Abstract
IgD is considered to be a recently evolved Ig, being previously found only in primates and rodents. Here we describe, from a teleost fish (the channel catfish, Ictalurus punctatus), a novel complex chimeric Ig heavy chain, homologous, in part, to the heavy chain (δ) of IgD. In addition to alternative secretory or membrane-associated C termini, this chimeric molecule contains a rearranged variable domain, the first constant domain of μ, and seven constant domains encoded by a δ gene homolog. Identification of the catfish gene as δ is based on the following properties: sequence relatedness to mammalian δ; a location within the IgH locus that is immediately downstream of the μ gene; separate terminal exons for the secretory and membrane forms; coexpression with the complete μ chain in some but not all B cells. These results (i) suggest that IgD is an ancient immunoglobulin that was present in vertebrates ancestral to both the mammals and the ray-finned fishes, and (ii) raise the possibility that this Ig isotype may have served an as yet unidentified important function early in the evolution of the immune system.
Keywords: evolution
Antibodies are characteristic of vertebrates above the level of the agnathan fish (1), but the early evolutionary history of these molecules is unclear. IgM is the only class of antibody universally found in all species that possess Igs, but its complexity and large size has led to some doubts that it was the primordial Ig. Interest in the origins of Igs has recently been stimulated by the observations (2–4) that elasmobranchs possess, in addition to IgM, novel classes of Ig. These large multidomain Igs, designated NAR, NARC, and IgW (and which are related to the previously described IgX of rajiformes, refs. 5 and 6), have been proposed as candidates for the primordial antibody (3, 4). In contrast, IgD is a class of antibody that is considered to have evolved relatively recently, having been described only in primates and rodents (7–10). Although the function, biochemistry and genetics of IgD have been the subject of intense interest (11–17) gene targeting (knockout) experiments revealed that IgD had a relatively minor functional role in immune responses in the mouse (18, 19). We report here the results of studies that identify a complex homolog of the IgD heavy (H) (δ) chain in the channel catfish (Ictalurus punctatus). This observation in a teleost fish suggests that this Ig isotype existed early in vertebrate evolution, and opens the possibility that it may have had an alternative function to that observed in the modern mammals.
MATERIALS AND METHODS
Library Screening.
A partial δ chain sequence was generated in 3′-rapid amplification of cDNA ends PCR protocols (20) of total RNA from catfish peripheral blood lymphocytes (PBLs). The forward primer, 5′-CAAAGCTTGCIACXCTIGTITGXCTIGT-3′ (I is inosine, X is T or C) corresponded to spacer nucleotides, a HindIII site and the conserved amino acid ATLVCLV sequence found in T cell receptor (TCR) β and Ig light chains. The 840-bp PCR fragment (containing δ exon 7, the membrane segment, and 3′ untranslated region) was cloned into the pCRII vector (Invitrogen) and sequenced on both strands by the chain termination method using Sequenase 2.0 (United States Biochemical). An unamplified catfish PBL cDNA library (5.1 × 106 recombinants in lambda ZAPII, Stratagene) was then screened with this PCR fragment of δ, and the filter lifts were washed three times for 20 min at high stringency (0.1× standard saline citrate, 0.1% SDS, 68°C). All cDNAs were sequenced on both strands using primers synthesized by the Medical University of South Carolina Nucleic Acid Synthesis Facility.
Phylogenetic Analysis of the Catfish δ Sequences.
The constant (C) region sequences were aligned using clustal v, with identity or PAM 250 weight tables, gap penalty of 10 and gap length penalty of 10. Alignments are available on request. Unrooted phylogenetic trees were generated from the sequence alignments employing the paup program (21). The most parsimonious trees were derived by heuristic search using bootstrapping and branch swapping (tree bisection-reconnection). The sequences used were from the following database entries: for μ, U27213U27213 (duck); X68700X68700 (axolotl); M20484M20484, JO3631 (frog); K00389K00389 (chicken); X14940X14940 (human); J00443J00443, V00822V00822, V00823V00823 (mouse); J00666J00666 (rabbit); X13920X13920 (shrew); M92050M92050 (pig); X59994X59994, S40921S40921 (sheep); U12456U12456 (bowfin); U12455U12455 (gar); X79482X79482, M27230M27230 (catfish); X58870X58870 (cod); S63348S63348, X65261X65261 (trout); S48652S48652 (salmon); M26182M26182 (ladyfish); X07782X07782, Y00840Y00840 (shark); M29679M29679, M35185M35185 (skate): for α, S40610S40610 (chicken); U27222U27222 (duck); X15045X15045 (gorilla); U12594U12594 (pig); V00785V00785 (mouse); X82116X82116 (rabbit): for γ, U03781U03781 (pig); X69797X69797 (sheep); J00453J00453 (mouse); X03604X03604, M12958M12958 (human): for υ, X65219X65219 (duck); X07174X07174 (chicken); X69492X69492 (axolotl); X15114X15114 (frog): for ɛ, U15150U15150 (horse); J00476J00476 (mouse); M84356M84356 (sheep): for NAR, U18701U18701: for NARC, U551450: for ω, U40560U40560: for skate IgX H chains, M29672M29672, M35185M35185: for frog IgX H chain, X13779X13779: for catfish δ, U67437U67437. The sequence of dog μ was from McCumber and Capra (22), the sequence of human α2 from Flanagan et al. (23), the sequence of mouse δ from Tucker et al. (11), and the sequence of human δ from White et al. (12).
Reverse Transcription (RT)–PCR Analysis.
Total RNA was isolated from catfish PBLs or cell lines by the guanidinium chloride method (24). To detect the expression of members of different VH families in δ transcripts, ≈1 μg of total RNA was converted into first strand cDNA using 50 ng of a reverse catfish δTM primer (5′-TCATCAAAGTATATCGTCTC-3′, corresponding to nucleotides 2876–2896 in cDNA M5) with 200 units of SuperScript II (GIBCO/BRL) according to the manufacturer’s recommended protocol. One percent of the first strand reaction was used in RT-PCR. The final reaction volume was 100 μl of 10 mM Tris⋅HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 200 mM dNTPs, and 40 mM tetramethylammonium chloride, containing 0.5 μg of specific forward and reverse primers. Four units of Taq DNA polymerase (Perkin–Elmer) was added, and 30 cycles of amplification (94°C for 1 min, 65°C for 2 min, 72°C for 3 min) were performed. Primers for the catfish variable region (V)H families 1–6 have been described previously (25). The VH7 primer was kindly provided by C. Lobb (Department of Microbiology, University of Mississippi Medical Center) and was a 19-mer spanning the leader/FR1 boundary of a VH7 sequence (26). The reverse δ primer, 5′-CACTTGCTCCATGTTTGACT-3′, corresponded to nucleotides 1006–1025 in cDNA M5. Fifty microliters of the PCR were electrophoresed on a 1% agarose gel and then analyzed by blot hybridization as described (25). The 291-bp Cμ1 probe spanned nucleotides 2134–2325 of the catfish H chain locus (accession no. X79482X79482). The probe was labeled to a specific activity of 2 × 109 cpm/μg with a random-primed labeling kit (Boehringer Mannheim). Hybridization and washing were as described above for the library screen, and exposure to Kodak BIOMAX MR film was for 25 min at 25°C. For the detection of μ and TCRα transcripts, first strand cDNA synthesis was carried out with an oligo T-adapter primer, 5′-TCTGAATTCTCGAGTCGACATC(dT17)-3′. The primers used to detect μ message have been described previously (27). Catfish TCRα primers were: forward, 5′-AGCCGTCAATTTACAAACTTC-3′ and reverse, 5′-TTGTGTCACCAATTCAAATGC-3′ (from catfish TCRα cDNA T8, accession no. U62043U62043). δ primers were: forward, 5′-AGCACACCATCTCTAAAACCA-3′ and reverse, 5′-TCATCAAAGTATATCGTCTC-3′ (encompassing nucleotides 2225–2885 of cDNA M5). All RT-PCR products were cloned and sequenced to verify their authenticity.
RESULTS AND DISCUSSION
In the course of an anchored PCR-based search for members of the Ig superfamily, primers based on the conserved sequence (ATLVCLA/V) around the first cysteine of Igλ and TCR β C regions, amplified a sequence from catfish PBLs that proved to be part of a novel H chain cDNA. This sequence contained an Ig-like transmembrane hydrophobic region and a short positively charged five-amino acid cytoplasmic tail. Use of this PCR product to probe a cDNA library from catfish PBLs yielded 29 positive cDNAs, which upon analysis were found to encode either the secreted or membrane-receptor forms of a previously undescribed fish Ig H chain. This novel full-length chimeric H chain contains a typical VH domain, the first C region domain of the catfish IgM H (μ) chain, seven additional C region domains, and alternative C-terminal segments for either the secreted or membrane forms of the molecule. We have termed this novel Ig H chain δ, since (as discussed below) it shares many features in common with mammalian Igδ.
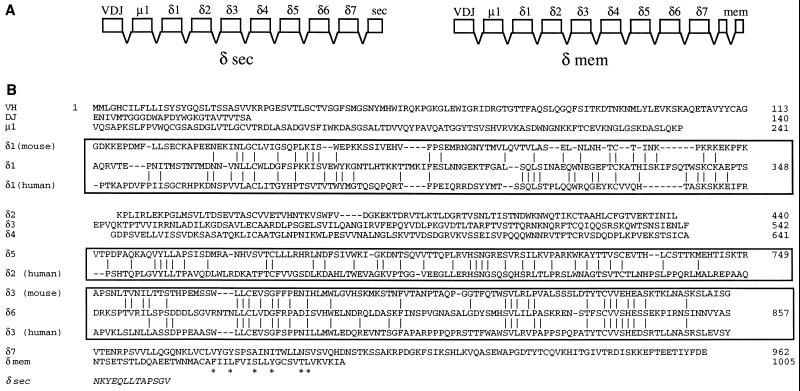
The inferred exon structures encoding the secreted and membrane forms of the catfish δ chain homolog are presented in Fig. 1A. Analysis of one complete and 28 partial cDNAs revealed that a splice donor site (29) was absent from the 3′ region of the secreted form of δ, and that the secreted and membrane forms are generated by utilization of alternative exons encoding the C termini of each. Fig. 1B shows the complete amino acid sequence of the catfish membrane form of δ, inferred from the full-length cDNA M5, compared with mouse and human δ C regions. This sequence shows a typical Ig-like membrane region containing the conserved antigen receptor transmembrane motif described by Campbell et al. (30). This motif is speculated to play a role in Ig receptor assembly.
Figure 1.
Structure and evolutionary relationships of the catfish IgD heavy chain. (A) Inferred exon structure encoding the secreted and membrane forms of the catfish δ chain. The exons are based on the deduced protein sequence of the Ig domain and on the identification of δ1 and δ2 exons in the germ line. (B) Complete amino acid sequence of the membrane form of catfish δ, inferred from the full-length cDNA M5 (accession no. U67437U67437). The sequence is shown by domain, and alignments with the homologous sequences of human and mouse δ C regions are boxed. The antigen receptor CART motif is marked with asterisks. The sequences of mouse δ are from ref. 11, and those of human δ are from ref. 12. The aligned regions (numbering of Kabat et al., ref. 28) are, for human δ1, 114–224; human δ2, 243–363; human δ3, 364–478; mouse δ1, 116–222; mouse δ3, 364–478. The sequence of the catfish δ secreted segment (δ sec) is taken from cDNA S1 (accession no. U67438U67438).
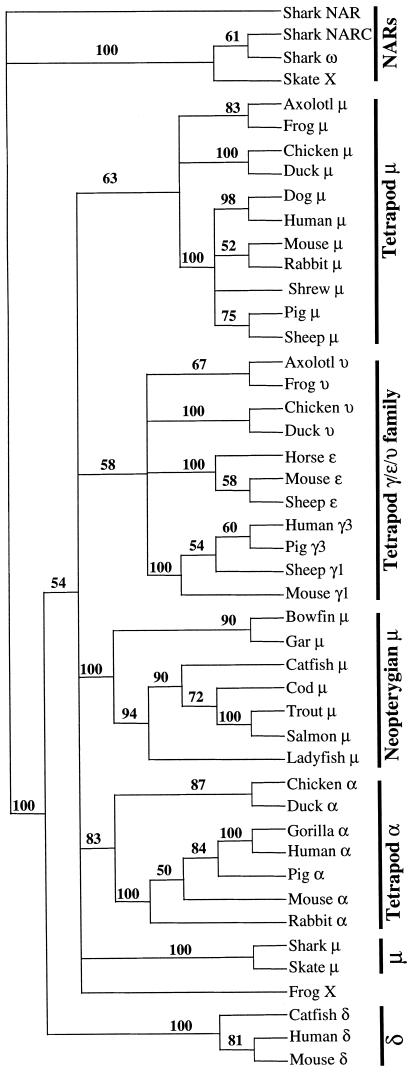
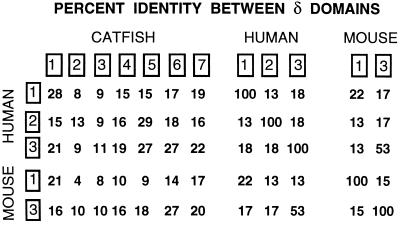
The phylogenetic relationship of catfish δ to other vertebrate Ig H chains was examined by analysis of H chain C region sequences using maximum-parsimony methods (paup, ref. 21). Alignments with variations in the number of sequences included, and of the alignment parameters (e.g., identity vs. PAM 250 matrix) were made, and trees were then generated using bootstrap methods in paup. The results were highly consistent with respect to δ: a typical tree is shown in Fig. 2, which illustrates clearly the clustering of the catfish δ with mouse and human δ. The delta branch is strongly supported by a bootstrap resampling confidence value of 100. Five different trees generated from different representative sequences of vertebrate H chain isotypes all showed a discrete δ branch, supported by bootstrap resampling values between 97–100, with a mean of 99 (data not shown, available on request). The tree shown in Fig. 2 contains three major branches: (i) the elasmobranch NAR/NARC/IgX/ω branch, (ii) the δ branch, and (iii) a large branch containing all μ, υ, γ, ɛ, and α sequences. Within this last branch can be seen the shorter branches reflecting the expected clustering of related sequences (a branch with all α sequences, a branch incorporating all γ, ɛ, and υ sequences and 3 m branches: bony fish, tetrapod, and elasmobranch). However, the relationship of the μ, α, and γ/ɛ/υ branches to one another was not resolved in this analysis. It has been observed (e.g., Mansikka, ref. 31) that Igs that are generally accepted as homologous often do not, after long evolutionary separation, share striking sequence similarities to one another, even when considered at the level of individual domains. Thus, although the relationship of catfish δ to mammalian δ is clearly indicated by parsimony-based (paup) analyses, the degree of amino acid sequence identity, in domain-by-domain comparisons, is relatively modest (Fig. 1B, Fig. 3). The highest values between human and catfish δ are 28% for the comparison of the δ1 domains, 29% for comparison of δ2 (human) and δ6 (catfish) domains, and 27% for comparison of human δ3 with catfish δ5 and δ6 domains. In the case of the mouse, the highest values were 21% for the comparison of the δ1 domains, and 27% for the comparison of mouse δ3 with catfish δ6. Interestingly, the sequence similarity between mouse and human δ1 domains is also low (22%), although the similarity between mouse δ3 and human δ3 is higher, at 53% identity. As shown in Fig. 1B, these results support the homology of catfish δ1 with the δ1 domains of both mouse (11) and human (12), the homology of the catfish δ5 domain with the human δ2 domain (the mouse lacks δ2) and the homology of catfish δ6 with the δ3 domains of both mouse and human.
Figure 2.
Phylogram of relationships between vertebrate Ig C regions. Sequences were aligned using clustal, with identity residue weight table, gap penalty 10, and gap-length penalty 10. The tree was generated from this alignment using paup (bootstrapping, with branch swapping/tree bisection-reconnection), and the values supporting each node are derived from 100 resamplings.
Figure 3.
Comparison of the amino acid sequences of δ chains. Percent identity values from alignments of the individual domains (boxed) of catfish, mouse and human δ chains are shown.
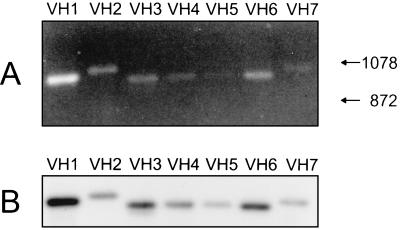
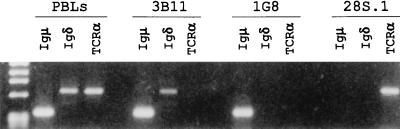
The cDNA clones showed that the catfish δ chain was expressed as a unique chimeric molecule containing the Cμ1 domain between the V and Cδ1 domains. This was unexpected, and analysis of the δ transcripts expressed in the pool of catfish B cells was undertaken to determine if δ mRNA could be expressed without the inclusion of Cμ1. Analysis by RT-PCR, using forward primers specific for each of the seven known catfish VH families, coupled with a reverse primer at the 3′ end of the Cδ1 exon (Fig. 4A), showed that (i) δ was expressed in combination with VH genes of each of the known families, and (ii) δ was expressed in mRNAs yielding products of the predicted sizes (≈980 bp) for messages containing Cμ1. The presence of Cμ1 in the PCR products was confirmed by blot transfer/hybridization using a Cμ1-specific probe (Fig. 4B), and by cloning and sequencing of the products from the RT-PCR reactions. At least three different PCR products were sequenced for each VH family and all contained typical catfish CDR3 regions (32). There appears to be no bias in diversity (D) or JH usage, i.e., cDNAs containing JH1, -2, -7, -8, and -9 segments (33) have been sequenced and short putative D sequences could be inferred by comparing the different cDNAs. No evidence was found for any catfish δ message that did not include Cμ1. Although these data showed the expression of δ mRNA in a pool of normal catfish B cells, they did not indicate if δ was coexpressed in the same cells as μ. To address this issue we utilized clonally derived long-term in vitro grown catfish B cell lines (25, 34). RT-PCR analysis clearly showed differential expression of μ and δ in these B cell lines. As shown in Fig. 5, B cell line 3B11 expressed mRNA for both μ and δ whereas the B cell line 1G8 expressed mRNA only for μ; similarly two other B cell lines (1B10 and 1D12) expressed μ only. The T cell line, 28S.1, did not express message for either μ or δ, but expressed message for TCRα as expected. As predicted, pooled freshly isolated PBLs expressed message for μ, δ, and TCRα. The δ and μ messages expressed by 3B11 cells were sequenced as cDNAs and found to share the identical VDJH rearrangements.
Figure 4.
Identification of catfish VH family usage in δ transcripts by RT-PCR. (a) RNA from freshly isolated PBLs was subjected to RT-PCR using forward primers specific for each of the seven described catfish VH families with a reverse primer at the 3′ end of the δ1 exon. The amplified products were electrophoresed on a 1% agarose gel and stained with ethidium bromide. The sizes of two HaeIII φX/174 DNA markers, 1078 and 872 (bp) are indicated by arrows. (b) Cμ1 sequences in the VH-δ1 PCR products were detected by blot hybridization analysis with a Cμ1 probe.
Figure 5.
Expression of μ, δ, and TCRα mRNA in freshly-isolated PBLs, B cell lines 3B11 and 1G8, and a T cell line 28S.1. The RT-PCR products were electrophoresed on a 1% agarose gel. The markers (HaeIII φX/174 DNA) shown from top to bottom at left are: 1353, 1078, 872, 601, 281/271, and 234 bp. The VDJH rearrangements for both μ and δ have been sequenced for the B cell line 3B11 (accession nos. U67440U67440 and U67439U67439).
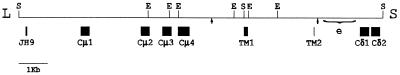
Although not directly shown, the coexpression of catfish δ and μ with the same VDJH, along with the inclusion of Cμ1 in all δ messages, suggests that δ is expressed by alternative pathways of pre-mRNA processing of a long primary transcript including both μ and δ exons. For such a transcript to be produced and processed, the δ gene must be situated adjacent to, and downstream of, the μ gene. Evidence that the δ gene is in such a position in the catfish IgH locus came from the genomic sequence downstream of the μ gene; exons δ1 and δ2 are 1580 and 1968 bp, respectively, 3′ of the TM2 exon of μ, as indicated in Fig. 6.
Figure 6.
Physical map of the catfish IgH locus showing the μ gene and exons 1 and 2 of the δ gene. A partial restriction endonuclease map of recombinant phage 12C (35) is shown, with the exons encoding the first four catfish μ C region domains (Cμ1–4), the alternative membrane-anchoring C terminus of μ (TM1 and -2), and the first two δ exons (Cδ1 and -2) marked by boxes below the line. S and E are SalI and EcoRI sites, respectively. The L and S outside of the map indicate long and short phage arms of the EMBL 3 vector. The cleavage/polyadenylylation signal sequences are indicated by arrows, and the brackets mark the region of the enhancer, e (36). Accession number for the sequence of phage 12C is X79482X79482.
The novel catfish gene identified here can thus be considered a homolog of mammalian Ig δ in terms of the following shared properties: (i) sequence relatedness, (ii) location immediately 3′ of μ in the IgH locus, (iii) separate C-terminal exons inferred for the secretory and membrane forms, and (iv) coexpression with μ in some, but not all B cells. However, catfish δ differs from its mammalian homologs in some significant ways. Structurally, it is unusual in that it includes the Cμ1 exon, and possesses a large number of Cδ domains i.e., seven (not including the secretory or membrane tail regions). We suggest that the presence of the Cμ1 domain in catfish δ permits normal assembly of catfish IgD with light chains. Although it has not been demonstrated directly that the catfish δ chain can associate with light chains to form an IgD molecule, it seems clear that Cδ1 would itself be incapable of covalently interacting with light chain. The cysteine expected to form the disulfide bond with light chain, which is found in the N-terminal region of Cμ1 domains, is missing in the catfish δ1 domain, being replaced with a serine at position 253 (Fig. 1B).
The reason for the large number of δ domains in the catfish is unknown: until we have some insight into the possible function of IgD in this species, it is difficult to speculate. Observations on IgD knockout mice have suggested that, in this species, its functional role is likely limited to features of immune regulation, such as accelerating and enhancing the maturation of a response (18). If IgD is relatively unimportant functionally, then the variation in the numbers of Cδ domains seen when comparing mouse, human, and catfish IgDs could result as a product of genetic drift. Certainly, IgD appears to have changed structurally more than any other Ig during vertebrate evolution. In the three groups known to express IgD (primates, rodents, and teleost fish) the δ chain has quite different structures, not only in terms of the number of C region domains, but in the absence of a hinge in the catfish molecule. This diversity of structures seen in IgD would be expected to affect its function as a receptor for antigen: while catfish IgD is predicted to have an elongated, stalk-like structure with relatively inflexible Fab regions, mammalian IgDs are much shorter with Fab arms that are predicted to be highly mobile.
If IgD has survived as an expressed Ig in certain mammals and fish, one question that remains is what has happened to the δ gene in other species that are intermediate in vertebrate evolution. Reports of antigenically or structurally novel Igs on the surface of B cells in species such as chicken and rabbit (37, 38) may indicate that δ genes exist but remain to be identified in other species. Further investigations are needed to determine if this is so, or if there have been deletions or inactivation of δ genes in those species from which IgD is believed to be absent. In this regard it should be noted that cross-hybridization has been observed using the channel catfish δ2 exon as a probe in Southern blot analyses with two closely related catfish species, the Brown Bullhead, I. nebulosus and the Blue Catfish, I. furcatus. In contrast no cross hybridizing bands were observed using DNA from mouse, turtle, Xenopus or non-Ictalurid fish species even under low stringency conditions (data not shown). The fish species assayed were the paddlefish (Polyodon spathula), bowfin (Amia calva), gar (Lepisosteus osseus), carp (Cyprinus carpio), Atlantic cod (Gadus morhua), Atlantic salmon (Salmo salar), chinook salmon (Oncorhynchus tshawytscha), coho salmon (O. kisutch), rainbow trout (O. mykiss), and the African lungfish (Protopterus sp.). The failure to detect d-related sequences in Southern blot analyses of DNA from distant species of fish may reflect either the evolutionary divergence of δ sequences, or the absence of the δ gene, in these species. Our identification of a δ homolog in a teleost fish is unlikely to shed light on the earliest origins of Igs. The presence of the δ gene immediately downstream of μ suggests that it arose as a duplication product of the μ gene. An additional question remains concerning IgD; if it was present early in vertebrate evolution, might its original function have been different from the one that remains in the mouse? Studies of IgD function in other species that express IgD, may provide answers to the question.
Acknowledgments
We thank Dr. Benjamin Normark of the Natural History Museum, London, for advice and comment on the phylogenetic analyses. The Basel Institute for Immunology was founded and is supported by F. Hoffmann La Roche AG Basel. This research was supported by grants from the National Institutes of Health (R37-AI-19530) and National Science Foundation (MCB-9406249).
ABBREVIATIONS
- H
heavy
- PBL
peripheral blood leukocyte
- V
variable region
- H
heavy chain
- C
constant
- D
diversity
- J
joining
- RT
reverse transcription
Footnotes
References
- 1.DuPasquier L. In: Fundamental Immunology. 3rd Ed. Paul W E, editor. New York: Raven; 1993. pp. 199–223. [Google Scholar]
- 2.Greenberg A S, Avila D, Hughes M, Hughes A, McKinney E C, Flajnik M F. Nature (London) 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 3.Greenberg A S, Hughes A L, Guo J, Avila D, McKinney E C, Flajnik M F. Eur J Immunol. 1996;26:1123–1129. doi: 10.1002/eji.1830260525. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein R M, Schluter S F, Shen S, Marchalonis J J. Proc Natl Acad Sci USA. 1996;93:3289–3293. doi: 10.1073/pnas.93.8.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding F A, Amemiya C T, Litman R T, Cohen N, Litman G W. Nucleic Acids Res. 1990;18:6369–6376. doi: 10.1093/nar/18.21.6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi K, Tomonaga S, Kajii T. Mol Immunol. 1984;21:397–404. doi: 10.1016/0161-5890(84)90037-3. [DOI] [PubMed] [Google Scholar]
- 7.Rowe D S, Fahey J L. J Exp Med. 1965;121:171–184. doi: 10.1084/jem.121.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abney E R, Parkhouse R M E. Nature (London) 1974;252:600–602. doi: 10.1038/252600a0. [DOI] [PubMed] [Google Scholar]
- 9.Martin L N, Leslie G A, Hindes R. Arch Allergy Appl Immunol. 1976;51:320–329. doi: 10.1159/000231606. [DOI] [PubMed] [Google Scholar]
- 10.Ruddick J H, Leslie G A. J Immunol. 1977;118:1025–1031. [PubMed] [Google Scholar]
- 11.Tucker P W, Liu C-P, Mushinski J F, Blattner F R. Science. 1980;209:1353–1360. doi: 10.1126/science.6968091. [DOI] [PubMed] [Google Scholar]
- 12.White M B, Shen A L, Word C J, Tucker P J, Blattner F R. Science. 1985;228:733–737. doi: 10.1126/science.3922054. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H-L, Blattner F R, Fitzmaurice L, Mushinski J F, Tucker P W. Nature (London) 1982;296:410–415. doi: 10.1038/296410a0. [DOI] [PubMed] [Google Scholar]
- 14.Blattner F R, Tucker P W. Nature (London) 1984;307:417–422. doi: 10.1038/307417a0. [DOI] [PubMed] [Google Scholar]
- 15.Thorbecke, G. J. & Leslie, G. A., eds. (1982) Ann. N.Y. Acad. Sci., 399–410.
- 16.Tisch R, Roifman C M, Hozumi N. Proc Natl Acad Sci USA. 1988;85:6914–6918. doi: 10.1073/pnas.85.18.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K M, Ishigami T, Hata D, Higaki Y, Morita M, Yamaoka K, Mayumi M, Mikawa H. J Immunol. 1992;148:29–34. [PubMed] [Google Scholar]
- 18.Roes J, Rajewsky K. J Exp Med. 1993;177:45–55. doi: 10.1084/jem.177.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitschke L, Kosco M H, Köhler G, Lamers M C. Proc Natl Acad Sci USA. 1993;90:1887–1891. doi: 10.1073/pnas.90.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swofford D L. paup: Phylogenetic Analysis Using Parsimony. Champagne, IL: Illinois Nat. Hist. Survey; 1993. , version 3.1.1. [Google Scholar]
- 22.McCumber L J, Capra J D. Mol Immunol. 1979;16:565–570. doi: 10.1016/0161-5890(79)90119-6. [DOI] [PubMed] [Google Scholar]
- 23.Flanagan J G, Lefranc M P, Rabbitts T H. Cell. 1984;36:681–688. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- 24.MacDonald R J, Swift G H, Przybyla A E, Chirgwin J M. Methods Enzymol. 1988;152:219–227. doi: 10.1016/0076-6879(87)52023-7. [DOI] [PubMed] [Google Scholar]
- 25.Miller N W, Rycyzyn M A, Wilson M R, Warr G W, Naftel J P, Clem L W. J Immunol. 1994;152:2180–2189. [PubMed] [Google Scholar]
- 26.Ventura-Holman T, Ghaffari S H, Lobb C. Eur J Immunogenet. 1996;23:7–14. doi: 10.1111/j.1744-313x.1996.tb00259.x. [DOI] [PubMed] [Google Scholar]
- 27.Warr G W, Miller N W, Clem L W, Wilson M R. Immunogenetics. 1992;35:253–256. doi: 10.1007/BF00166830. [DOI] [PubMed] [Google Scholar]
- 28.Kabat E A, Wu T T, Perry H M, Gottesman K S, Foeller G. Sequences of Proteins of Immunological Interest. Natl. Inst. Health, Washington, DC: U.S. Dept. Health Hum. Serv.; 1991. [Google Scholar]
- 29.Peterson M L, Perry R P. Mol Cell Biol. 1989;9:726–738. doi: 10.1128/mcb.9.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campbell K S, Backstrom B T, Tiefenthaler G, Palmer E. Semin Immunol. 1994;6:393–410. doi: 10.1006/smim.1994.1049. [DOI] [PubMed] [Google Scholar]
- 31.Mansikka A. J Immunol. 1992;149:855–861. [PubMed] [Google Scholar]
- 32.Ghaffari S H, Lobb C J. J Immunol. 1991;146:1037–1046. [PubMed] [Google Scholar]
- 33.Hayman J R, Ghaffari S H, Lobb C J. J Immunol. 1993;151:3587–3596. [PubMed] [Google Scholar]
- 34.Miller N W, Chinchar V G, Clem L W. J Tissue Cult Methods. 1994;16:117–123. [Google Scholar]
- 35.Wilson M R, Marcuz A, van Ginkel F, Miller N W, Clem L W, Middleton D, Warr G W. Nucleic Acids Res. 1990;18:5227–5233. doi: 10.1093/nar/18.17.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magor B G, Wilson M R, Miller N W, Clem L W, Middleton D L, Warr G W. J Immunol. 1994;153:5556–5563. [PubMed] [Google Scholar]
- 37.Chen C-L H, Lehmeyer J E, Cooper M D. J Immunol. 1982;129:2580–2585. [PubMed] [Google Scholar]
- 38.Wilder R L, Yuen C C, Coyle S A, Mage R G. J Immunol. 1979;122:464–468. [PubMed] [Google Scholar]