Abstract
Cap formation is the first step of pre-mRNA processing in eukaryotic cells. Immediately after transcription initiation, capping enzyme (CE) is recruited to RNA polymerase II (Pol II) by the phosphorylated carboxyl-terminal domain of the Pol II largest subunit (CTD), allowing cotranscriptional capping of the nascent pre-mRNA. Recent studies have indicated that CE affects transcription elongation and have suggested a checkpoint model in which cotranscriptional capping is a necessary step for the early phase of transcription. To investigate further the role of the CTD in linking transcription and processing, we generated a fusion protein of the mouse CTD with T7 RNA polymerase (CTD-T7 RNAP). Unexpectedly, in vitro transcription assays with CTD-T7 RNAP showed that CE promotes formation of DNA·RNA hybrids or R loops. Significantly, phosphorylation of the CTD was required for CE-dependent R-loop formation (RLF), consistent with a critical role for the CTD in CE recruitment to the transcription complex. The guanylyltransferase domain was necessary and sufficient for RLF, but catalytic activity was not required. In vitro assays with appropriate synthetic substrates indicate that CE can promote RLF independent of transcription. ASF/SF2, a splicing factor known to prevent RLF, and GTP, which affects CE conformation, antagonized CE-dependent RLF. Our findings suggest that CE can play a direct role in transcription by modulating displacement of nascent RNA during transcription.
Keywords: RNA displacement, RNA polymerase II, RNA processing
RNA polymerase II (Pol II) plays a critical role not only in the transcription of mRNA precursors, but also in their subsequent processing. The best characterized example is cotranscriptional cap formation, which is achieved by the physical interaction of capping enzymes (CEs) with the phosphorylated carboxyl-terminal domain of the Pol II largest subunit (CTD) (1–5). The CTD, composed mainly of a repeated heptapeptide motif, YSPTSPS, is extensively phosphorylated, and its regulation is tightly controlled during the transcription cycle (6–8). Phosphorylation of serine-5 in the heptad repeats, by Cdk7 in human and Kin28 in yeast, occurs shortly after transcription initiation and is required for cotranscriptional recruitment of CE (9, 10).
The formation of the 5′-terminal m7G(5′)ppp(5′)N cap is the first step of pre-mRNA processing and involves a series of three enzymatic reactions. RNA triphosphatase (RTPase) removes a phosphate from the 5′ end of the nascent transcript, RNA guanylyltransferase (GTase) transfers GMP from GTP to the diphosphate RNA terminus, and RNA (guanine-N7) methyltransferase (MTase) adds a methyl group (11). In budding yeast, capping is carried out by three polypeptides that are encoded separately (12). Cet1 (RTPase) interacts with Ceg1 (GTase), and this interaction is critical for stimulating Ceg1 activity (13). Abd1 (MTase) and Ceg1 bind directly to the phosphorylated CTD (2, 3). Mammalian CEs consist of two components, a bifunctional CE (with an N-terminal RTPase domain and C-terminal GTase domain) and an MTase. The CE GTase domain binds directly to the phosphorylated CTD (1).
Several proteins interact with CE and affect its catalytic activities. Although the CTD phosphorylated at either serine-2 or serine-5 is capable of binding CE, only the serine-5-phosphorylated CTD stimulates GTase catalytic activity (14). A Pol II elongation factor, Spt5, interacts with CE and also stimulates GTase activity (15). HIV-encoded Tat interacts with both RTPase and GTase domains of CE and stimulates their catalytic activities (5).
A number of studies suggesting that CE and/or MTase function in transcription have led to models involving interplay between 5′ capping and transcription elongation near the promoter (7). Early in vitro experiments suggested that cap formation, specifically methylation, could play a stimulatory role in transcription (16). More recent studies have shown that CE can act as a transcriptional regulator. In vitro experiments in yeast demonstrated that Cet1 represses transcription reinitiation (17), whereas mutation of ceg1 and abd1 affects transcription elongation (18, 19). Finally, human CE is capable of relieving transcription repression by the negative elongation factor (NELF) (20). These data suggest that CE plays a role in regulating an early phase of transcription elongation and provides an attractive explanation for coupling cap formation to transcription: to provide a checkpoint to ensure that nascent RNA is properly modified and that only such transcripts are extended further (7). However, the precise mechanisms by which CE contributes to transcription elongation remain enigmatic. Several studies have indicated that both wild-type and catalytically inactive CEs are capable of affecting transcription elongation (17, 19, 20). These data support the hypothesis that recruitment of CE to the transcription machinery, rather than formation of the cap structure, is critical for its function in elongation. It is then possible that other properties of CE (e.g., an ability to interact with RNA) (17, 21) play a role in transcription.
Here, we describe an unexpected function for mammalian CE. Our findings arose from experiments designed initially to investigate properties of the CTD. For these studies, we created a mouse CTD-T7 RNA polymerase (CTD-T7 RNAP) fusion protein for use in in vitro transcription assays. We found that when the CTD moiety was phosphorylated, addition of human CE specifically promoted hybridization of the nascent transcript with template DNA, forming transcriptional R loops. The GTase domain, but not catalytic activity, was found to be necessary and sufficient for R-loop formation (RLF). Assays with presynthesized RNA and DNA indicate that CE can facilitate RLF independent of transcription. This activity was inhibited by GTP, but not other nucleotides, suggesting that the Gp-GTase and/or GTP-bound conformation of CE is inactive, and ASF/SF2, an SR protein splicing factor that maintains genomic stability by preventing RLF (22), antagonized CE-dependent RLF. These experiments suggest an unexpected function for CE during transcription.
Results
Preparation of Mouse CTD-T7 RNA Polymerase Fusion Proteins.
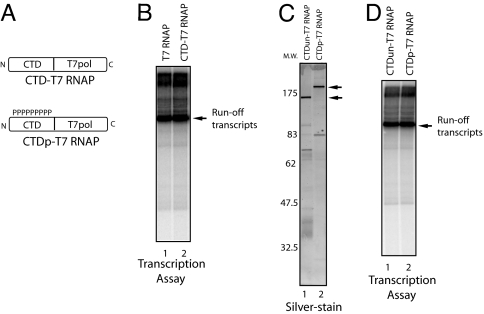
T7 RNAP, unlike most other RNA polymerases, is a single subunit polymerase, and a relatively short promoter element is sufficient to initiate accurate transcription (23). Nonetheless, the mechanism of T7 RNAP transcription shares significant similarities with multisubunit RNA polymerases, and it is likely that fundamental aspects of transcription are maintained between T7 RNAP and multisubunit RNA polymerases (24, 25). To take advantage of its simplicity to study the functions of the phosphorylated CTD in transcription and RNA processing, we constructed a plasmid that expresses His-tagged T7 RNAP (26) fused with the mouse Pol II CTD. In an initial experiment, T7 RNAP fused to the CTD at its C terminus and purified from Escherichia coli had no detectable transcription activity (data not shown). This finding might be due to protein misfolding caused by the fusion of the CTD in close proximity to the catalytic domain (27). We next fused the CTD to the N terminus of T7 RNAP (CTD-T7 RNAP) (Fig. 1A). Bacterially expressed and purified T7 RNAP and CTD-T7 RNAP produced comparable levels of runoff transcripts (Fig. 1B), indicating that fusion of the CTD to the N terminus caused no apparent defect in transcription. Therefore, we used this fusion protein in subsequent experiments. We next prepared phosphorylated CTD-T7 RNAP (CTDp-T7 RNAP) by incubation with HeLa nuclear extract as a source of kinases. We found that CTD-T7 RNAP, but not T7 RNAP, was phospholabeled, indicating that the CTD moiety is specifically phosphorylated, containing both serine-2 and -5 phosphorylation (data not shown) (Fig. 1A) (44). To prepare unphosphorylated CTD-T7 RNAP (CTDun-T7 RNAP), Mg2+ and ATP were omitted from the kinase reaction. These preparations were repurified on Ni-NTA beads (Fig. 1C). Repurified CTDun-T7 and CTDp-T7 RNAP produced similar levels of runoff transcripts (Fig. 1D).
Fig. 1.
T7 RNAP-CTD fusion proteins. T7 RNAP fused with mouse CTD. (A) Schematic representation of T7 RNAP fused with the CTD (CTD-T7 RNAP) and phosphorylated CTD (CTDp-T7 RNAP) are shown. (B) In vitro transcription by T7 RNAP and CTD-T7 RNAP (≈100 ng, respectively) with linearized plasmid DNA. Purified RNAs were resolved by denaturing PAGE. Runoff transcripts are indicated by arrow. (C) The 120-ng protein samples were resolved by SDS/PAGE and silver-stained. Molecular marker is indicated on the left. (D) In vitro transcription by CTDun-T7 RNAP and CTDp-T7 RNAP (≈120 ng, respectively) with linearized plasmid DNA.
Human CE Promotes Transcriptional R Loops in Vitro.
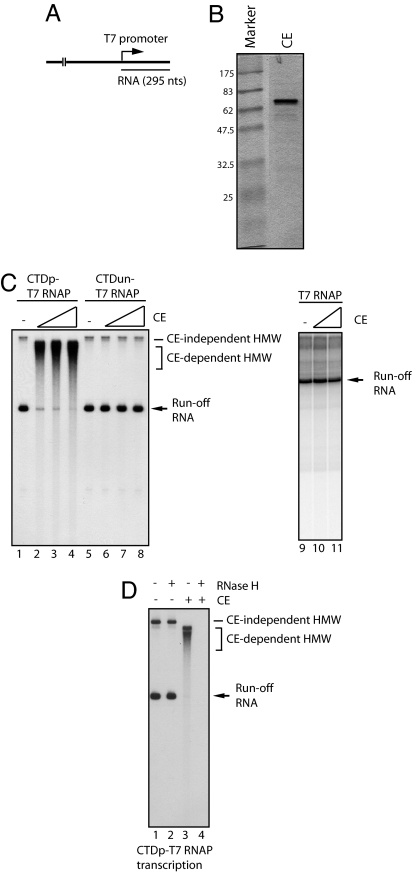
To investigate functions of CE during transcription and RNA processing, we performed in vitro transcription assays with CTDp-T7 RNAP and a linearized plasmid (Fig. 2A). Unexpectedly, we found that increasing amounts of human CE (see Fig. 2B and ref. 21) led to the accumulation of high-molecular-weight (HMW) transcripts and a reduction of runoff transcripts (Fig. 2C, lanes 1–4). Noting that the mobility of HMW transcripts induced by CE differed significantly from those of HMW transcripts formed without CE (compare lanes 1 and 2–4; see CE-independent and CE-dependent HMW transcripts), CE in some way stimulates accumulation of specific HMW transcripts. Furthermore, the CE-dependent HMW transcripts were not observed in assays with CTDun-T7 RNAP (Fig. 2C, lanes 5–8) or T7 RNAP (lanes 9–11). These results suggest that CE bound to the phosphorylated CTD caused accumulation of the HMW transcripts in a cotranscriptional manner.
Fig. 2.
Human CE promotes cotranscriptional RLF. (A) Template DNA for in vitro transcription driven by T7 promoter. Plasmid DNA was linearized by restriction enzyme digestion. The length of runoff transcripts is 295 nt. (B) The 1.2 μg of purified human CE was resolved by SDS/PAGE and visualized by Coomassie staining. (C) CE promotes the formation of HMW transcripts. In vitro transcription with CTDp-T7 RNAP was performed with increasing amounts (0, 60, 150, and 300 ng) of CE (lanes 1–4). Transcription by CTDun-T7 RNAP with increasing amounts (0, 60, 150, and 300 ng) of CE is shown in lanes 5–8. Transcription by T7 RNAP with increasing amounts (0, 60, and 300 ng) of CE is shown in lanes 9–11. CE-independent and CE-dependent HMW are indicated. The arrow indicates runoff transcripts. (D) HMW transcripts are DNA·RNA hybrids. Transcription of CTDp-T7 RNAP was performed in the presence (lanes 3 and 4) or absence (lanes 1 and 2) of 300 ng of CE. Transcription was terminated by heat (70°C, 15 min). Subsequently reaction mixtures were treated with (lanes 2 and 4) or without (lanes 1 and 3) RNase H, and RNAs were resolved by denaturing PAGE.
We next wished to characterize the CE-dependent HMW transcripts. Given that CTDp-T7 RNAP initiated transcription specifically from the T7 promoter (Fig. 2C, lane 1), it seemed unlikely that synthesis of HMW RNA resulted from nonspecific transcription. One possibility we considered was that the nascent transcripts hybridized with the template DNA strand, thereby migrating slower than expected, even in the denaturing gels used. To test this idea, reaction mixtures were treated with RNase H to degrade the RNA moiety of putative RNA·DNA hybrids. Strikingly, the HMW transcripts were completely digested by RNase H (Fig. 2D, lane 4), whereas the amount of runoff transcripts was unchanged (Fig. 2D, lane 2). These experiments indicate that the CE-dependent HMW transcripts consist of DNA·RNA hybrids or R loops (22, 28). The CE-dependent HMW transcripts are not sequence-dependent because they were observed with both orientations of the template (data not shown). Additionally, the SR protein ASF/SF2, which also is known to interact with the CTD, was unable to induce RLF and could indeed prevent it (22), indicating that not any CTD-binding protein can induce R loops.
RLF by Human CE.
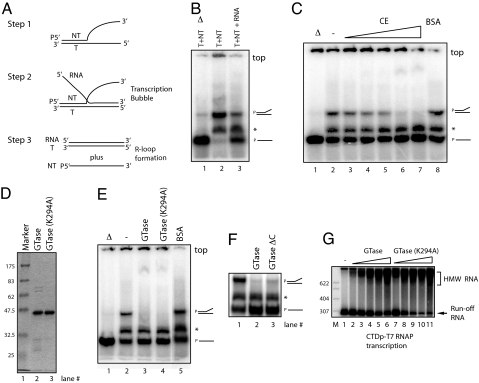
The prior experiments suggested that CE is capable of stimulating RLF during transcription. To obtain additional evidence for this, we developed an assay for RLF that would be independent of transcription. Transcription-dependent R loops can be formed when RNA polymerase fails to displace the nascent transcripts from the template DNA strand (29). As a consequence, a DNA·RNA duplex and the separated nontemplate strand remain after passage of the polymerase. RLF also can take place at a locally denatured DNA duplex that encloses an RNA·DNA hybrid (or bubble) (30). To construct a bubble-like structure independent of de novo transcription, we prepared two short stretches of an oligonucleotide (template and nontemplate strands) that partially hybridize in the first half of the DNA strands, whereas the second half of the two strands is entirely noncomplementary (Fig. 3A, step 1). RNA was then added to hybridize with the second half of the template strands (step 2). The resultant substrates have an ability to form an R-loop-like structure because the RNA can form a duplex with the full-length template strand if the nontemplate strand is displaced (step 3).
Fig. 3.
The GTase domain of human CE promotes RLF. (A) Experimental strategy for the RLF assay. Template strand (T), nontemplate strand (NT), and RNA are indicated. NT strand was labeled at its 5′ end (P). (B) RLF assay. Partially hybridized T and NT strands are shown (lane 2). Addition of RNA resulted in spontaneous RLF, converting ≈60% of NT into single-stranded NT (ssNT, lane 3). Heat-denatured products as molecular makers are shown in lane 1. Partially hybridized T and NT strands and ssNT are indicated on the right side. Asterisk indicates products made by aberrant T and NT hybridization. Top of the gel is indicated. (C) CE promotes RLF. RLF assays were performed with increasing amounts (0, 20, 50, 100, 200, and 400 ng) of CE (lanes 2–7) or 400 ng of BSA (lane 8). (D) Purified wild-type human GTase and GTase(K294A) (2 μg, respectively) were resolved by SDS/PAGE and visualized by Coomassie staining. (E) RLF assays were performed with 200 ng of wild-type human GTase (hGTase, lane 3), hGTaseK294A (lane 4), or BSA (lane 5). (F) RLF assays were performed with 200 ng of each GTase (lane 2) and GTase-active mutant having a C-terminal truncation of 28 aa (lane 3). (G) In vitro transcription with CTDp-T7 RNAP was performed with increasing amounts (30, 60, 90, 120, and 150 ng) of hGTase (lanes 2–6) or hGTaseK294A (lanes 7–11). Samples were resolved by nondenaturing PAGE in B, C, E, and F and by denaturing PAGE in G.
We next asked whether CE could enhance RLF in the prior assay. We first observed that addition of the RNA alone resulted in reduction of partially hybridized DNA strands and accumulation of a single-stranded nontemplate strand, indicating that spontaneous RLF occurred (Fig. 3B, lanes 2 and 3). This finding might be because RNA·DNA hybrids are typically more stable than the corresponding DNA·DNA hybrid (31). We did not find triplet structures (Fig. 3A, step 2) in this assay (Fig. 3B, lane 3). However, we found that addition of the splicing factor ASF/SF2 appears to stabilize such structures. We also observed minor products (indicated by asterisks) presumably made by aberrant T and NT hybridization. These products were unaffected in subsequent assays. Importantly, and consistent with the transcription assays we showed earlier (Fig. 2C), we found that addition of CE promoted RLF in a concentration-dependent manner (Fig. 3C, lanes 2–7), whereas equivalent amounts of BSA had no effect (Fig. 3C, lane 8). As expected, CE-dependent RLF required RNA and could also be observed with labeled RNA instead of DNA (data not shown). These experiments suggest that this assay recapitulates the results obtained with the transcription-coupled assay, and we therefore further analyzed CE-dependent RLF by using this assay system.
GTase Domain of CE Is Sufficient to Facilitate RLF.
To investigate which domains of CE are minimally required for RLF, we examined truncated CE derivatives in RLF assays. The previous data indicating that CTD phosphorylation is necessary for CE-dependent RLF during transcription suggested that the GTase domain is likely involved in RLF. Indeed, we also observed that a bacterially expressed and purified GTase domain from CE (1, 21) was sufficient to induce RLF (Fig. 3E, lane 3) in a concentration-dependent manner (data not shown). A GTase-active mutant lacking 28 C-terminal residues also stimulated RLF (Fig. 3F). To determine whether GTase activity is required, we next analyzed a catalytically inactive mutant. The GTase derivative GTaseK294A possesses an alanine mutation at lysine-294, which is the residue that forms the covalent enzyme–GMP intermediate (1). Significantly, recombinant GTaseK294A also supported RLF (Fig. 3E, lane 4). Consistent with these experiments, transcription assays with CTDp-T7 RNAP showed that both GTase and GTaseK294A caused comparable levels of RLF (Fig. 3G). These experiments indicate that the GTase domain of CE is sufficient to facilitate RLF, and the lysine residue that covalently links to GMP is dispensable.
CE-Dependent RLF Is Suppressed by ASF/SF2.
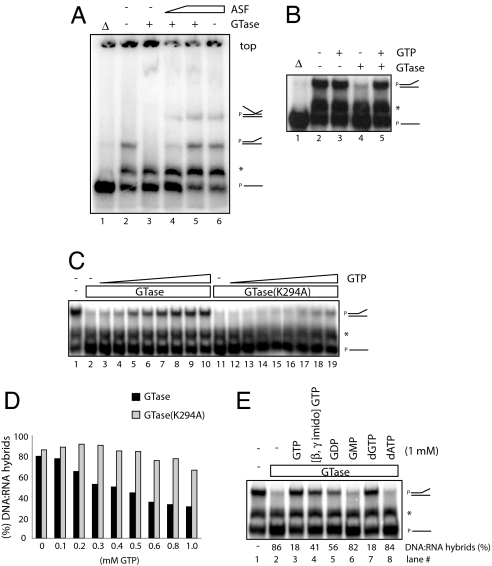
We next asked whether other proteins might affect RLF, either positively or negatively, in the RLF assay. ASF/SF2, a splicing factor that possesses RNA-binding activity (32), maintains genomic stability in vivo by preventing RLF (22). Therefore, it is plausible that ASF/SF2 might antagonize the effect of CE. Indeed, CE-dependent RLF was prevented by recombinant ASF/SF2 (33) in a dose-dependent manner (Fig. 4A, lanes 3–5; see the second lowest mobility products). Interestingly, we found that addition of ASF/SF2 resulted in accumulation of distinct products (the lowest mobility products; lanes 4–6), which likely correspond to stabilized triplet structures. These experiments indicate that the ability of CE to promote RLF is not common to other, more typical RNA-binding proteins, such as ASF/SF2, and indeed this activity can be suppressed by ASF/SF2.
Fig. 4.
Characterization of CE-dependent RLF. (A) ASF/SF2 prevents CE-dependent RLF. RLF assays were performed with 200 ng of hGTase in the presence of increasing amounts (0, 70, and 140 ng) of ASF/SF2 (lanes 3–5). The effect of 140 ng of ASF-SF2 alone is shown in lane 6. Putative triplet structures containing all three nucleic acid strands correspond to the lowest mobility products. (B) GTP inhibits CE-dependent RLF. RLF assays were performed with 200 ng of wild-type GTase alone (lane 4) or plus 1 mM GTP (lane 5); 1 mM GTP alone is shown in lane 3. (C) Wild-type, but not mutant, GTase is a target for GTP-mediated inhibition of RLF. RLF assays were performed with 200 ng of hGTase (lanes 2–10) or hGTaseK294A (lanes 11–19) in the presence of increasing amounts (0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.8, and 1.0 mM) of GTP. (D) Quantitation of DNA·RNA hybrids shown in C. The amounts of DNA·RNA hybrids caused by CE were quantitated by the ratio of hybridized template strands in the presence or absence of hGTase. (E) (Upper) RLF assays with 200 ng of hGTase plus 1 mM GTP, β,γ-imido-GTP, GDP, GMP, dGTP, and dATP were examined. (Lower) Quatitations of RLF are shown.
CE-Dependent RLF Is Suppressed by GTP.
Crystal structures of the Chlorella virus RNA GTase have illustrated the molecular mechanism for catalysis, in which a large conformational change occurs upon GTP binding, shifting the structure from an open to a closed state (34). To investigate whether such a conformational change might affect CE activity in RLF, we performed RLF assays in the presence of substrates required for GTase catalysis. Strikingly, we found that 1 mM GTP prevented RLF with wild-type GTase (Fig. 4B, lanes 4 and 5), whereas GTP alone had no effect (lanes 2 and 3). Increasing amounts of GTP showed that half-saturation was achieved at ≈0.6 mM GTP (Fig. 4C, lanes 2–10). We note that the GTP concentration in the transcription assays we performed earlier (e.g., Fig. 2B) was 50 μM, which is significantly less than in the RLF assays, and thus would not be sufficient to inhibit RLF. Importantly, GTP prevented RLF with the GTaseK294A mutant significantly less than with wild-type GTase (Fig. 4D and lanes 5–10 and 14–19 in Fig. 4C). These data suggest that the GTase catalytic site at least partially overlaps the region required for RLF.
Although the substrate specificity of GTase for catalyzing the transfer of GMP from GTP to the 5′-diphosphate end of RNA has been established (12), the substrate specificity for nucleotide triphosphate binding to GTase has not been investigated. Nonetheless, we next asked whether the ability of GTP to block CE-dependent RLF is unique to GTP. We found that, although both GTP and dGTP strongly inhibited CE-dependent RLF (Fig. 4E, lanes 3 and 7), nonhydrolyzable GTP (β,γ-imido-GTP) and GDP inhibited partially (lanes 4 and 5), and GMP and dATP had no inhibitory effect (lanes 6 and 8). ATP inhibited partially, whereas CTP and UTP were nearly inactive (data not shown). These data suggest that the overall structure of GTP is important for the inhibition of CE-dependent RLF, whereas GTP hydrolysis is not essential but stimulatory. Taken together, these results suggest that CE bound to GTP is inactive in RLF possibly because of the conformational change from an open to a closed state.
Discussion
Numerous studies have provided evidence that the Pol II CTD plays a central role in the coordination of transcription and pre-mRNA processing events (35). This function is accompanied by direct or indirect associations with a variety of mRNA processing factors, including 5′ CEs, splicing factors, and several components of the 3′ end-processing machinery (2, 3, 36, 37). We initially designed an assay employing CTD-T7 RNAP to investigate properties of CTD-associating factors during transcription and pre-mRNA processing. In this study, we described a function of CE in which the GTase domain promotes formation of transcriptional R loops in vitro. Furthermore, phosphorylation of the CTD is required for CE-dependent transcriptional RLF, providing evidence that the CTD plays an important role in CE recruitment to the transcription complex. Next we discuss the possible mechanisms by which RLF is stimulated by CE and physiological roles of this activity in Pol II transcription.
A transcriptional R loop is a structure in which nascent transcripts are partially or completely hybridized with the template strand of a double-stranded DNA, leaving the nontemplate strand unpaired. An R-loop structure was indeed observed in early studies of Pol II transcription. Purified calf thymus Pol II was found to generate extensive RNA·DNA hybrids during in vitro transcription (29). Additionally, the structure of the template DNA (i.e., supercoiled vs. linear) significantly influences the formation and size of RNA·DNA hybrids within and outside of the transcription bubble (38, 39). G-rich transcripts also are known to facilitate RLF (28, 40, 41). These data suggest that newly synthesized RNA can have an inherent capacity to form DNA·RNA hybrids extending from the transcription active site of the RNA polymerase. However, structural studies have shown that the nascent transcript that has emerged from the exit channel of the polymerase has already separated from the template DNA strand (42, 43), implying that R loops would not be directly extended from the transcription bubble. Therefore, the mechanism by which transcriptional R loops are generated remains unclear.
Enzymes that facilitate formation of R loops appear to be rare. To our knowledge, only one other enzyme has been shown to possess this activity, the E. coli RecA protein (44, 45). However, this activity almost certainly involves a different mechanism than that used by CE, in which RecA polymerizes along double-strand DNA and the nucleoprotein structure then pairs with and exchanges a complementary RNA. How CE facilitates RLF is unknown, but it is intriguing that it reflects an activity of the GTP-free, open form of the enzyme (34). This finding suggests that capping and RLF activities are mutually exclusive, consistent with the fact that they both use the GTase domain.
CE promotes formation of transcriptional R loops that are stable even during denaturing PAGE, which suggests that the RNA·DNA hybrids are relatively extensive. Given that transcriptional R loops are deleterious to genome stability in vivo (22, 46) except in the exceptional case involving transcription of S regions during class-switch recombination (28), it is unlikely that CE generally leads to extensive RLF in vivo. Although the physiological roles of CE-dependent RLF remain to be elucidated, one possibility is that CE modulates RNA displacement from template DNA during transcription in a way that affects Pol II elongation. Given that overextended DNA·RNA hybrid formation can negatively regulate the stability and processivity of both T7 RNAP and the Pol II elongation complexes (ECs) (39, 47), CE would thus possess an ability to destabilize, stall, or in some other way modulate the Pol II EC.
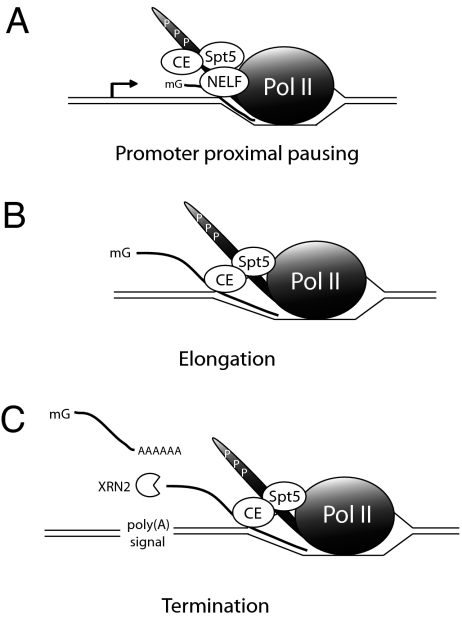
Our data have shown that a phosphorylated CTD and, as a result, CE recruitment to the transcription complex are critical for RLF during transcription. In yeast, ChIP experiments indicate that Ceg1, the counterpart of mammalian GTase, is released from the transcription machinery soon after cotranscriptional capping (9). Thus, CE-dependent RLF in yeast would only occur, if at all, at an early phase of transcription. Indeed, a ceg1 mutant strain with diminished GTase activity showed reduced Pol II elongation at promoter-proximal regions (18). It is conceivable that this process in some way involves RLF. However, temperature-sensitive mutants of Ceg1 that prevent capping had no change in the amount of Pol II recruitment to the 5′ ends of several genes (19). Although the Spt5–CE interaction is conserved in yeast (48), in mammals the Spt5-containing DSIF complex works together with the multisubunit NELF complex to modulate transcriptional pausing (49). An intriguing, albeit speculative, possibility is that the DSIF complex might modulate RLF by CE (see Fig. 5).
Fig. 5.
Possible roles of CE-dependent RLF during transcription in mammals. (A) The Spt5-containing DSIF complex, with NELF, functions to modulate transcriptional pausing at promoter-proximal regions. The DSIF complex and/or cap formation at the pre-mRNA 5′ end modulates RLF by CE. (B) Soon after promoter clearance, NELF dissociates from the Pol II EC, and CE acquires an ability to facilitate transient RLF to modulate elongation. (C) After transcription past the poly(A) site, RLF by CE induces transcriptional pausing. This finding could help to facilitate both cotranscriptional polyadenylation as well as subsequent transcription termination
Recent results raise the possibility that CE also can function downstream during elongation or even termination. Specifically, ChIP experiments have shown that CE, unlike Ceg1 in yeast, associates with active genes not only at the promoter, but also along the length of the gene, with a significant accumulation near the 3′ end (K. Glover and D. Bentley, personal communication). DSIF (but not NELF) also has been found in downstream regions (50). These findings raise the possibility that CE might facilitate transient RLF during transcription as a mechanism of modulating elongation (see Fig. 5).
The presence of CE at the 3′ ends of genes raises the intriguing possibility that CE functions in transcription termination. A precedent for this finding is provided by the vaccinia virus CE, which functions, together with a virus-encoded ATPase, to terminate the synthesis of viral transcripts by vaccinia RNA polymerase (51). How CE and RLF might contribute to termination is unclear. One possibility is that formation of R loops downstream of the poly(A) signal induces Pol II pausing (Fig. 5). Considerable evidence suggests that such pausing is important for subsequent termination (52, 53).
In any event, our findings provide an unexpected role for CE, in which its GTase domain promotes formation of transcriptional R loops in vitro. Recent data have indicated that CE plays roles in Pol II transcription and have provided a checkpoint model in which capping of the pre-mRNA 5′ end acts as a safeguard to ensure that only properly modified nascent transcripts can extend further. Now our data reveal potential roles for CE in transcription elongation, in which CE would regulate the processivity of the Pol II EC by modulating RNA displacement. Thus, our observations provide an unanticipated activity for CE, as well as a possible mechanism by which transcription and pre-mRNA processing events are connected.
Materials and Methods
Plasmids.
The plasmid (pBH161)-expressing His-tagged T7 RNAP was a gift from W. McAllister (State University of New York Downstate Medical Center, Brooklyn, NY) (26). For expression of CTD-T7 RNAP, which consists of the His-tagged CTD from mouse Pol II fused to the N terminus of T7 RNAP, the mouse CTD generated by PCR was inserted into pBH161 with XhoI sites (pBH161-CTD). Template DNA, which contains the human complement gene C2 poly(A) signal and MAZ termination signal, was generated by PCR. PCR products were inserted into pBluescript SK (Stratagene, La Jolla, CA) in both orientations.
Expression and Purification of Recombinant Proteins.
E. coli BL21 transformed with pBH161-CTD was grown at 15°C in LB medium containing 200 μg/ml ampicillin and induced by addition of 0.5 mM isopropyl β-d-thiogalactoside after the culture reached A600 = ≈0.4. Cells were harvested after 18 h and disrupted by sonication in binding buffer [50 mM Tris (pH 7.9)/100 mM NaCl/0.5% Nonidet P-40/5 mM imidazole/0.5 mM PMSF]. Soluble proteins were incubated with Ni-NTA agarose (Qiagen, Valencia, CA) equilibrated with binding buffer. The agarose was washed with Wash buffer [50 mM Tris (pH 7.9)/0.5 M NaCl/20 mM imidazole/0.5 mM PMSF]. Bound proteins were eluted with elution buffer [50 mM Tris (pH 7.9)/0.5 M NaCl/100 mM imidazole/0.5 mM PMSF]. Eluted proteins were dialyzed against buffer D [20 mM Hepes (pH 7.9)/100 mM KCl/0.25 mM EDTA/0.5 mM DTT/0.5 mM PMSF/20% glycerol]. Purified CTD-T7 RNAP was phosphorylated by incubation with HeLa nuclear extract and repurified under native conditions described in ref. 54. Eluted fractions of CTDun-T7 RNAP and CTDp-T7 RNAP were further purified by ion exchange chromatography by using DEAE-5PW (Tosoh, Tokyo, Japan). Expression and purification of CE, human GTase, GTaseK294A (1, 21), and ASF/SF2 (33) were previously described.
Transcription Assays.
The 12.5-μl transcription assays were carried out in reaction mixtures containing 1 μg of linearized plasmid DNA, 2.5% poly(vinyl alcohol), 6 mM MgCl2, 1 mM DTT, 50 μM NTPs, 4 μCi (1 Ci = 37 GBq) of 400 Ci/mmol [α-32P]UTP, 6.4 mM Hepes (pH 7.9), 6.4% glycerol, 32 mM KCl, 80 μM PMSF, 80 μM EDTA (pH 8.0), ≈100 ng of CTDp-T7 RNAP, and indicated amounts of CEs. The reaction mixtures were preincubated for 15 min at 4°C and incubated for an additional 30 min at 30°C. In Fig. 2D, transcription was terminated by heat (70°C, 15 min). Subsequently, reaction mixtures were treated with 0.75 units of RNase H (Promega, Madison, WI) for 15 min at 37°C. After proteinase K treatment, RNA products were extracted by phenol/chloroform and fractionated on 6% polyacrylamide/8.3 M urea gels. Radioactive products were analyzed by PhosphorImager.
RLF Assay.
The oligonucleotide sequences used were 5′-GAATACAAGCTTGCATGCCTGCAGAGTACTGTGCACAACGTTTT-3′ (nontemplate DNA) and 5′-GGGGATCCTCTAGAGTCGACCTCGAGGCATGCAAGCTTGTATTC-3′ (template DNA). RNA was synthesized by in vitro transcription with SP6 RNA polymerase (Promega) of SmaI-digested pGEM3 (Promega) and purified by PAGE. Nontemplate DNA was labeled at its 5′ end with 32P by T4 DNA polynucleotide kinase (New England Biolabs, Ipswich, MA) and [γ-32P]ATP. To prepare substrates, the mixture containing 25 nM nontemplate DNA, 25 nM template DNA, and annealing buffer [10 mM Tris (pH 7.9)/100 mM NaCl] was heated at 70–75°C for 2 min and allowed to cool slowly to room temperature. Then 10-μl reaction mixtures were preincubated with 25 fmol of prehybridized template and nontemplate DNA/25 fmol of RNA/2% poly(vinyl alcohol)/5 mM DTT/8 mM Hepes (pH 7.9)/8% glycerol/40 mM KCl/0.1 mM PMSF/0.1 mM EDTA (pH 8.0) for 10 min at 4°C. After the addition of indicated amounts of CEs, the reaction mixtures were incubated for 20 min at 30°C. The reaction was stopped by SDS. Samples were analyzed by 5% nondenaturing PAGE and analyzed by PhosphorImager.
Acknowledgments
We thank W. McAllister, C. David, and S. Millhouse for providing reagents, and I. Boluk for help with the manuscript. This work was supported by National Institutes of Health Grant GM28983 to (J.L.M.).
Abbreviations
- CE
capping enzyme
- CTD
carboxyl-terminal domain
- EC
elongation complex
- GTase
RNA guanylyltransferase
- HMW
high-molecular-weight
- NELF
negative elongation factor
- RLF
R-loop formation
- Pol II
RNA polymerase II.
Footnotes
The authors declare no conflict of interest.
References
- 1.Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho EJ, Takagi T, Moore CR, Buratowski S. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shatkin AJ, Manley JL. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 5.Chiu YL, Ho CK, Saha N, Schwer B, Shuman S, Rana TM. Mol Cell. 2002;10:585–597. doi: 10.1016/s1097-2765(02)00630-5. [DOI] [PubMed] [Google Scholar]
- 6.Dahmus ME. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 7.Orphanides G, Reinberg D. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 8.Phatnani HP, Greenleaf AL. Genes Dev. 2006;20:2922–2936. doi: 10.1101/gad.1477006. [DOI] [PubMed] [Google Scholar]
- 9.Schroeder SC, Schwer B, Shuman S, Bentley D. Genes Dev. 2000;14:2435–2440. doi: 10.1101/gad.836300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komarnitsky P, Cho EJ, Buratowski S. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furuichi Y, Shatkin AJ. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuman S. Prog Nucleic Acid Res Mol Biol. 2001;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- 13.Cho EJ, Rodriguez CR, Takagi T, Buratowski S. Genes Dev. 1998;12:3482–3487. doi: 10.1101/gad.12.22.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho CK, Shuman S. Mol Cell. 1999;3:405–411. doi: 10.1016/s1097-2765(00)80468-2. [DOI] [PubMed] [Google Scholar]
- 15.Wen Y, Shatkin AJ. Genes Dev. 1999;13:1774–1779. doi: 10.1101/gad.13.14.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jove R, Manley JL. Proc Natl Acad Sci USA. 1982;79:5842–5846. doi: 10.1073/pnas.79.19.5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers LC, Lacomis L, Erdjument-Bromage H, Tempst P. Mol Cell. 2002;10:883–894. doi: 10.1016/s1097-2765(02)00644-5. [DOI] [PubMed] [Google Scholar]
- 18.Kim HJ, Jeong SH, Heo JH, Jeong SJ, Kim ST, Youn HD, Han JW, Lee HW, Cho EJ. Mol Cell Biol. 2004;24:6184–6193. doi: 10.1128/MCB.24.14.6184-6193.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley D. Mol Cell. 2004;13:377–387. doi: 10.1016/s1097-2765(04)00007-3. [DOI] [PubMed] [Google Scholar]
- 20.Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Proc Natl Acad Sci USA. 2004;101:7572–7577. doi: 10.1073/pnas.0401493101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wen Y, Yue Z, Shatkin AJ. Proc Natl Acad Sci USA. 1998;95:12226–12231. doi: 10.1073/pnas.95.21.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X, Manley JL. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Rosa MD. Cell. 1979;16:815–825. doi: 10.1016/0092-8674(79)90097-7. [DOI] [PubMed] [Google Scholar]
- 24.Steitz TA. EMBO J. 2006;25:3458–3468. doi: 10.1038/sj.emboj.7601211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cramer P. Bioessays. 2002;24:724–729. doi: 10.1002/bies.10127. [DOI] [PubMed] [Google Scholar]
- 26.He B, Rong M, Lyakhov D, Gartenstein H, Diaz G, Castagna R, McAllister WT, Durbin RK. Protein Expr Purif. 1997;9:142–151. doi: 10.1006/prep.1996.0663. [DOI] [PubMed] [Google Scholar]
- 27.Sousa R, Chung YJ, Rose JP, Wang BC. Nature. 1993;364:593–599. doi: 10.1038/364593a0. [DOI] [PubMed] [Google Scholar]
- 28.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. Nat Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 29.Kadesch TR, Chamberlin MJ. J Biol Chem. 1982;257:5286–5295. [PubMed] [Google Scholar]
- 30.Trinh V, Langelier MF, Archambault J, Coulombe B. Microbiol Mol Biol Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts RW, Crothers DM. Science. 1992;258:1463–1466. doi: 10.1126/science.1279808. [DOI] [PubMed] [Google Scholar]
- 32.Tacke R, Manley JL. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Millhouse S, Manley JL. Mol Cell Biol. 2005;25:533–544. doi: 10.1128/MCB.25.2.533-544.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hakansson K, Doherty AJ, Shuman S, Wigley DB. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 35.Hirose Y, Manley JL. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 36.Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 38.Drolet M, Bi X, Liu LF. J Biol Chem. 1994;269:2068–2074. [PubMed] [Google Scholar]
- 39.Gopal V, Brieba LG, Guajardo R, McAllister WT, Sousa R. J Mol Biol. 1999;290:411–431. doi: 10.1006/jmbi.1999.2836. [DOI] [PubMed] [Google Scholar]
- 40.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tian M, Alt FW. J Biol Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- 42.Jiang M, Ma N, Vassylyev DG, McAllister WT. Mol Cell. 2004;15:777–788. doi: 10.1016/j.molcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 43.Westover KD, Bushnell DA, Kornberg RD. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 44.Kasahara M, Clikeman JA, Bates DB, Kogoma T. Genes Dev. 2000;14:360–365. [PMC free article] [PubMed] [Google Scholar]
- 45.Zaitsev EN, Kowalczykowski SC. Genes Dev. 2000;14:740–749. [PMC free article] [PubMed] [Google Scholar]
- 46.Huertas P, Aguilera A. Mol Cell. 2003;12:711–721. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 47.Kireeva ML, Komissarova N, Kashlev M. J Mol Biol. 2000;299:325–335. doi: 10.1006/jmbi.2000.3755. [DOI] [PubMed] [Google Scholar]
- 48.Pei Y, Shuman S. J Biol Chem. 2002;277:19639–19648. doi: 10.1074/jbc.M200015200. [DOI] [PubMed] [Google Scholar]
- 49.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 50.Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Mol Cell Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deng L, Shuman S. Genes Dev. 1998;12:538–546. doi: 10.1101/gad.12.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gromak N, West S, Proudfoot NJ. Mol Cell Biol. 2006;26:3986–3996. doi: 10.1128/MCB.26.10.3986-3996.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosonina E, Kaneko S, Manley JL. Genes Dev. 2006;20:1050–1056. doi: 10.1101/gad.1431606. [DOI] [PubMed] [Google Scholar]
- 54.Hirose Y, Manley JL. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]