Abstract
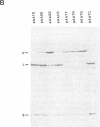
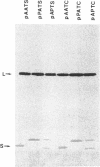
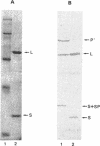
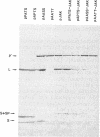
Penicillin G acylase from Escherichia coli ATCC 11105 is synthesized from its precursor polypeptide into a catalytically active heterodimer via a complex posttranslational processing pathway. Substitutions in the pair of aminoacyl residues at the cleavage site for processing the small and large subunits were made. Their processing phenotypes and penicillin G acylase activities were analyzed. By the introduction of a prolyl residue at either position, the processing of the small subunit was blocked without a change in enzymatic activity. Four other substitutions had no effect. At the site for processing the large subunit, four substitutions out of the seven examined blocked processing. In general, penicillin G acylase activity seemed to be proportional to the efficiency of the large-subunit-processing step. Ser-290 is an amino acid critical for processing and also for the enzymatic activity of penicillin G acylase. In the mutant pAATC, in which Ser-290 is mutated to Cys, the precursor is processed, but there is no detectable enzymatic activity. This suggests that there is a difference in the structural requirements for the processing pathway and for enzymatic activity. Recombination analysis of several mutants demonstrated that the small subunit can be processed only when the large subunit is processed first. Some site-directed mutants from which signal peptides were removed showed partial processing phenotypes and reduced enzymatic activities. Their expression showed that the prerequisite for penicillin G acylase activity is the efficient processing of the large subunit and that the maturation of the small subunit does not affect the enzymatic activity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BOMSTEIN J., EVANS W. G. AUTOMATED COLORIMETRIC DETERMINATION OF 6-AMINOPENICILLANIC ACID IN FERMENTATION MEDIA. Anal Chem. 1965 Apr;37:576–578. doi: 10.1021/ac60223a034. [DOI] [PubMed] [Google Scholar]
- Barbero J. L., Buesa J. M., González de Buitrago G., Méndez E., Péz-Aranda A., García J. L. Complete nucleotide sequence of the penicillin acylase gene from Kluyvera citrophila. Gene. 1986;49(1):69–80. doi: 10.1016/0378-1119(86)90386-0. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Daumy G. O., Danley D., McColl A. S., Apostolakos D., Vinick F. J. Experimental evolution of penicillin G acylases from Escherichia coli and Proteus rettgeri. J Bacteriol. 1985 Sep;163(3):925–932. doi: 10.1128/jb.163.3.925-932.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass J., Civelli O., Herbert E. Polyprotein gene expression: generation of diversity of neuroendocrine peptides. Annu Rev Biochem. 1984;53:665–715. doi: 10.1146/annurev.bi.53.070184.003313. [DOI] [PubMed] [Google Scholar]
- Fisher J. M., Scheller R. H. Prohormone processing and the secretory pathway. J Biol Chem. 1988 Nov 15;263(32):16515–16518. [PubMed] [Google Scholar]
- Garten W., Bosch F. X., Linder D., Rott R., Klenk H. D. Proteolytic activation of the influenza virus hemagglutinin: The structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981 Dec;115(2):361–374. doi: 10.1016/0042-6822(81)90117-3. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larder B., Purifoy D., Powell K., Darby G. AIDS virus reverse transcriptase defined by high level expression in Escherichia coli. EMBO J. 1987 Oct;6(10):3133–3137. doi: 10.1002/j.1460-2075.1987.tb02623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan P. B. Penicillin acylases. An update. Appl Biochem Biotechnol. 1984 Oct-Dec;9(5-6):537–554. doi: 10.1007/BF02798404. [DOI] [PubMed] [Google Scholar]
- Matsuda A., Komatsu K. I. Molecular cloning and structure of the gene for 7 beta-(4-carboxybutanamido)cephalosporanic acid acylase from a Pseudomonas strain. J Bacteriol. 1985 Sep;163(3):1222–1228. doi: 10.1128/jb.163.3.1222-1228.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda A., Toma K., Komatsu K. Nucleotide sequences of the genes for two distinct cephalosporin acylases from a Pseudomonas strain. J Bacteriol. 1987 Dec;169(12):5821–5826. doi: 10.1128/jb.169.12.5821-5826.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S. J., Kim Y. C., Park Y. W., Min S. Y., Kim I. S., Kang H. S. Complete nucleotide sequence of the penicillin G acylase gene and the flanking regions, and its expression in Escherichia coli. Gene. 1987;56(1):87–97. doi: 10.1016/0378-1119(87)90161-2. [DOI] [PubMed] [Google Scholar]
- Ohashi H., Katsuta Y., Hashizume T., Abe S. N., Kajiura H., Hattori H., Kamei T., Yano M. Molecular cloning of the penicillin G acylase gene from Arthrobacter viscosus. Appl Environ Microbiol. 1988 Nov;54(11):2603–2607. doi: 10.1128/aem.54.11.2603-2607.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson R. G., Harris T. J., Lamb R. A. Fusion protein of the paramyxovirus simian virus 5: nucleotide sequence of mRNA predicts a highly hydrophobic glycoprotein. Proc Natl Acad Sci U S A. 1984 Nov;81(21):6706–6710. doi: 10.1073/pnas.81.21.6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid A., Choppin P. W. Identification of biological activities of paramyxovirus glycoproteins. Activation of cell fusion, hemolysis, and infectivity of proteolytic cleavage of an inactive precursor protein of Sendai virus. Virology. 1974 Feb;57(2):475–490. doi: 10.1016/0042-6822(74)90187-1. [DOI] [PubMed] [Google Scholar]
- Schumacher G., Sizmann D., Haug H., Buckel P., Böck A. Penicillin acylase from E. coli: unique gene-protein relation. Nucleic Acids Res. 1986 Jul 25;14(14):5713–5727. doi: 10.1093/nar/14.14.5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnick T. M., Lerner R. A., Sutcliffe J. G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981 Oct 15;293(5833):543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- Sizmann D., Keilmann C., Böck A. Primary structure requirements for the maturation in vivo of penicillin acylase from Escherichia coli ATCC 11105. Eur J Biochem. 1990 Aug 28;192(1):143–151. doi: 10.1111/j.1432-1033.1990.tb19207.x. [DOI] [PubMed] [Google Scholar]
- Slade A., Horrocks A. J., Lindsay C. D., Dunbar B., Virden R. Site-directed chemical conversion of serine to cysteine in penicillin acylase from Escherichia coli ATCC 11105. Effect on conformation and catalytic activity. Eur J Biochem. 1991 Apr 10;197(1):75–80. doi: 10.1111/j.1432-1033.1991.tb15884.x. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]