Abstract
As a promiscuous dimerization partner the retinoid X receptor (RXR) can contribute to signaling by multiple nuclear receptors. However, the impact of RXR cosignaling and the possible existence of an RXR homodimer signaling pathway are largely unexplored. We report here on the separation of RXR homo- and heterodimerization as an essential step towards the elucidation of the roles of RXR homo- and heterodimers in retinoid-rexinoid signaling. RXR homodimerization was specifically disrupted by single mutations in the RXR dimerization interface. In contrast, even multiple mutations did not fully impair RXR heterodimerization with retinoic acid receptor (RAR). Importantly, the mutation of mouse RXRα (mRXRα) Tyr402 substantially weakened RAR heterodimerization while concomitantly increasing homodimerization. Not only did this lead to cooperatively enhanced RXR homodimer binding to DR1 or DR5 elements, but unexpectedly, the mutant acquired significant binding efficiency for noncognate DR3 or DR4 elements as well. The increased stability of RXR homodimers on DR1 correlated with increased transcriptional activity of mRXRαY402A on DR1-based reporter genes. Weak, if any, heterodimerization was observed with thyroid, vitamin D3, or peroxisome proliferator-activating receptors. A model accounting for the structural impact of the Tyr402 mutation on dimerization is discussed. These results provide the basis for a genetic replacement of wild-type RXRs by mutants like mRXRαY402A to elucidate the physiological impact of RXR homo- and heterodimerization.
Three retinoid X receptors (RXRα, -β, and -γ), members of the nuclear hormone receptor superfamily, act as ligand-inducible transcription factors. RXRs are promiscuous dimerization partners for a large number of nuclear (orphan) receptors. Notably, RXRs are also able to activate transcription from cognate reporter genes as homodimers. The selectivity of the transcriptional response of RXR homo- and heterodimers is generally believed to be the consequence of regulation at multiple levels. These comprise (i) a receptor-specific DNA response element repertoire (however, this is highly degenerate for natural target genes and multiple receptors or dimers may share common response elements); (ii) the generation and/or availability of the cognate ligand(s); and (iii) the formation, dynamics, and recruitment of cellular coregulator complexes (for reviews see references 5, 14, 16, and 19 and references cited therein).
It is well established that rexinoid agonists (9-cis-retinoic acid or a synthetic rexinoid) can confer transcriptional activity on RXR homodimers, while RXR heterodimers are transcriptionally silent, unless the heterodimer partner of RXR is liganded. The mechanistic basis of this phenomenon, generally referred to as “RXR subordination” (15, 25-27), has been solved recently (12). Binding of agonists or certain antagonists by the dimerization partner leads to transcriptional synergy with rexinoid agonists (6, 24). Whether some seemingly “permissive” heterodimers, such as those with peroxisome proliferator-activating receptor (PPAR) or nerve growth factor IB, can transactivate in the presence of rexinoid agonists, or whether this permissiveness is due to low levels of endogenous ligands that synergize with the rexinoid remains still to be established.
Two types of dimerization functions mediate homo- and heterodimerization of RXR. One involves several surfaces in the DNA binding domain (DBD) that establish weak response element-specific interfaces with corresponding surfaces in the partner DBD. The second is a single strong dimerization function in the receptor ligand binding domains (LBDs) of both partners and differs between homo- and heterodimers, and to some extent between the partners of RXR. In vitro studies have shown that the RXR LBD forms homodimers with relatively low affinity (13) compared to its heterodimeric association with retinoic acid receptor (RAR) (7). While the response element repertoire of nuclear receptor homo- and heterodimers is dictated by DNA binding specificity and dimerization characteristics of the DBDs, the LBD essentially stabilizes the predetermined full-length receptor dimers on these elements by increasing the cooperativity of DNA binding (4, 17, 28, 29).
A comparison of the two types of dimerization interfaces reveals that the RXR DBD homodimer buries 400 Å2 of solvent-exposed area and requires the DNA scaffold to form (23). The three-dimensional structure of the RXR LBD homodimer (3, 4) reveals a much larger dimerization interface of 1,830 Å2, involving residues from helices H7 to H10 and loops L8-9 and L9-10. The residues at the interface organize as a hydrophobic cluster surrounded by charged and polar amino acids. Globally the same RXR structures and amino acids contribute to both the RXR homodimer and the RAR-RXR heterodimer interfaces (4).
The aim of the present study was to (i) identify RXR residues specific to RXR homo- and heterodimerization surfaces and (ii) understand the molecular basis of the higher heterodimer stability relative to RXR homodimers. We report that RXR homodimerization can be readily disrupted by single RXR mutations introduced into the homodimerization interface, while RAR-RXR heterodimerization could be diminished but not eliminated by single or multiple alanine substitutions. This study led to the identification of RXR mutants that exhibit enhanced homodimerization efficiency and simultaneous loss of heterodimerization ability.
MATERIALS AND METHODS
Mutant expression vectors.
RXR and VP16 fusion constructs were generated by PCR-assisted site-directed mutagenesis, with Deep Vent DNA polymerase (Biolabs) and the following plasmids as template: pSG5-mRXRα (20) for substitution of all residues at the dimerization interface and VP16-CAS for all VP16 fusion proteins. GAL-mRXRα mutants were obtained by subcloning the BamHI-BglII fragment from the pSG5-mRXRα constructs into BamHI and BglII sites of pG4MpolyII-mRXRα(DE). Sequences of all recombinants and of the PCR primers used for the various constructions are available on request. All constructions were verified by DNA sequencing. The expression levels and the sizes of all mutants were compared and verified by Western blotting (see also Fig. 2).
FIG.2.
Increase-gain of binding affinity of RXRY402A homodimers on DR1 (A), DR5 (B), DR4 (C), and DR3 (D) response elements and parallel loss-impairment of heterodimerization of RXRY402A with PPARγ (A), RARα (B), TRα (C), and VDR (D) as shown by EMSA. The indicated volume (microliters) of RXR proteins was incubated with or without 0.2 μl of in vitro-translated mPPARγ (A), 50 ng of purified histidine-tagged mRARαΔAB (B), 100 ng of purified full-length cTRα (C), or 0.2 μl of in vitro-translated hVDR (D) for 15 min at 4°C before 32P-labeled DR1, DR5, DR4, or DR3 oligonucleotide was added. The autoradiograms shown are representative of two independent experiments. Upper bands, homodimers; lower bands, heterodimers.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were done as described by Zechel et al. (28, 29). Recombinant proteins used for EMSAs were in vitro-translated (TNT T7 coupled transcription-translation system; Promega) mRXRα or mutants, human vitamin D3 receptor (hVDR), and mPPARγ; Escherichia coli-expressed and purified histidine-tagged mRARαΔAB (residues 81 to 462); and chicken thyroid receptor α (cTRα).
Cell culture and transfection.
Cos1 cells plated at a density of 105 cells per well of 24-well plates were transfected as described previously (2) by the calcium phosphate method. Precipitates contained 10 ng of GAL-mRXRα wild-type (wt) or mutant expression vectors; 50 ng of VP16-hRARα, VP16-cTRα, VP16-hVDR, and VP16-mPPARγ expression vectors; or, as indicated for VP16-mRXRα and VP16-mRXRαY402A, 100 ng of 17m-tk-Luc or (17m)x5-Glob-CAT reporter gene and 50 ng of cytomegalovirus-β-galactosidase (CMV-β-Gal; used as an internal control to normalize for variations in the transfection efficiency). The total quantity of DNA was adjusted at 1 μg with pBluescript. The luciferase and chloramphenicol acetyltransferase (CAT) activities were measured using Luc-lite and CAT enzyme-linked immunosorbent assay reagents from Packard and Roche, respectively, according to the manufacturer's recommendations.
Western blot analysis.
The expression of the various mutants for RXR was verified by immunoblotting using either in vitro-translated proteins or proteins transiently expressed in Cos1 cells. Protein were separated on a sodium dodecyl sulfate-10% polyacrylamide gel and transferred onto a nitrocellulose membrane. The membranes were blocked for 1 h at room temperature (RT) in 5% nonfat dry milk (Bio-Rad) containing phosphate-buffered saline and then incubated for 2 h at RT or overnight at 4°C with rabbit polyclonal antibody directed against RXR (Santa Cruz) in Tris-buffered saline (TBS)-0.1% Tween-5% bovine serum albumin. Following three washes in TBS-0.1% Tween, the secondary antibody, anti-rabbit-horseradish peroxidase (Amersham), was incubated with the membrane for 1 h at RT in TBS-0.1% Tween. The chemiluminescence reaction was performed according to the manufacturer's recommendations.
RESULTS
RXR mutants with altered homo- and heterodimerization functions.
To investigate the (differential) contribution of residues at the RXR dimerization surface(s) to homo- and heterodimerization, we used three-dimensional structure-guided (3, 4) alanine scanning mutagenesis (Table 1). The abilities of the various mutants to form homo- and/or heterodimers were analyzed by EMSAs, as the efficiency of dimer binding to DNA response elements correlates with the strength of dimerization (17, 28, 29). Remarkably, RXR homodimerization could be completely prevented by introducing single mutations in either helix H7 (K361, m2), loop L8-9 (N382, m5; D384, m6), helix H9 (E395, m7), or helix H10 (K422, m17; L424, m18; R426, m19) (Table 1). Also, double and triple mutants were found that exhibited completely dissociated EMSA patterns indicating disabled homo- but not heterodimerization (m8, m14, m15, and m20; Table 1 and Fig. 1A). Note that differential expression or stability of mutant RXR does not contribute to these results, as all mutants were similarly expressed (Fig. 1B and data not shown). Remarkably, none of the single or compound mutations completely eliminated RAR-RXR heterodimer formation. Rather, depending on the residue, a decreased efficiency of heterodimerization was observed (Table 1). The strongest effect was noted for the helix H9 mutation RXRY402A (m11; Table 1 and Fig. 2B, compare lanes 9 to 11 with lane 8).
TABLE 1.
Homo- and heterodimerization abilities of the RXR mutants as assayed by EMSAa
| Location | Mutant | Amino acid(s) mutated | Activity with dimerization partner
|
|
|---|---|---|---|---|
| RXR | RAR | |||
| H7 | m1 | E357A | 135 ± 15 | 102 ± 1 |
| m2 | K361A | <5 | 115 ± 13 | |
| Loop L7-8 | m3 | R363A | 90 ± 10 | 127 ± 10 |
| m4 | D364A | 70 ± 5 | 105 ± 7 | |
| Loop L8-9 | m5 | N382A | <5 | 119 ± 7 |
| m6 | D384A | <5 | 98 ± 8 | |
| H9 | m7 | E395A | <5 | 110 ± 5 |
| m8 | D384A/E395A | <5 | 102 ± 6 | |
| m9 | R398A | 160 ± 40 | 106 ± 25 | |
| m10 | E399A | 24 ± 4 | 136 ± 15 | |
| m11 | Y402A | 228 ± 15 | 23 ± 7 | |
| m12 | E406A | 30 ± 10 | 120 ± 15 | |
| m13 | E399A/Y402A/E406A | 230 ± 30 | 30 ± 5 | |
| m14 | R363A/D384A/R426A | <5 | 115 ± 13 | |
| m15 | R363A/E395A/R426A | <5 | 90 ± 8 | |
| H10 | m16 | F420A | >300 | 81 ± 21 |
| m17 | K422A | <5 | 105 ± 7 | |
| m18 | L424A | <5 | 125 ± 16 | |
| m19 | R426A | <5 | 70 ± 8 | |
| m20 | K422A/R426A | <5 | 68 ± 5 | |
| m21 | L427A | 8 ± 1 | 122 ± 8 | |
| m22 | R431A | 145 ± 15 | 117 ± 13 | |
| m23 | L435A | 15 ± 10 | 108 ± 15 | |
| Loop L10-11 | m24 | E439A | 68 ± 8 | 72 ± 5 |
| m25 | D364A/D384A/E399A/ Y402A/E406A/ R426A/E439A | 98 ± 3 | 43 ± 7 | |
After autoradiography, the EMSAs were quantified by densitometry and the results were expressed as the percentages of RXR mutant dimerization activities compared with the wtRXR activity, which was arbitrarily defined as 100%. The results are the means ± standard deviations of two to three independent experiments.
FIG. 1.
Altered dimerization in RXR mutants. (A) EMSA illustrating loss or gain of homodimerization potential of RXR mutants. While the homodimerization of mutants m3 and m24 on DR1 was unaffected or partially impaired, the mutants m8, m10, m12, m14, m15, and m20 had a total or near-total loss of homodimerization function. In contrast, the mutants m11 and m13 showed enhanced homodimerization on DR1 elements. Identical results were obtained in three independent experiments. (B) Western blot analysis of in vitro-translated wt and mutant RXRs. An 0.1-μl amount of in vitro-translated full-length RXRs was separated on a sodium dodecyl sulfate-10% polyacrylamide gel, transferred onto a nitrocellulose membrane, and immunoblotted with monoclonal mRXR 4RX3A2 antibodies. The blot shown is a representative selection of the RXR mutants included in this study; all other mutants are expressed at similar levels.
Interestingly, this RXR mutation not only decreased RAR heterodimerization efficiency but also strengthened homodimerization on both DR1 and DR5 response elements (Table 1, Fig. 1A, and Fig. 2A and B, compare lanes 4 to 6 with lanes 1 to 3). Note in this respect that RXR homodimer binding to DR5 elements is well established (18). In keeping with these results, the presence of Y402A in the compound mutant m13 not only compensated for the loss of DR1 binding efficiency by the mutations m10 and m12 but further enhanced RXR homodimerization efficiency (Fig. 1A and Table 1). Similarly to RXRY402A the triple mutant m13 displayed a substantially weakened heterodimerization function. The dominant homodimer-stabilizing effect of the Y402A mutation was most remarkable for the compound mutant m25 (Table 1). Although the mutant carries six mutations, each of which on its own destabilizes RXR homodimer binding to DR1 elements, the presence of alanine at position 402 fully compensates for these effects.
Taken together, these data indicate that, despite their globally very similar dimerization surfaces, the RXR residues creating this surface contribute very differently to (the stabilization of) homo- and heterodimerization interfaces.
RXRY402A homodimers exhibit an altered response element repertoire.
The RXR mutation Y402A impaired not only the heterodimerization with RAR but also that with TR, VDR, and PPARγ. In all cases RXRY402A efficiently formed homodimers on DR1-, DR3-, DR4-, and DR5-based response elements (Fig. 2). Indeed, the systematic comparison of receptor dose-dependent DNA binding of wt and Y402A mutant RXRs revealed that the mutant not only exhibited altered DNA binding efficiency but, moreover, acquired a distinct response element repertoire. Increased DR1-binding efficiency of the mutant is obvious from the observations that at least 1 μl of in vitro-translated RXR is required to detect binding (Fig. 2A, lane 2), while under identical conditions 0.5 μl of RXRY402A-expressing lysate suffices for maximal binding (lane 4). In the presence of PPARγ, RXRY402A did not form any significant amount of heterodimers (lanes 11 to 13), while heterodimers were the main species formed with the wt counterpart under identical conditions (lanes 8 to 10).
Similarly, RXRY402A homodimers bound to DR5 elements more efficiently than did RXR (Fig. 2B, compare lanes 1 to 3 with lanes 4 to 6), while RXR-RAR heterodimerization was strongly disfavored when RXR was replaced by RXRY402A (compare lane 8 with lanes 9 to 11). No RXR homodimer complexes were formed with DR3 or DR4 response elements (Fig. 2C and D), while unexpectedly, RXRY402A homodimers bound to both DR4 (Fig. 2C, lanes 2 to 4) and DR3 (Fig. 2D, lanes 2 to 4). Moreover, heterodimerization of the RXR mutant with TR or VDR was dramatically reduced, if not absent, compared with the wt receptor (Fig. 2C, compare lanes 6 to 8 with lanes 9 to 11; Fig. 2D, compare lanes 6 and 7 with lanes 8 to 10).
Together these results support the notion that mutating tyrosine 402 in helix H9 of RXR to alanine eliminates the well-established stabilization of DNA binding by RXR heterodimerization. Therefore, this tyrosine apparently plays a pivotal role allowing RXR to act as a promiscuous partner of a great number of nuclear receptors.
Mutation of RXR Y402 strengthens RXR homodimerization in vivo.
To analyze directly the dimerization and transactivation functions of RXRY402A in intact cells, we used a mammalian two-hybrid system comprising GAL-RXR or GAL-RXRY402A coexpressed with VP16-fused RXRΔAB (hereafter termed “VP16-RXR”) or its Y402A mutant homologue. The coexpression of GAL-RXR with VP16-RXR or VP16-RXRY402A yielded three- to fourfold activation of transcription, respectively, of the cognate reporter gene (Fig. 3). Similarly, GAL-RXRY402A cotransfected with VP16-RXR resulted in a twofold-induction of the CAT reporter gene expression (Fig. 3). In contrast, the combination of GAL-RXRY402A with VP16-RXRY402A gave a robust dose-dependent induction of transcription, reaching a maximum of 22- to 25-fold (Fig. 3). This demonstrates that the Y402A mutation greatly enhances RXR homodimerization efficiency provided that the mutation is present in both subunits. In keeping with this conclusion the full-length RXRY402A-induced transactivation in the presence of the RXR-selective agonist SR11237 was significantly more efficient than the wt RXR (Fig. 4A). Importantly, the wt and mutant RXRs were expressed in Cos cells at similar levels (Fig. 4B).
FIG. 3.
In vivo enhanced mRXRY402A homodimerization by mammalian two-hybrid analysis. Ten nanograms of GAL-RXRα or GAL-RXRY402A was cotransfected with increasing concentrations of VP16-RXRY402A as indicated or 200 ng of VP16-RXR, along with 100 ng of the (17m)x5-Glob-CAT reporter gene. The results shown are the averages ± standard errors of the means of four independent experiments.
FIG. 4.
The preferentially homodimer-forming mRXRY402A mutant displays higher transcription activation potential than the wt counterpart on DR1-based reporters. (A) Indicated quantities of full-length mRXRα wt (black columns) or mRXRY402A (gray columns) were cotransfected in Cos1 cells with 200 ng of the DR1-tk-CAT reporter gene and 50 ng of CMV-β-Gal as internal control. After transfection, cells were treated with 1 μM RXR-selective agonist SR11237. The results are expressed as fold induction over the basal activity (white columns) of the reporter gene observed in the presence of SR11237. The data presented are the averages ± standard errors of the means of four independent experiments. (B) The same level of expression of the full-length mRXRα wt and mRXRY402A was observed after transient expression in Cos1 cells as assayed by Western blotting. In these experiments, 50 ng of each plasmid was expressed for 24 h in Cos1 cells plated in 24-well plates before being immunoblotted with a polyclonal antibody to RXRα (Santa Cruz). Note that with longer exposure of the blot the endogenous RXR can be detected.
Taken together, the above results show that a single mutation of tyrosine 402 to alanine in helix H9 enhances RXR homodimer formation, leading to an increased transactivation potential on cognate DR1-based reporter genes.
Impairment of RXRY402A heterodimerization in vivo.
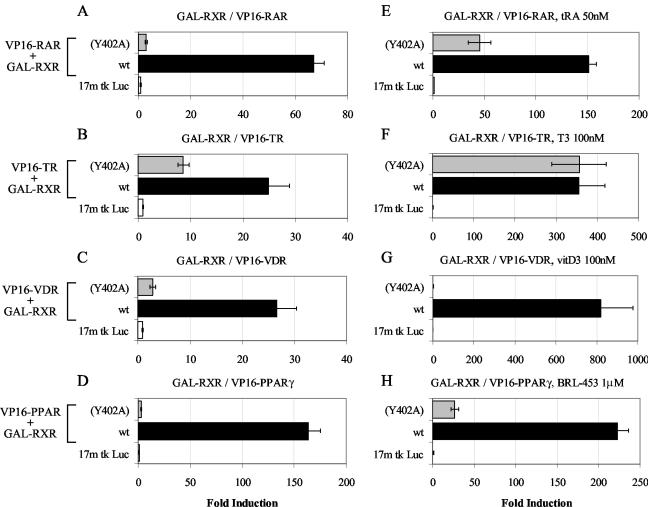
To assess the heterodimerization ability of RXRY402A in vivo, we used a mammalian two-hybrid system consisting of GAL-RXR or GAL-RXRY402A expressed together with VP16 fusions of RAR, TR, VDR, or PPAR LBDs and the 17-mer-tk-Luc reporter gene.
While in the absence of ligand the heterodimerization of RXR-RAR (Fig. 5A), RXR-TR (Fig. 5B), RXR-VDR (Fig. 5C), and RXR-PPAR (Fig. 5D) was obvious from the strong two-hybrid signals (wt bars), this signal was 90 to 98% decreased when GAL-RXRY402A was used (Y402A bars). Only TR showed a significant, albeit 65% reduced, interaction with the Y402 mutant (Fig. 5B). Interestingly, the cognate ligands of the various dimerization partners exerted a very different effect on heterodimerization with RXR and RXRY402A. ATRA stabilized RAR interaction with both wt and mutant RXRs, but even in the presence of the ligand the dimerization efficiency of RXRY402A was largely inferior to that of its wt counterpart (Fig. 5E). The same was true for the effect of BRL-453 on RXR-PPARγ interaction (Fig. 5H). Thyroid hormone (T3), which has a strong effect on TR-RXR dimerization, also stabilized interaction with RXRY402A such that mutant and wt RXRs interacted similarly efficiently with holo-TR (Fig. 5F). In striking contrast, vitamin D3 did not at all stabilize the VDR-RXRY402A heterodimer, even though it had a very strong positive effect on dimerization with wt RXR (Fig. 5G).
FIG. 5.
Mammalian two-hybrid analysis reveals impaired heterodimerization of mRXRY402A with RAR, TR, VDR, or PPAR in vivo. Ten nanograms of wt mRXRα (black columns) or mRXRY402A (gray columns) was cotransfected with 50 ng of VP16-hRARα, VP16-cTRα, VP16-hVDR, or VP16-mPPARγ and 100 ng of the 17-mer-tk-Luc reporter gene in the absence (A to D) or presence of 50 nM all-trans retinoic acid (tRA) (E), 100 nM thyroid hormone (T3) (F), 100 nM vitamin D3 (vitD3) (G), and 1 μM BRL-453 (H). The transcriptional activity exerted by each heterodimer was expressed as fold induction of reporter gene activity, in the absence (A to D) or presence (E to H) of the cognate ligand. The results shown are the averages ± standard errors of the means of three independent experiments.
To investigate the impact of those different ligand-mediated dimerization effects on transcription activation by the corresponding heterodimers, transient-transactivation assays with retinoic acid and vitamin D3 reporter genes were performed. Because Cos1 cells express endogenous retinoid receptors, we analyzed the effect of the Y402A mutation on RXR heterodimerization with endogenous RAR. The rationale of this experiment is that the addition of exogenous wt RXR (which is usually limiting in such assays) should increase the number of transcriptionally competent RAR-RXR heterodimers and augment the level of transcription activation seen with the endogenous RAR-RXR heterodimers. In contrast, no or much less stimulation should be seen on this reporter with the heterodimerization-incompetent RXRY402A. Indeed, wt RXR dose-dependently stimulated ATRA-induced transcription further by nearly threefold, while only minimal stimulation was seen with RXRY402A (Fig. 6A). Furthermore, no impairment of heterodimer-dependent DR5 activation was seen, confirming that RXRY402A does not act as a dominant-negative RXR and therefore cannot compete in vivo with the endogenous RXR for binding to the endogenous RAR partner. In the case of VDR, no significant transactivation of a DR3-based reporter gene was observed in the presence of vitamin D3, demonstrating the absence of significant levels of endogenous VDR in Cos cells (DR3 in Fig. 6B). Expression of exogenous VDR resulted only in a marginal stimulation of transcription. This was apparently due to limiting levels of endogenous RXR, as coexpression of exogenous wt RXR strongly increased vitamin D3-induced transcription from the DR3-based reporter (Fig. 6B). Importantly, this increase was drastically compromised when the RXRY402A was coexpressed in place of the wt RXR (compare gray and black bars in Fig. 6B).
FIG. 6.
Impaired heterodimerization in the presence of mRXRY402A blocks transcription activation of the cognate reporter genes by RAR (A) and VDR (B) by the corresponding agonists in vivo. Cos1 cells were transfected with 200 ng of DR5 (A)- or DR3 (B)-tk-CAT reporter genes, and VDR, RXR wt (black bars), or mRXRY402A (gray bars) expression vectors as indicated. CMV-β-Gal (25 ng) was used as an internal control to normalize for variations in transfection efficiencies. Note that in panel A the endogenous retinoid receptors were used as activators. After transfection, 50 nM ATRA (A) or 100 nM vitamin D3 (B) was added to the cells for an additional 24 h. The data presented are the averages ± standard errors of the means of three to four independent experiments and represent the fold induction over the activity of each reporter gene observed in the absence of added ligand. In panel A, the endogenous retinoid receptors induce the DR5-tk-CAT about sixfold in the absence of exogenously expressed RXR, as determined with a DR5-less reporter recombinant (data not shown). In the case of VDR (B) no endogenous activity was detected.
Together the above results demonstrate that the RXR residue Y402 in helix H9 is critical for the ability of RXR to heterodimerize both in vitro and in vivo and that impaired heterodimerization efficiency largely compromises RAR-RXR- and VDR-RXR-mediated transcription activation. Thus, RXRY402A should be a very useful reagent to reveal by genetic replacement not only RXR homodimer-dependent transcription but also the extent of heterodimer-dependent signaling for receptors that can form both homodimers and RXR heterodimers.
The three-dimensional structure analysis reveals divergent roles of Y402 in RXR homo- and heterodimers.
The above analysis demonstrates the ability of RXR to preferentially form heterodimers rather than homodimers and reveals that mutation of a single residue, RXR Y402 to alanine, suffices to generate an RXR variant with the inverse dimerization characteristic. We therefore studied the possible role of this residue in the formation of homo- and heterodimerization interfaces on the basis of the previously established crystal structures of RXR homo- and heterodimers (3, 4, 10, 11). Most of the RXR dimerization surface residues do not undergo major side chain rearrangements upon homo- or heterodimerization. Interestingly, one important exception is helix H9 residue Y402, which adopts a completely different conformation in the two types of RXR dimers (Fig. 7A). In both RXRα-RARα and RXRα-PPARγ heterodimers, Y402 displays a similar conformation and makes several stabilizing contacts with RXR partners. The hydrophobic part of Y402 is involved in van der Waals interactions with RARα or PPARγ residues along H10 (P375, M379 and Q430, A433, Q437, respectively), while the hydroxyl moiety points towards a small hydrophilic cavity where it is involved in a network of water-mediated hydrogen bonds stabilizing the heterodimer interfaces (Fig. 7B). Consequently, the replacement of Y402 by alanine removes many cross-subunit interactions stabilizing the heterodimer, in keeping with a 76% decreased heterodimer formation of RXRY402A (Table 1). Upon RXR self-association, Y402 adopts a distinct conformation (Fig. 7A). Indeed, in helix H10 of RXRα, the presence of the branched residue L425 at the position occupied by the linear residue M379 in RARα or Q437 in PPARγ prevents Y402 of the interacting protomer from adopting the conformation found in heterodimers (Fig. 7C and D). Due to the steric restrictions exerted by L425 of the homodimeric partner, the Y402 side chain rotates around the Cα-Cβ bond by 147°. In this position, Y402 interacts with G418, A421, and L425 of the second RXR protomer (Fig. 7C). Therefore, it appears that Y402 contributes differently to the RXR homo- and heterodimer interfaces, in good agreement with the differential impairment of RXR homo- and heterodimerization by the Y402A mutation.
FIG. 7.
Structural comparison of RXR homo- and heterodimer interfaces. (A) Superposition of the RXRα homodimerization (orange) and the RXRα heterodimerization (light blue) surfaces. The side chains involved in both interfaces are depicted and labeled (mRXRα numbering), as are the secondary structural elements. (B) Part of the RXRα-RARα heterodimerization interface showing Y402 and its interactions with the RARα LBD. Helices H9 and H10 are depicted as Cα traces with different colors for each protomer. Side chain atoms are colored by atom type (carbon, yellow; oxygen, red; sulfur, green; nitrogen, blue). Water molecules are depicted as red spheres. (C) Part of the RXRα-RXRα homodimerization interface showing Y402 and its interactions with the interacting RXRα LBD. Helices H9 and H10 are depicted as Cα traces with different colors for each protomer. Side chain atoms are colored by atom type (carbon, yellow; oxygen, red; nitrogen, blue). (D) Superposition of the RXRα homo- and heterodimerization interfaces described for panels B and C showing the steric clash between RXRα L425 in the RXRα-RXRα homodimer and Y402 of RXRα in the RARα-RXRα heterodimer. The figure was generated with SETOR (8).
Notably, nuclear receptor sequence alignments reveal that receptors with a leucine at the position corresponding to RXRα L425 do not form heterodimers with RXR (Fig. 8). Conversely, receptors that are well-known heterodimeric partners of RXR contain a smaller (A or G) or more flexible (M or Q) residue at this position. Taken together, these observations suggest that the nature of the residue at the position analogous to RXRα L425 is an important determinant of the capacity of a given receptor to interact with RXR. It appears that the presence of a branched residue (i.e., a leucine) at this position forces Y402 to adopt a conformation that reduces the ability of RXR to dimerize. The origin of Y402's destabilizing influence on RXR homodimerization is not obvious on the basis of the presently available crystal structures, and further structural studies (i.e., the RXRY402A homodimer crystal structure) will be required to fully understand the impact of Y402 on RXR dimerization function. Nevertheless, our observation that the Y402A mutation greatly enhances RXR homodimerization efficiency provided that the mutation is present in both subunits (Fig. 3) supports the idea of a structural rearrangement of the dimer interface which can occur only if both RXR protomers are mutated. In the symmetric homodimer, the two Y402 residues may prevent the optimal relative orientation of the constituent LBDs. Of note, in contrast to the twofold symmetry of the RXR LBD homodimer, RXRα-RARα and RXRα-PPARγ heterodimers are asymmetric, with each subunit deviated roughly 10° from the twofold axis as illustrated in Fig. 7D (10).
FIG. 8.
Alignment of the H10 helices of some nuclear receptors. Receptors with a leucine at the position corresponding to mRXRα L425 generally do not form strong heterodimers with RXR, whereas receptors that are known heterodimeric partners of RXR contain a smaller or more flexible residue at this position.
DISCUSSION
Two interfaces are established between RXR homo- and heterodimers, a strong one in the LBD and a weak one in the DBD (for a comprehensive description see reference 16). While the LBD interface stabilizes direct repeat response element binding through cooperative DNA binding, the DBD interface dictates the response element repertoire of the corresponding homo- or heterodimer (14, 22, 23, 28, 29). We have scrutinized the previously defined (3, 4) LBD interface to assess the roles of the RXR residues constituting the dimerization surfaces in homo- and/or heterodimerization. Alanine scanning mutagenesis revealed that single mutations of charged, hydrophilic, or hydrophobic residues located in helices H7, H9, and H10 and loop L8-9 are sufficient to disrupt RXR homodimers as revealed by the cooperativity of binding to DR1 response elements. In contrast, none of the single or compound mutants could disrupt RXR-RAR heterodimerization completely, as deduced from DR5-based EMSAs. These data show that, despite the similar and largely overlapping dimerization surfaces, individual RXR residues contribute very differently to homo- and heterodimerization.
Remarkably, the single alanine mutation of RXR tyrosine 402 in helix H9 decreases heterodimer stability (with RAR, VDR, or TR), while simultaneously increasing homodimerization. Moreover, the Y402A mutation could restore RXR homodimerization capacity in other dimerization-defective mutants and thus acts as a dominant-positive homodimerization mutant.
The above mutational analysis and the structural interpretation of these data identified Y402 as an mRXRα residue that is critical for the preferential formation of RXR heterodimers by decreasing the potential of RXR to homodimerize. Mutation of this residue to alanine destabilizes heterodimers because a number of key contacts in the interface are lost while concomitantly the negative effect of Y402 on homodimer formation is relieved. We believe that this mutation enables the RXR homodimer to adopt a different dimeric interface with a much stronger association between the two protomers.
How to explain that a single mutation in the LBD can increase RXR homodimer stability on noncognate response elements such as DR3 or DR4? The DNA-response element primarily used by RXR homodimers consists in a direct repetition of the consensus half-site 5′-AGGTCA-3′ spaced by 1 bp (DR1). An insertion of one or several base pairs in the spacing (to generate DR2, DR3, DR4, or DR5 response elements) has severe consequences for receptor DBD-DNA interaction because the distance and thus spacing between the two subunits, as well as their relative orientation, would have to be readjusted to allow optimal DNA binding. Therefore, two (nonexclusive) possibilities could explain the extended response element repertoire of the Y402A mutant homodimer. One possibility is that the mutant no longer establishes a dimerization interface at the level of the DBDs. This would result in a loss of cooperative binding, which may be compensated for by the increased homodimerization of the LBDs. If this were the case, the DBD could no longer dictate the response element repertoire and the increased LBD dimerization, which is rather independent of the detailed DNA binding, would allow relaxed response element recognition until the response element spacing created a problem for LBD dimerization itself. The second possibility is based on the flexibility of RXR in forming favorable interactions with various partners in such a way that the resulting dimers can bind to differently spaced direct repeats. It is possible that the RXR DBD might adapt, at least to some extent, its conformation to permit, in the context of a suitable partner, the various interaction surfaces required. In the context of a highly stabilized Y402A homodimer mutant, the RXR DBD may have sufficient flexibility to generate (some) DBD-DBD contacts allowing the recognition of noncognate response elements. In the case of the wt RXR, the strength of the interaction between the two LBD subunits is probably too weak to generate sufficient cooperativity for DNA binding and/or may not sufficiently force the DBD subunits to adapt a conformation that allows binding to noncognate response elements.
Irrespective of the importance of structural considerations, the present work provides for the first time a tool to separate RXR homo- and heterodimerization and thus the possibility to design genetic strategies to study the possible existence of RXR homodimer signaling. While there is evidence for RXR signaling in the absence of RAR ligands (for an example see reference 1), it has been impossible in these studies to rule out the possibility that, even though RXR is usually nonresponsive to its ligand (12), so-called “permissive heterodimers” (9, 21) with nondefined partners may mediate the observed effects. Mutants like mRXRY402A may allow generation of cells (or even animals) in which an RXR-selective ligand will preferentially activate RXR homodimer signaling.
Acknowledgments
We are grateful to Cathie Erb and Séverine Schloegel for the construction of VP16-RXRY402A. We thank Cécile Rochette-Egly for antibodies and Jean-Marie Wurtz and Souphatta Sasorith for nuclear receptor alignments.
This work was supported by the Institut Nationale de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg, the European Community (QLG3-CT2000-00844 and QLG1-CT2001-01935), and Bristol-Myers Squibb.
REFERENCES
- 1.Benoit, G. R., M. Flexor, F. Besancon, L. Altucci, A. Rossin, J. Hillion, Z. Balajthy, L. Legres, E. Segal-Bendirdjian, H. Gronemeyer, and M. Lanotte. 2001. Autonomous rexinoid death signaling is suppressed by converging signaling pathways in immature leukemia cells. Mol. Endocrinol. 15:1154-1169. [DOI] [PubMed] [Google Scholar]
- 2.Bocquel, M. T., V. Kumar, C. Stricker, P. Chambon, and H. Gronemeyer. 1989. The contribution of the N- and C-terminal regions of steroid receptors to activation of transcription is both receptor and cell-specific. Nucleic Acids Res. 17:2581-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourguet, W., M. Ruff, P. Chambon, H. Gronemeyer, and D. Moras. 1995. Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-alpha. Nature 375:377-382. [DOI] [PubMed] [Google Scholar]
- 4.Bourguet, W., V. Vivat, J. M. Wurtz, P. Chambon, H. Gronemeyer, and D. Moras. 2000. Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell 5:289-298. [DOI] [PubMed] [Google Scholar]
- 5.Chen, H., M. Tini, and R. M. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218-224. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. Y., J. Clifford, C. Zusi, J. Starrett, D. Tortolani, J. Ostrowski, P. R. Reczek, P. Chambon, and H. Gronemeyer. 1996. Two distinct actions of retinoid-receptor ligands. Nature 382:819-822. [DOI] [PubMed] [Google Scholar]
- 7.Dong, D., and N. Noy. 1998. Heterodimer formation by retinoid X receptor: regulation by ligands and by the receptor's self-association properties. Biochemistry 37:10691-10700. [DOI] [PubMed] [Google Scholar]
- 8.Evans, S. V. 1993. SETOR: hardware-lighted three-dimensional solid model representations of macromolecules. J. Mol. Graph. 11:127-128, 134-138. [DOI] [PubMed]
- 9.Forman, B. M., K. Umesono, J. Chen, and R. M. Evans. 1995. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541-550. [DOI] [PubMed] [Google Scholar]
- 10.Gampe, R. T., Jr., V. G. Montana, M. H. Lambert, A. B. Miller, R. K. Bledsoe, M. V. Milburn, S. A. Kliewer, T. M. Willson, and H. E. Xu. 2000. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerisation among nuclear receptors. Mol. Cell 5:545-555. [DOI] [PubMed] [Google Scholar]
- 11.Gampe, R. T., V. G. Montana, M. H. Lambert, G. B. Wisely, M. V. Milburn, and H. E. Xu. 2000. Structural basis for autorepression of retinoid X receptor by tetramer formation and the AF-2 helix. Genes Dev. 14:2229-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Germain, P., J. Iyer, C. Zechel, and H. Gronemeyer. 2002. Coregulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 415:187-192. [DOI] [PubMed] [Google Scholar]
- 13.Kersten, S., D. Kelleher, P. Chambon, H. Gronemeyer, and N. Noy. 1995. Retinoid X receptor alpha forms tetramers in solution. Proc. Natl. Acad. Sci. USA 92:8645-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khorasanizadeh, S., and F. Rastinejad. 2001. Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 26:384-390. [DOI] [PubMed] [Google Scholar]
- 15.Kurokawa, R., J. Di Renzo, M. Boehm, J. Sugarman, B. Gloss, M. G. Rosenfeld, R. A. Heyman, and C. K. Glass. 1994. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature 371:528-531. [DOI] [PubMed] [Google Scholar]
- 16.Laudet, V., and H. Gronemeyer. 2002. The nuclear receptor facts book. Academic Press, San Diego, Calif.
- 17.Mader, S., J. Y. Chen, Z. Chen, J. White, P. Chambon, and H. Gronemeyer. 1993. The patterns of binding of RAR, RXR and TR homo- and heterodimers to direct repeats are dictated by the binding specificities of the DNA binding domains. EMBO J. 12:5029-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mader, S., P. Leroy, J. Y. Chen, and P. Chambon. 1993. Multiple parameters control the selectivity of nuclear receptors for their response elements. Selectivity and promiscuity in response element recognition by retinoic acid receptors and retinoid X receptors. J. Biol. Chem. 268:591-600. [PubMed] [Google Scholar]
- 19.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 20.Nagpal, S., S. Friant, H. Nakshatri, and P. Chambon. 1993. RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerisation in vivo. EMBO J. 12:2349-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perlmann, T., and L. Jansson. 1995. A novel pathway for vitamin A signaling mediated by RXR heterodimerisation with NGFI-B and NURR1. Genes Dev. 9:769-782. [DOI] [PubMed] [Google Scholar]
- 22.Rastinejad, F., T. Perlmann, R. M. Evans, and P. B. Sigler. 1995. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature 375:203-211. [DOI] [PubMed] [Google Scholar]
- 23.Rastinejad, F., T. Wagner, Q. Zhao, and S. Khorasanizadeh. 2000. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. 19:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy, B., R. Taneja, and P. Chambon. 1995. Synergistic activation of retinoic acid (RA)-responsive genes and induction of embryonal carcinoma cell differentiation by an RA receptor α (RARα)-, RARβ-, or RARγ-selective ligand in combination with a retinoid X receptor-specific ligand. Mol. Cell. Biol. 15:6481-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulman, I. G., C. Li, J. W. Schwabe, and R. M. Evans. 1997. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 11:299-308. [DOI] [PubMed] [Google Scholar]
- 26.Vivat, V., C. Zechel, J. M. Wurtz, W. Bourguet, H. Kagechika, H. Umemiya, K. Shudo, D. Moras, H. Gronemeyer, and P. Chambon. 1997. A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J. 16:5697-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westin, S., R. Kurokawa, R. T. Nolte, G. B. Wisely, E. M. McInerney, D. W. Rose, M. V. Milburn, M. G. Rosenfeld, and C. K. Glass. 1998. Interactions controlling the assembly of nuclear-receptor heterodimers and co-activators. Nature 395:199-202. [DOI] [PubMed] [Google Scholar]
- 28.Zechel, C., X. Q. Shen, P. Chambon, and H. Gronemeyer. 1994. Dimerisation interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 13:1414-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zechel, C., X. Q. Shen, J. Y. Chen, Z. P. Chen, P. Chambon, and H. Gronemeyer. 1994. The dimerisation interfaces formed between the DNA binding domains of RXR, RAR and TR determine the binding specificity and polarity of the full-length receptors to direct repeats. EMBO J. 13:1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]