] [Google Scholar]
- 36.Tse, C., E. I. Georgieva, A. B. Ruiz-Garcia, R. Sendra, and J. C. Hansen. 1998. Gcn5p, a transcription-related histone acetyltransferase, acetylates nucleosomes and folded nucleosomal arrays in the absence of other protein subunits. J. Biol. Chem. 273:32388-32392. [DOI] [PubMed] [Google Scholar]
- 37.Ugai, H., K. Uchida, H. Kawasaki, and K. K. Yokoyama. 1999. The coactivators p300 and CBP have different functions during the differentiation of F9 cells. J. Mol. Med. 77:481-494. [DOI] [PubMed] [Google Scholar]
- 38.Wang, X., C. He, S. C. Moore, and J. Ausio. 2001. Effects of histone acetylation on the solubility and folding of the chromatin fiber. J. Biol. Chem. 276:12764-12768. [DOI] [PubMed] [Google Scholar]
- 39.Wang, X., S. C. Moore, M. Laszckzak, and J. Ausio. 2000. Acetylation increases the alpha-helical content of the histone tails of the nucleosome. J. Biol. Chem. 275:35013-35020. [DOI] [PubMed] [Google Scholar]
- 40.Widom, J. 1989. Toward a unified model of chromatin folding. Annu. Rev. Biophys. Biophys. Chem. 18:365-395. [DOI] [PubMed] [Google Scholar]
- 41.Yamauchi, T., Y. Oike, J. Kamon, H. Waki, K. Komeda, A. Tsuchida, Y. Date, M. X. Li, H. Miki, Y. Akanuma, R. Nagai, S. Kimura, T. Saheki, M. Nakazato, T. Naitoh, K. Yamamura, and T. Kadowaki. 2002. Increased insulin sensitivity despite lipodystrophy in Crebbp heterozygous mice. Nat. Genet. 30:221-226. [DOI] [PubMed] [Google Scholar]
- 42.Yang, X. J., V. V. Ogryzko, J. Nishikawa, B. H. Howard, and Y. Nakatani. 1996. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature 382:319-324. [DOI] [PubMed] [Google Scholar]
- 43.Yao, T. P., S. P. Oh, M. Fuchs, N. D. Zhou, L. E. Ch'ng, D. Newsome, R. T. Bronson, E. Li, D. M. Livingston, and R. Eckner. 1998. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93:361-372. [DOI] [PubMed] [Google Scholar]
- 44.Yuan, Z. M., Y. Huang, T. Ishiko, S. Nakada, T. Utsugisawa, H. Shioya, Y. Utsugisawa, Y. Shi, R. Weichselbaum, and D. Kufe. 1999. Function for p300 and not CBP in the apoptotic response to DNA damage. Oncogene 18:5714-5717. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y., and D. Reinberg. 2001. Transcription regulation by histone methylation: interplay between different covalent modifications of the core histone tails. Genes Dev. 15:2343-2360. [DOI] [PubMed] [Google Scholar]