Abstract
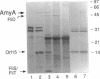
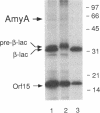
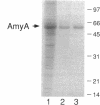
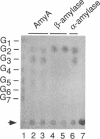
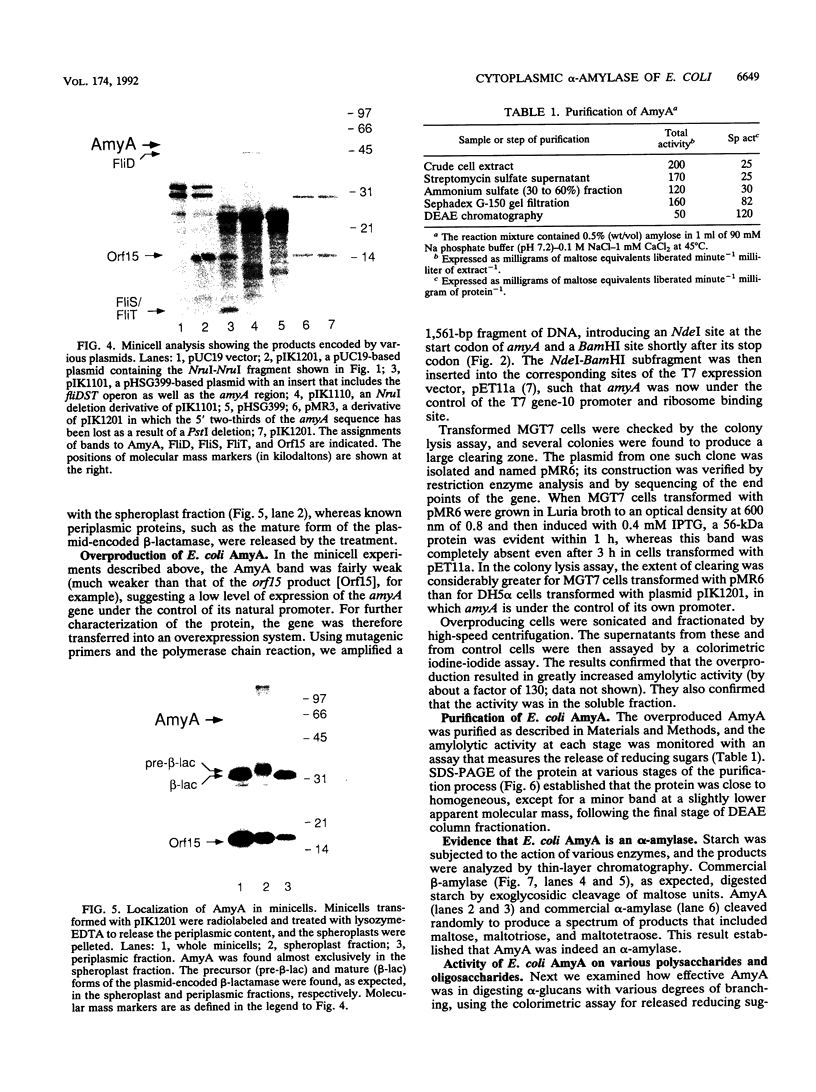
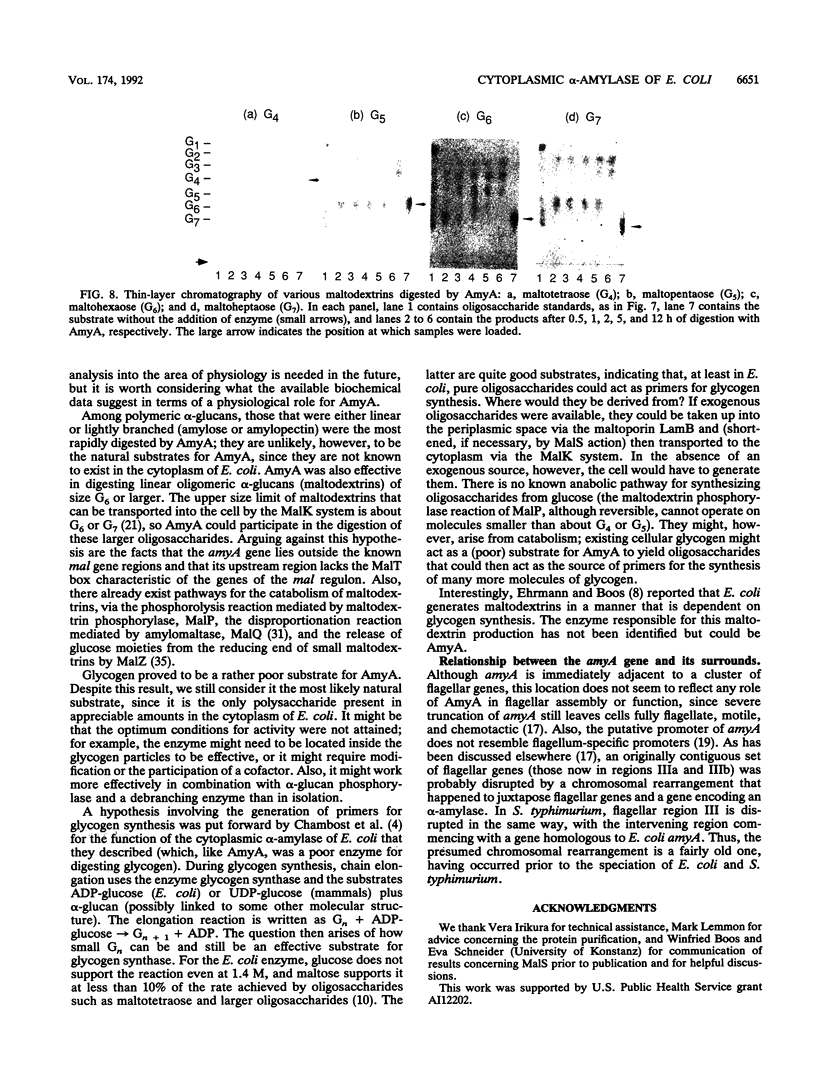
In the gap between two closely linked flagellar gene clusters on the Escherichia coli and Salmonella typhimurium chromosomes (at about 42 to 43 min on the E. coli map), we found an open reading frame whose sequence suggested that it encoded an alpha-amylase; the deduced amino acid sequences in the two species were 87% identical. The strongest similarities to other alpha-amylases were to the excreted liquefying alpha-amylases of bacilli, with > 40% amino acid identity; the N-terminal sequence of the mature bacillar protein (after signal peptide cleavage) aligned with the N-terminal sequence of the E. coli or S. typhimurium protein (without assuming signal peptide cleavage). Minicell experiments identified the product of the E. coli gene as a 56-kDa protein, in agreement with the size predicted from the sequence. The protein was retained by spheroplasts rather than being released with the periplasmic fraction; cells transformed with plasmids containing the gene did not digest extracellular starch unless they were lysed; and the protein, when overproduced, was found in the soluble fraction. We conclude that the protein is cytoplasmic, as predicted by its sequence. The purified protein rapidly digested amylose, starch, amylopectin, and maltodextrins of size G6 or larger; it also digested glycogen, but much more slowly. It was specific for the alpha-anomeric linkage, being unable to digest cellulose. The principal products of starch digestion included maltotriose and maltotetraose as well as maltose, verifying that the protein was an alpha-amylase rather than a beta-amylase. The newly discovered gene has been named amyA. The natural physiological role of the AmyA protein is not yet evident.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baecker P. A., Furlong C. E., Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose synthetase as deduced from the nucleotide sequence of the glg C gene. J Biol Chem. 1983 Apr 25;258(8):5084–5088. [PubMed] [Google Scholar]
- Baecker P. A., Greenberg E., Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli 1,4-alpha-D-glucan:1,4-alpha-D-glucan 6-alpha-D-(1, 4-alpha-D-glucano)-transferase as deduced from the nucleotide sequence of the glg B gene. J Biol Chem. 1986 Jul 5;261(19):8738–8743. [PubMed] [Google Scholar]
- Bartlett D. H., Matsumura P. Identification of Escherichia coli region III flagellar gene products and description of two new flagellar genes. J Bacteriol. 1984 Nov;160(2):577–585. doi: 10.1128/jb.160.2.577-585.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambost J. P., Favard A., Cattanéo J. Purification et propriétés d'une alpha-amylase endocellulaire d'Escherichia coli. Bull Soc Chim Biol (Paris) 1967 Nov 10;49(10):1231–1246. [PubMed] [Google Scholar]
- Chen G. S., Segel I. H. Escherichia coli polyglucose phosphorylases. Arch Biochem Biophys. 1968 Sep 20;127(1):164–174. doi: 10.1016/0003-9861(68)90213-0. [DOI] [PubMed] [Google Scholar]
- Choi Y. L., Kawamukai M., Utsumi R., Sakai H., Komano T. Molecular cloning and sequencing of the glycogen phosphorylase gene from Escherichia coli. FEBS Lett. 1989 Jan 30;243(2):193–198. doi: 10.1016/0014-5793(89)80128-0. [DOI] [PubMed] [Google Scholar]
- Dubendorff J. W., Studier F. W. Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J Mol Biol. 1991 May 5;219(1):45–59. doi: 10.1016/0022-2836(91)90856-2. [DOI] [PubMed] [Google Scholar]
- Ehrmann M., Boos W. Identification of endogenous inducers of the mal regulon in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3539–3545. doi: 10.1128/jb.169.8.3539-3545.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox J., Kawaguchi K., Greenberg E., Preiss J. Biosynthesis of bacterial glycogen. Purification and properties of the Escherichia coli B ADPglucose:1,4-alpha-D-glucan 4-alpha-glucosyltransferase. Biochemistry. 1976 Feb 24;15(4):849–857. doi: 10.1021/bi00649a019. [DOI] [PubMed] [Google Scholar]
- Freundlieb S., Boos W. Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J Biol Chem. 1986 Feb 25;261(6):2946–2953. [PubMed] [Google Scholar]
- Homma M., Komeda Y., Iino T., Macnab R. M. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol. 1987 Apr;169(4):1493–1498. doi: 10.1128/jb.169.4.1493-1498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J Bacteriol. 1985 Aug;163(2):464–471. doi: 10.1128/jb.163.2.464-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara H., Sasaki T., Tsuboi A., Yamagata H., Tsukagoshi N., Udaka S. Complete nucleotide sequence of a thermophilic alpha-amylase gene: homology between prokaryotic and eukaryotic alpha-amylases at the active sites. J Biochem. 1985 Jul;98(1):95–103. doi: 10.1093/oxfordjournals.jbchem.a135279. [DOI] [PubMed] [Google Scholar]
- Jeanningros R., Creuzet-Sigal N., Frixon C., Cattaneo J. Purification and properties of a debranching enzyme from Escherichia coli. Biochim Biophys Acta. 1976 Jun 7;438(1):186–199. doi: 10.1016/0005-2744(76)90235-7. [DOI] [PubMed] [Google Scholar]
- Jones C. J., Homma M., Macnab R. M. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989 Jul;171(7):3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi I., Müller V., Williams A. W., Irikura V. M., Macnab R. M. Subdivision of flagellar region III of the Escherichia coli and Salmonella typhimurium chromosomes and identification of two additional flagellar genes. J Gen Microbiol. 1992 Jun;138(6):1051–1065. doi: 10.1099/00221287-138-6-1051. [DOI] [PubMed] [Google Scholar]
- Kumar A., Larsen C. E., Preiss J. Biosynthesis of bacterial glycogen. Primary structure of Escherichia coli ADP-glucose:alpha-1,4-glucan, 4-glucosyltransferase as deduced from the nucleotide sequence of the glgA gene. J Biol Chem. 1986 Dec 5;261(34):16256–16259. [PubMed] [Google Scholar]
- Kutsukake K., Ohya Y., Iino T. Transcriptional analysis of the flagellar regulon of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Matsumura P., Silverman M., Simon M. Synthesis of mot and che gene products of Escherichia coli programmed by hybrid ColE1 plasmids in minicells. J Bacteriol. 1977 Dec;132(3):996–1002. doi: 10.1128/jb.132.3.996-1002.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller V., Jones C. J., Kawagishi I., Aizawa S., Macnab R. M. Characterization of the fliE genes of Escherichia coli and Salmonella typhimurium and identification of the FliE protein as a component of the flagellar hook-basal body complex. J Bacteriol. 1992 Apr;174(7):2298–2304. doi: 10.1128/jb.174.7.2298-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss J. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol. 1984;38:419–458. doi: 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- SIGAL N., CATTANEO J., SEGEL I. H. GLYCOGEN ACCUMULATION BY WILD-TYPE AND URIDINE DIPHOSPHATE GLUCOSE PYROPHOSPHORYLASE-NEGATIVE STRAINS OF ESCHERICHIA COLI. Arch Biochem Biophys. 1964 Dec;108:440–451. doi: 10.1016/0003-9861(64)90425-4. [DOI] [PubMed] [Google Scholar]
- STEIN H. [Cholecystography in young infants]. Arch Kinderheilkd. 1961 Nov;165:27–33. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E., Freundlieb S., Tapio S., Boos W. Molecular characterization of the MalT-dependent periplasmic alpha-amylase of Escherichia coli encoded by malS. J Biol Chem. 1992 Mar 15;267(8):5148–5154. [PubMed] [Google Scholar]
- Takkinen K., Pettersson R. F., Kalkkinen N., Palva I., Söderlund H., Käriäinen L. Amino acid sequence of alpha-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J Biol Chem. 1983 Jan 25;258(2):1007–1013. [PubMed] [Google Scholar]
- Tapio S., Yeh F., Shuman H. A., Boos W. The malZ gene of Escherichia coli, a member of the maltose regulon, encodes a maltodextrin glucosidase. J Biol Chem. 1991 Oct 15;266(29):19450–19458. [PubMed] [Google Scholar]
- Tsukagoshi N., Ihara H., Yamagata H., Udaka S. Cloning and expression of a thermophilic alpha-amylase gene from Bacillus stearothermophilus in Escherichia coli. Mol Gen Genet. 1984;193(1):58–63. doi: 10.1007/BF00327414. [DOI] [PubMed] [Google Scholar]
- Vihinen M., Mäntsälä P. Microbial amylolytic enzymes. Crit Rev Biochem Mol Biol. 1989;24(4):329–418. doi: 10.3109/10409238909082556. [DOI] [PubMed] [Google Scholar]
- Yang M., Galizzi A., Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. doi: 10.1093/nar/11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Jen Y., Takeuchi E., Inouye M., Nakayama H., Tagaya M., Fukui T. Alpha-glucan phosphorylase from Escherichia coli. Cloning of the gene, and purification and characterization of the protein. J Biol Chem. 1988 Sep 25;263(27):13706–13711. [PubMed] [Google Scholar]