Abstract
Vibrio cholerae persists in aquatic environments predominantly in a nonculturable state. In this study coccoid, nonculturable V. cholerae O1 in biofilms maintained for 495 days in Mathbaria, Bangladesh, pond water became culturable upon animal passage. Culturability, biofilm formation, and the wbe, ctxA, and rstR2 genes were monitored by culture, direct fluorescent antibody (DFA), and multiplex PCR. DFA counts were not possible after formation of biofilm. Furthermore, wbe, but not ctxA, were amplifiable, even after incubation for 54 and 68 days at room temperature (≈25°C) and 4°C, respectively, when no growth was detectable. Slower biofilm formation and extended culturability were observed for cultures incubated at 4°C, compared with ≈25°C, suggesting biofilm production to be temperature dependent and linked to loss of culturability. Small colonies appearing after incubation in microcosms for 54 and 68 days at 25°C and 4°C, respectively, were wbe positive and ctxA and rstR2 negative, indicating loss of bacteriophage CTXΦ. The coccoid V. cholerae O1 observed as free cells in microcosms incubated for 495 days could not be cultured, but biofilms in the same microcosms yielded culturable cells. It is concluded that biofilms can act as a reservoir for V. cholerae O1 between epidemics because of its long-term viability in biofilms. In contrast to biofilms produced in Mathbaria pond water, V. cholerae O1 in biofilms present in cholera stools and incubated under identical conditions as the Mathbaria pond water biofilms could not be cultured after 2 months, indicating that those V. cholerae cells freshly discharged into the environment are significantly less robust than cells adapted to environmental conditions.
Keywords: Bangladesh, bacteriophage CTXΦ, DFA, multiplex-PCR, ctxA
Cholera continues to pose a serious health threat globally, notably in those countries where clean drinking water is not available to local populations. Vibrio cholerae serogroups O1 and O139 are associated with epidemic and pandemic cholera. Cholera is endemic in the Ganges delta, occurring twice yearly in epidemic form (1). It is also a major health problem for countries of Africa, Latin America, and Asia (2). V. cholerae O1 is native to both marine and fresh water environments and exists in association with plankton (3). In general, it can be isolated from only 1% of water samples collected during epidemic periods and rarely, if ever, between epidemics (4). However, fluorescent antibody-based studies show that V. cholerae O1 is, nevertheless, present in aquatic environments throughout the year (5). Furthermore, the presence of nonculturable V. cholerae O1 is confirmed by molecular methods (6). The question of whether such nonculturable cells in aquatic environments are capable of returning to an actively growing state to initiate cholera epidemics has been debated.
Extensive studies have shown that V. cholerae O1 becomes coccoid and enters into a nonculturable state in the environment when conditions are not conducive to active growth (5, 7). Some of the coccoid nonculturable cells retain metabolic activity (5, 8, 9), although little is known about the reservoir of V. cholerae between epidemics and the mechanism whereby nonculturable cells become culturable to initiate seasonal epidemics of cholera. Results of laboratory-based studies suggest biofilms provide a powerful mechanism for the persistence of vibrios in the environment (10). Nonculturable cells undergo reduction in size, becoming coccoid in morphology and unable to grow on bacteriological growth media (7, 11). Although viability (12) and infective potential (13) of nonculturable cells of many species and genera of bacteria have been documented (14), how reversion to culturability occurs remains unclear. Animal passage, employing ligated rabbit ileal loops (RILs), has been found to yield culturable V. cholerae O1 (13) and, in a human volunteer study, culturable V. cholerae O1 cells were recovered in stool samples 48 h after nonculturable V. cholerae O1 were ingested (15).
An epidemiological and ecological survey of V. cholerae in the coastal ecosystem of the Bay of Bengal provided firm evidence that nonculturable V. cholerae O1 cells are present between epidemics in biofilms in samples collected from bodies of water serving as drinking water source for the Mathbaria area of Bangladesh, where outbreaks of cholera occur annually (16). In the study reported here, biofilms in laboratory microcosms prepared with Mathbaria water (MW) were found to harbor nonculturable coccoid V. cholerae O1 cells that became culturable even after 1 year, but only via animal passage. Evidence is provided here that V. cholerae O1 persists in the nonculturable state in biofilms in the natural aquatic environment, leading to the conclusion that biofilms play a significant role in the annual cycle of V. cholerae O1 in the environment and notably in annual epidemics of cholera in Bangladesh.
Results
Isolation of V. cholerae O1 from Environmental Samples.
A total of 480 water and plankton samples were analyzed (July 2005 through January 2007) for V. cholerae O1 (1, 16). For this study, enriched [6–8 h in alkaline peptone water (APW)] samples were plated on taurocholate/tellurite/gelatin/agar (TTGA) with streptomycin (Sm) (70 μg/ml) and also without Sm (17). V. cholerae O1 was isolated from 22 of 480 samples plated on TTGA without Sm, whereas only 11 yielded culturable V. cholerae O1 when plated on TTGA containing Sm [supporting information (SI) Table 1]. The frequency of isolation of V. cholerae O1 was significantly higher from Mathbaria site 2, which is connected to a flowing river, and Bakerganj site 5, also a river site, compared with sites 3, 4, and 8, which are ponds (SI Table 1). Also, the only V. cholerae O1 isolated by exploiting Sm resistance phenotype occurred in May 2005 at Mathbaria site 2, a pond connected to a flowing river that, with or without antibiotic in the isolation medium, had yielded V. cholerae O1 in an immediate past sampling round in April 2005 (SI Table 1). All strains isolated on TTGA without antibiotic were resistant to Sm and also to trimethoprim, sulfamethoxazole, and furazolidone. The difference in efficiency of the two methods (with and without Sm) in isolating V. cholerae O1 from water samples, 2% of samples plated on TTGA with Sm and 5% of samples plated on TTGA without Sm, was statistically significant (paired t test; P = 0.002).
Water collected from Mathbaria site 2, which contained V. cholerae O1, as demonstrated by both enrichment culture and direct fluorescent antibody (DFA), was used for the laboratory microcosm experiments. DFA staining of the water revealed clusters of biofilm fragments containing V. cholerae O1 (1). Multiplex-PCR (M-PCR) using a DNA template prepared from water of the same microcosm amplified primers for the genes wbe and ctxA (SI Table 2), confirming that MW is, in fact, an environmental reservoir for potentially toxigenic V. cholerae O1.
V. cholerae O1 Cells in Pond Water Microcosms.
Autoclaved MW microcosms inoculated with V. cholerae O1 yielded initial counts of 107 cfu/ml when plated on TTGA and Luria agar (LA), whereas counts on thiosulfate/citrate/bile salts/sucrose(TCBS) agar were 105 cfu/ml (SI Table 2). Initial DFA counts (day 1) were 8.0 × 108 cells per ml. M-PCR with template DNA prepared from the two microcosms on day 1 amplified the primers for wbe and ctxA (SI Table 2). Plate counts of MW microcosms maintained at room temperature (MW-RT) varied significantly from those maintained at 4°C (MW-4C). Both microcosms showed a gradual reduction in plate counts on TTGA and LA with passage of time. However, an even faster decline in plate counts on TCBS agar was observed, with no counts after 15 days. Conversely, counts for both MW-RT and MW-4C microcosms varied significantly on TTGA and LA at day 15, and subsequently, with respect to time of incubation, continued to decline; eventually no V. cholerae O1 counts were obtained after 40 and 85 days, respectively (SI Table 2). As the monitoring for culturable and nonculturable V. cholerae O1 in the microcosms continued over several weeks after nonculturability, i.e., as the decline in number of culturable cells occurred, it was observed that those fewer and fewer colonies appearing on TTGA and LA, and notably at 40 days, were tiny (<1.0 mm), grew very slowly, and did not grow on TCBS.
Interestingly, DNA templates supported amplification of primers for genes wbe and ctxA by M-PCR in cells incubated up to 40 days (SI Table 2). After 68 days, MW-4C yielded only a few colonies on TTGA and LA, and the DNA template from the same microcosm amplified primers for wbe but not ctxA (SI Table 2). At this stage of extended incubation, some colonies among many tiny colonies on TTGA and LA were confirmed as V. cholerae O1 (1, 16). However, the tiny colonies did not appear on TTGA and LA at day 85 and thereafter, even when concentrates of larger volumes (>10 ml) from microcosms were enriched and plated. Furthermore, template DNA from these preparations did not amplify the genes rstR2, wbe, and ctxA.
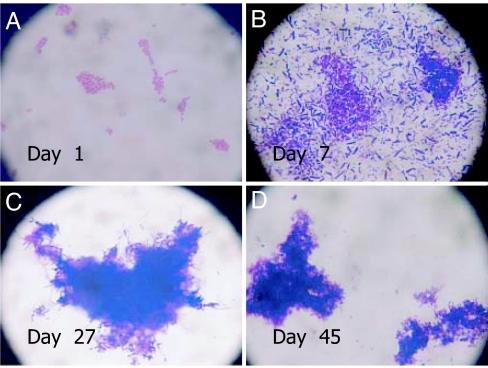
Unlike plate counts, which declined sharply with time of incubation, the DFA counts did not change for both the MW-RT and MW-4C microcosms, until day 15. As shown in SI Table 2, the counts for MW-4C microcosm further remained unchanged until day 26, whereas those for MW-RT were not countable because the cells had aggregated within biofilm (Fig. 1). After incubation for 2 days, the MW-RT microcosm revealed clusters of cells assembled in a coordinated pattern (Fig. 1A), with a large number of cells in clusters on day 7 (Fig. 1B), finally transforming to much larger biofilms after 26 days (Fig. 1C). With time, the curved rods of V. cholerae O1, culturable at day 1, transformed into coccoid cells, gradually becoming smaller and finally appearing as dense clusters of biofilms (Fig. 1D).
Fig. 1.
Micrographs showing V. cholerae O1 in MW microcosms at different stages of incubation at room temperature (RT) (MW-RT). Cells were collected from microcosms, and smears were prepared on clean glass slides, air-dried, stained with crystal violet, washed, and visualized by using a phase-contrast microscope (model BX2; Olympus, Tokyo, Japan). Cells at day 1 were observed to be arranged in microcolonies (A), with larger aggregations in dark blue, an indication of biofilm, at day 7 (B). Cells appeared as multiple layers, tightly attached to thick biofilm surrounding the aggregations of cells at day 27 (C) and as aggregates of thick biofilm intensifying with time at day 45 (D).
In contrast to results obtained for the MW-RT microcosm, DFA counts of the MW-4C microcosm remained unchanged until after incubation for 26 days (SI Table 2). In addition, cell assemblages were not observed early during incubation, as was the case for V. cholerae O1 cells in MW-RT. Formation of biofilm was not obvious, even after 40 days, but did begin to occur and increase with time thereafter, up to day 99 (SI Fig. 4), indicating that biofilm formation in V. cholerae O1 is temperature dependent and negatively linked to culturability.
The initially culturable curved rods typical of V. cholerae O1 (SI Fig. 5A) transformed into small coccoid cells (SI Fig. 5 B–D), and eventually become nonculturable. These cells, occurring either as single cells (SI Fig. 5 B and D) or in biofilms (separated by filtration), did not grow on agar or in broth, even after prolonged enrichment in APW. Nonculturable V. cholerae O1 in biofilms when viewed under a microscope, i.e., cells detectable by DFA but not by culture or M-PCR (SI Table 2), continued to show reduction in size with the time of incubation until barely visible by DFA when examined by phase-contrast microscopy (SI Fig. 5D).
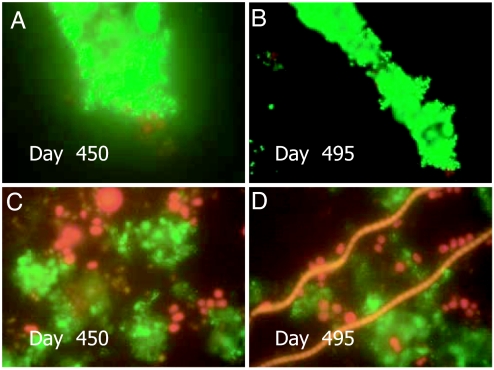
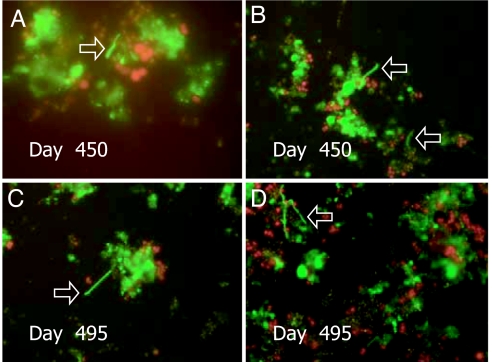
Epifluorescent micrographs of cells from the MW-RT and MW-4C microcosms are shown in Figs. 2 and 3, respectively. The physical appearance of V. cholerae O1 cells in successive stages of transformation to nonculturability in the MW-RT and MW-4C microcosms, i.e., altered morphology, reduced size, and accompanying biofilm formation, occurring after initial inoculation up to 450 and 495 days, are shown in Figs. 2 and 3.
Fig. 2.
Epifluorescence micrographs of nonculturable V. cholerae O1 in microcosms MW-RT (A and B) and MW-4C (C and D). Samples were stained with DFA reagent specific for V. cholerae O1 (New Horizon Diagnostics) and visualized with an epifluorescence microscope (Olympus, model BX51, BX2 Series) (4). Microscopic images were captured digitally (Olympus, DP 20) and processed by using Photoshop Version 5 (Adobe Systems, San Jose, CA). (A and B) Coccoid V. cholerae O1 cells attached to a thin biofilm at 450 days and 495 days after inoculation into MW-RT, respectively. (C and D) Coccoid V. cholerae O1 cells in biofilm 450 days and 495 days after inoculation into MW-4C, respectively.
Fig. 3.
Epifluorescence micrographs of nonculturable V. cholerae O1 in MW-4C microcosms. Samples were pretreated with 0.025% yeast extract and 0.002% nalidixic acid, stained by using cholera DFA (New Horizon Diagnostics), visualized under an epifluorescence microscope (Olympus, model BX51) (1, 4, 17). Images were captured digitally (Olympus DP 20) and processed by using Photoshop Version 5. (A and B) Elongation of V. cholerae O1 in a biofilm that had been incubated for 450 days after initial inoculation. (C and D) Elongation of coccoid V. cholerae O1 cells in a biofilm 495 days after inoculation into MW-4C.
To determine whether nonculturable cells in the microcosms could be induced to culturability, a variety of procedures were used (12). Coccoid cells in biofilms were poorly responsive to viability experiment, as reported elsewhere (17). That is, they did not become elongated in response to yeast extract and nalidixic acid. The process of elongation, which occurred in a relatively large number of cells early in incubation (Fig. 1 and SI Fig. 4 B–D), took longer for cells that had been nonculturable for a longer time. That is, only a few of these cells within biofilms responded to viability after incubation for 15 months (Fig. 3).
M-PCR results showed that DNA templates prepared from MW-RT and MW-4C microcosms amplified all of the primers tested, i.e., wbe, ctxA, and rstR2, even after incubation for 40 days (SI Table 2). However, after 54 days, only wbe was amplified from the MW-RT microcosm, whereas all primers were amplified for MW-4C (SI Table 2). M-PCR did not amplify all primers for both MW-RT and MW-4C microcosms incubated for 68 days, with the exception being the MW-4C, which amplified only wbe (SI Table 2). Further analysis of both MW-RT and MW-4C microcosms by M-PCR at day 85 did not amplify any of the three primer pairs tested (SI Table 2). Note especially that ctxA and rstR2 of bacteriophage CTXΦ were negative for both MW-RT and MW-4C microcosms at day 54 and 68, respectively (SI Table 2). These results suggest, with high probability, that the entire CTXΦ genome is lost during extended incubation in the microcosms. Significantly, in contrast, wbe was amplified, even after the CTXΦ gene was no longer detectable in either MW-RT or MW-4C, at days 54 and 68, respectively.
MW Microcosms Inoculated with Cholera Stool.
Culturable V. cholerae O1 in cholera stool samples inoculated into dialysis bags, which were suspended in 1 liter of autoclaved MW and maintained at room temperature (25–33°C), were examined at selected time intervals by culture, DFA, and M-PCR for genes wbe, ctxA, and rstR2. As shown in SI Table 2, cholera stools in dialysis bags suspended in autoclaved MW at day 1 yielded plate counts of 105 cfu/ml, direct counts of ≈107 cells per ml, and amplified primers for wbe, ctxA, and rstR2. The DFA results showed that the majority of the cells had already become coccoid by the first day of incubation (SI Fig. 6A) and that the cells occurred as clusters in biofilm. These cholera stool microcosms amplified primers for wbe, ctxA, and rstR2 at day 15, but did not produce colonies when plated. Direct counting by DFA was not possible because of the extensive biofilms that had formed by 15 days (SI Fig. 6). As shown in SI Table 2, results of monitoring up to 68 days revealed the cholera stool microcosm was consistently negative by enrichment culture, with coccoid cells in the biofilm further reduced in size with extended incubation (SI Fig. 5D).
MW Microcosms.
Microcosms prepared with unfiltered MW that had tested positive for V. cholerae O1 revealed biofilm aggregates containing both curved rods and coccoid cells of V. cholerae O1 when examined by DFA. Also, M-PCR, using template DNA from the same microcosm, amplified primers for the genes wbe and ctxA (1). See SI Table 2. However, samples at day 15 and up to day 99 were negative for V. cholerae O1 when tested by both enrichment culture and M-PCR. In sharp contrast, biofilm aggregates containing coccoid cells were positive by DFA.
Ileal Loop Analysis.
The nonculturable V. cholerae O1 cells from various microcosms described above were harvested, and equal volumes of cell pellets were used in animal challenge experiments. None of the harvested nonculturable coccoid cells grew on agar even after prolonged enrichment in APW. Adult RILs were inoculated (18) with nonculturable coccoid cells in biofilm aggregates, confirmed as such by DFA (18). After incubation for 18 h, the contents in RIL were removed aseptically, and the volume was measured to determine fluid accumulation. Contents from each of the RILs, separately enriched in APW for 6–8 h, and ≈5–10 μl from the APW streaked onto TTGA agar and incubated for 18 h at 37°C, yielded significantly large numbers of colonies on TTGA, confirmed by serotyping and M-PCR (1) to be V. cholerae O1.
The nonculturable V. cholerae O1 cells from originally cholera-positive stools and maintained in MW microcosms at room temperature for 59 days were culturable after passage in RIL assay. Resuscitation by animal passage in the RILs was also achieved for V. cholerae O1 in biofilm aggregates collected from pond water and maintained at room temperature for up to 54 days. Nonculturable V. cholerae O1 in biofilm from MW-RT and MW-4C microcosms were recoverable by means of RILs up to 495 days. Interestingly, none of these nonculturable cells inoculated into the RILs produced significant fluid accumulation at 18 h, even though they were able to be cultured.
Filtration Experiment.
Water samples from microcosms containing nonculturable cells were filtered through 0.22-μm membrane filters (1). The V. cholerae O1 cells retained on the 0.22-μm membranes were coccoid in morphology but occurred as clusters of cells within thick biofilms (SI Fig. 5C). The few cells observed in the filtrates were single, coccoid, and only rarely within a biofilm and usually in a biofilm fragment (SI Fig. 5D).
Animal passage of free cells in the filtrates did not yield culturable cells. In sharp contrast to the free cells, the clusters of nonculturable cells in biofilm collected by filtration were able to be cultured after animal passage.
Discussion
V. cholerae and many related Gram-negative bacteria have been shown to become nonculturable under specific experimental conditions, although the time required for these cells to become nonculturable is variable (8, 9, 19–21). In this study, V. cholerae O1 cells in all microcosms became nonculturable on TCBS agar within 10–15 days, as has been reported by other investigators (19–21). V. cholerae O1 in biofilms collected from MW and in the biofilm in clinical specimens, when suspended in autoclaved MW that had tested positive for V. cholerae O1 by both culture and DFA, became nonculturable within 15 days. Conversely, MW-RT and MW-4C microcosms inoculated with freshly grown V. cholerae O1 showed culturability on TTGA and LA for 40 and 68 days, respectively. Miller et al. (22) suggested that toxigenic V. cholerae O1 could remain culturable for longer periods at a salinity of 0.25–3.0%, a pH of 8.0, and 25°C. The temperature, pH, and salinity of MW used in the studies reported here were not very different from those conditions used previously.
Initial plate counts on TCBS for both MW-RT and MW-4C microcosms were only 1% of the counts obtained on TTGA and LA. The counts on TCBS agar declined sharply within 2 weeks with no growth obtained for either microcosm on TCBS thereafter. However, TCBS, a medium widely used for selective growth of vibrios, has long been known to be inhibitory to a significant portion of a V. cholerae population (23) and is not an appropriate medium for direct counting or isolation of V. cholerae.
Although the curved rods typical of V. cholerae O1 observed at day 1 gradually became coccoid, the DFA counts did not change significantly, evidence for intact nonculturable cells, induced, being present but not culturable. Moreover, highly significant was the observation by microscope that individual planktonic cells in the MW microcosm did form aggregates and produce biofilm that enclosed the aggregates, with those cells thus enclosed becoming coccoid. The clusters of coccoid cells were unequivocally visible within the biofilms, confirming the reports of Kierec and Watnick (24) and Watnick et al. (10). Many microorganisms produce polysaccharides, either as capsules or as extracellular matrices, in their immediate environment (25), a function of which is to provide protection against adverse external conditions (26).
The genes ctxA and ctxB, encoding cholera toxin (CT) subunits A and B, respectively, are constituents of a lysogenic bacteriophage, CTXΦ, whose 7.0-kb genome has the structure of a compound transposable element. The CTXΦ genome consists of a core region encoding CT and other genes that code for virion morphogenesis, as well as an RS2 region encoding proteins for regulation, replication, and integration of the CTXΦ genome (27). In the V. cholerae chromosome, the lysogenic CTXΦ genome frequently is flanked by a related genetic element, RS1 (28), the genome of a satellite phage that utilizes CTXΦ machinery to produce RS1Φ particles (29). RS1Φ encodes at least four ORFs (rstABCR) that jointly determine expression of a site-specific recombination catalyzing integration of plasmids carrying portions of the CTX element into the genome of nontoxigenic V. cholerae strains at a 17-bp target sequence, attRS1 (28). Under appropriate conditions, CTXΦ is self-transmissible and replicates as a plasmid (27). The filamentous CTXΦ propagates by horizontally infecting susceptible strains of V. cholerae and Vibrio mimicus (30).
Strains of V. cholerae O1 lacking CTXΦ prophage have been reported to occur in the aquatic environment (31). However, it is not known whether ctx− strains exist in the environment as a separate clonal lineage, originally lacking CTXΦ, or whether CTXΦ previously had been excised. Evidence is provided here that the ctxA and rstR2 genes of CTXΦ can be lost simultaneously from toxigenic V. cholerae O1, whereas the wbe gene encoding the O1 antigen is retained, confirmed by M-PCR. It was not clear, however, from results obtained in this study, whether M-PCR for ctxA and rstR2 was unsuccessful because the entire CTXΦ genome was excised or because truncations of these loci had occurred. Because ctxA and rstR2 are located at two opposite termini of the CTXΦ genome (27), it is concluded that the entire CTXΦ was excised.
Existence of ctx progenitors of pandemic strains in the aquatic environment of Mathbaria has been documented (1). Evidence for the loss of CTXΦ prophage, resulting in ctx V. cholerae O1, as presented here, indicates that toxigenic strains in the environment can become nontoxigenic during interepidemic periods. Thus, our results suggest a mechanism whereby ctx V. cholerae O1 can occur in the environment and serve as progenitor, because it has long been known that environmental isolates of V. cholerae O1 rarely carry the genes for cholera toxin (31).
Because the nonculturable state has been considered to be a survival strategy for many bacterial pathogens (4, 5, 7–9, 19, 32), the mechanism by which such cells return to culturability is important to define. Resuscitation from the dormant state has been reported for an array of bacterial pathogens, mainly by induction by temperature upshift (9), but other explanations include increased nutrient concentration (13, 33, 34), a combination of temperature upshift and increased nutrient concentration (20), or the action of a heat-stable enterobacterial autoinducer (35). However, recovery of culturable bacterial cells by temperature upshift alone has also been suggested to represent regrowth of either one or a few cells remaining culturable (36) or insensitive to H2O2 (37).
What is not disputed is the fact that cholera epidemics occur seasonally in endemic areas, but during interepidemic periods, nonculturable cholera bacteria are detectable in water only when molecular and immunochemical methods are used. That is, culturable V. cholerae O1 rarely can be isolated. It is recognized that, in addition to naturally occurring V. cholerae O1 that give rise to initial diarrheal epidemics in the spring months of the year, the aquatic environment receives large quantities of these bacteria released into the water during diarrheal epidemics. From an ecological point of view, if return from the nonculturable state to culturability were simply a temperature and/or nutrient response, the characteristic seasonality of cholera would be expected to result in a series of sporadic outbreaks, particularly during the summer months, when temperature and nutrient levels of shallow water bodies in tropical cholera-endemic areas of the world are higher than in spring and fall, the two seasons of annual cholera, which actually occurs in a distinctive bimodal pattern (38) related to blooms of the zooplankton host of V. cholerae O1 (2, 3, 7).
Despite nonculturability having long been proposed as a survival strategy for cholera bacteria in the aquatic environment between epidemics, the reservoir and the mechanism by which nonculturable cells regain culturability to initiate seasonal cholera are not clear. In this study, nonculturable V. cholerae O1 cells in MW microcosms were observed to occur in aggregates within biofilms. Also, in animal challenge experiments, nonculturable V. cholerae cells in biofilms converted to actively growing V. cholerae O1, even after having been nonculturable for more than 1 year, in microcosms from which culturable cells could not be recovered. The results presented in this study explicitly demonstrate that those nonculturable cells, after exposure to MW microcosms for 495 days, that proved viable occurred only within biofilms. It is concluded that biofilm-bound, nonculturable cells in the natural aquatic environment play a significant role in the seasonal epidemics of cholera (1, 16).
A recent study carried out in Dhaka, Bangladesh, reported enhanced infectivity of V. cholerae O1 in human stools being largely caused by the presence of V. cholerae O1 in biofilms formed in vivo (39). Furthermore, a Sm-supplemented primary isolation medium was used to obtain improved recovery of cholera vibrios and those that were isolated by using the Sm-supplemented medium were referred to as “conditionally viable environmental cells (CVEC)” (17), the inference being that conditions of conventional culture methods underestimate the numbers of V. cholerae O1, because the antibiotic-supplemented medium suppresses competition from other aquatic bacteria. In the study reported here, a component of a 10-year, more extensive program of research on the ecology and pathogenicity of V. cholerae O1 in selected coastal environmental sites within cholera endemic areas located adjacent to the Bay of Bengal (1), both Sm-supplemented and nonsupplemented media were used. Poor recovery of V. cholerae O1 was achieved when the drug was used, in sharp contrast to results reported for a study carried out at sites in Dhaka City, Bangladesh (17), where the yield of V. cholerae O1 from the antibiotic medium was >2-fold compared with the standard culture medium without antibiotic. Unlike the Dhaka City study, where V. cholerae O1 was isolated from the environment consistently and for an extended period (August–December) (17), isolation of culturable V. cholerae O1 from the Mathbaria and Bakerganj environments was successful only within a narrow time period, namely spring and fall, the very times when seasonal epidemics of cholera occur in Bangladesh. Also, noteworthy was the observation in this study that the frequency of isolation of V. cholerae O1 was higher for environmental sites linked to a river. The only sample in the present study that yielded V. cholerae O1 when antibiotic media were used was collected at the Mathbaria site no. 2, which is linked to a flowing river, analogous to the site in the Dhaka City study (17). The Mathbaria site yielded V. cholerae O1 in two preceding samplings regardless of whether antibiotic-supplemented media were used. In fact, only when sampling was done immediately after the seasonal peak of endemic cholera did the antibiotic medium prove successful in isolating V. cholerae O1. This difference is most probably attributable to the ecology of the coastal Bay of Bengal, which is significantly different from Dhaka City, the most populated city in Bangladesh, where epidemics of seasonal cholera are intense and continue for extended periods of time. The bodies of water in and immediately surrounding Dhaka City are heavily contaminated with domestic sewage.
In this study, we were able to confirm that cholera stool, indeed, can contain aggregates of cells in biofilm fragments that remain infective, revive, and become culturable upon animal challenge. However, the aggregates of cells in biofilms in cholera stool that were present immediately after release to the environment contained cells that remained infective only for a short period, thereby having a temporal limitation with respect to contributing to the intensity of cholera epidemics and most probably only for those areas in and around a major city like Dhaka, where conditions of poor sewage treatment expedite the fecal–oral mode of transmission. It cannot, however, account for the annual seasonality of cholera that occurs with significant regularity in the spring and fall months in the remote coastal villages that Mathbaria and Bakerganj, Bangladesh, represent.
In previous studies, we have shown conclusively that biofilms form in situ in the aquatic environment and that such biofilms for most of the year will contain cholera bacteria in the nonculturable state (1, 16). Analogous to the proposal of Faruque et al. (39), namely that in vivo-formed clumps of V. cholerae O1 cells shed by cholera patients increase transmissibility of cholera, such in vivo-formed clumps of V. cholerae O1, when inoculated into MW retained infectivity for at most 2 months. In contrast, V. cholerae O1 inoculated into MW microcosms also formed biofilms (1, 16), but those biofilms are more likely to play a significant role in the overall ecology of V. cholerae and to account for seasonal epidemics of cholera in cholera-endemic areas of Bangladesh. Interepidemic periods of seasonal cholera may comprise a few or several months, during which V. cholerae O1 is not detectable by culture but is present and detectable when molecular methods are used (1, 16). Seasonal epidemics of cholera in Bangladesh characteristically commence when water harboring cholera bacteria is ingested (2). Therefore, understanding that the cholera vibrios are present throughout the year in the aquatic environment of an endemic area is critical to recognizing the basis of the periodicity of cholera outbreaks that is both predictable and similar year to year.
Preepidemic enrichment of nonculturable V. cholerae in the human host as a method of amplification of an epidemic V. cholerae strain immediately before onset of a epidemic (17) can contribute to transmission via the fecal–oral route of infectivity of human-amplified clones, but it cannot account for the natural environmental reservoir, nor for the centuries-long consistent and persistent pattern of cholera epidemics that have been historically documented for this disease in Bangladesh (38). However, biofilms formed in situ offer significantly greater implication for endemic cholera (1) and provide yet another environmental factor to be considered in understanding and preventing cholera epidemics. In addition, environmental factors driven by climate, including blooms of plankton for which vibrios are commensal and/or symbiotic, play a key role in the endemicity of cholera that cannot be ignored in public health planning and response (3, 7, 13).
Materials and Methods
Collection of Environmental Samples.
Samples were collected and analyzed by employing methods described previously (1, 16). This study, part of which includes sample collection from the environment, made use of the availability of samples routinely collected in an ongoing major study of the epidemiology and ecology of V. cholerae O1 in Mathbaria and Bakerganj, where cholera is endemic (16). Water and plankton samples collected biweekly from three ponds (one connected to a flowing river) in Mathbaria, and monthly from one river and one pond in Bakerganj (July 2004 to January 2007), were included in this study. All samples were collected while following aseptic techniques described elsewhere (1, 16).
Processing of Environmental Samples.
Samples were inoculated into APW (Difco, Detroit, MI) and incubated at 37°C for 6–8 h before plating on TCBS agar (Eiken, Tokyo, Japan) and TTGA (Difco), as described elsewhere (1, 16). To test the selective efficiency of TTGA, with and without antibiotic, samples enriched in APW were streaked on TTGA plates with Sm (70 μg/ml) and also without Sm (17). Inoculated plates were incubated at 37°C for 18–24 h. Presumptive V. cholerae colonies were confirmed, as described elsewhere (1, 16).
Collection and Analysis of Clinical Samples.
Stool samples from cholera patients who had not taken antibiotics before reporting to Dhaka hospital of the International Center for Diarrhoeal Disease Research, Bangladesh, were collected in sterile 4-oz (118-ml) vials. Aliquots of the samples were used to inoculate TTGA plates. After incubation overnight at 37°C, suspected V. cholerae colonies were confirmed, as described elsewhere (1, 16).
Preparation of Laboratory Microcosms.
Pond water microcosms.
Microcosms were prepared with MW positive for V. cholerae O1 when tested by both culture and DFA (1). The MW microcosms (1 liter in 2.5-liter conical flasks) were autoclaved and V. cholerae O1 (EM-226) El Tor cells of exponential phase, grown in Luria–Bertani (LB) broth at 37°C and washed with PBS (pH 8) and finally MW, were inoculated at a final concentration of 107 cfu/ml into duplicate microcosms. One was maintained at room temperature (25–33°C), designated MW-RT, and the other at 4°C (MW-4C). At selected time intervals, samples from microcosms were examined by plating on TTGA, TCBS, and LB agar, and DFA counts and M-PCR (as described below) were also done.
Clinical sample microcosms.
Microcosms, prepared as above, were treated as follows. Aliquots of V. cholerae O1 culture positive stool samples were transferred to a series of PBS-presoaked and washed 25 mm × 16 mm dialysis tubes (Sigma, St. Louis, MO) containing MW, prepared as described above. That is, the tubes were sealed and placed in MW microcosms incubated at room temperature. Each of the dialysis tubes was aseptically sampled periodically for culturable V. cholerae O1 and also tested by both DFA and M-PCR.
MW and plankton microcosms.
Microcosms were constructed in sterile 2.5-liter conical flasks by using 1-liter MW aliquots (not autoclaved) seeded with plankton fractions collected by filtering 100 liters of MW (1, 16) through a 20-μm filter, the 1 liter of water being from the pond that tested positive for V. cholerae O1 by culture, DFA, and M-PCR. DFA slides of MW, prepared before collection of plankton and examined under the microscope, revealed biofilm clusters containing cells typical of V. cholerae O1 (1). The mouth of each flask was sealed aseptically with sterilized aluminum foil and then incubated at room temperature. At selected time intervals, V. cholerae O1 total culturable counts, DFA, and M-PCR were determined.
DFA.
V. cholerae O1 counts by DFA were made with samples preincubated with, or without, 0.025% yeast extract (Difco) and 0.002% nalidixic acid (Sigma–Aldrich, St. Louis, MO), overnight in the dark. Samples were centrifuged and pellets were stained with cholera DFA reagents (New Horizon Diagnostics, Columbia, MD). V. cholerae O1 in stool samples were counted in samples diluted 10-fold in PBS before fixing on slides for staining with DFA (40). Fluorescent stained cells were counted by using an epifluorescence microscope (Olympus BX51) and recorded with a digital camera attachment (Olympus DP20).
M-PCR.
The genes wb encoding O antigens of V. cholerae O1 (wbe) and O139 (wbf), ctxA encoding cholera toxin subunit A, and rstR2, encoding a repressor for El Tor CTXΦ, were amplified by using M-PCR (1). Samples preincubated overnight with 0.025% yeast extract and 0.002% nalidixic acid were subjected to M-PCR, using the procedure described in ref. 41. Primer sequences for genes, wbe, ctxA, and rstR2 are shown in SI Table 3.
Animal Passage of Nonculturable V. cholerae O1.
Cells of nonculturable V. cholerae O1 in MW microcosms were collected by filtering 10 ml through a 0.22-μm membrane to a final volume of 2 ml. Bacteria washed from the surface of the membrane filters were aseptically transferred to sterile Eppendorf tubes in duplicate (1.0 ml each) of which 1.0 ml was enriched in APW for 6–8 h at 37°C to determine that no culturable cells were present. An aliquot was also inoculated into RILs, following methods described elsewhere (19). The rabbits were killed after 18–20 h and contents of the RIL were collected aseptically and enriched in APW at 37°C for 6–8 h before plating (4). A loopfull of enriched APW was streaked onto TCBS agar and TTGA and incubated at 37°C for 18–24 h. Presumptive V. cholerae colonies were confirmed by biochemical, serological, and, finally, molecular methods (1, 16).
Filtration.
Nonculturable V. cholerae O1 planktonic cells (free cells in the water phase of the microcosms) and cells bound within biofilm in the microcosms were separated by filtration employing 0.22-μm membrane filters (1). The filtrate (planktonic-phase cells) and cells retained on the membrane filters (biofilm-bound cells) were separately subjected to enrichment culture and animal passage.
Supplementary Material
Acknowledgments
This research was funded by National Oceanic and Atmospheric Administration Grant S0660009 and by National Institutes of Health Research Grant 1R01A13912901 under collaborative agreements between the Johns Hopkins Bloomberg School of Public Health; the International Center for Diarrhoeal Disease Research, Bangladesh (ICDDR,B); and the University of Maryland. The ICDDR,B is supported by donor countries and agencies, which provide unrestricted support to the center for its operation and research.
Abbreviations
- APW
alkaline peptone water
- DFA
direct fluorescent antibody
- LA
Luria agar
- M-PCR
multiplex-PCR
- MW
Mathbaria water
- RIL
rabbit ileal loop
- Sm
streptomycin
- TCBS
thiosulfate/citrate/bile salts/sucrose
- TTGA
taurocholate/tellurite/gelatin/agar.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705599104/DC1.
References
- 1.Alam M, Sultana M, Nair GB, Sack RB, Sack DA, Siddique AK, Ali A, Huq A, Colwell RR. Appl Environ Microbiol. 2006;72:2849–2855. doi: 10.1128/AEM.72.4.2849-2855.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sack RB, Siddique AK, Longini IM, Nizam A, Yunus M, Islam MS, Morris JG, Ali A, Huq A, Nair GB, et al. J Infect Dis. 2003;187:96–101. doi: 10.1086/345865. [DOI] [PubMed] [Google Scholar]
- 3.Colwell RR, Spira WM. In: The Ecology of Vibrio cholerae. Barua D, Greenough WBI, editors. New York: Plenum; 1992. pp. 107–127. [Google Scholar]
- 4.Huq A, Rahaman RR, Ali A, Chowdhury MAR, Parveen S, Sack DA, Cohen ER. Appl Environ Microbiol. 1990;56:2370–2373. doi: 10.1128/aem.56.8.2370-2373.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu HS, Roberts N, Singleton FL, Atwell RW, Grimes DJ, Colwell RR. Microb Ecol. 1982;8:313–323. doi: 10.1007/BF02010671. [DOI] [PubMed] [Google Scholar]
- 6.Rivera ING, Lipp EK, Choopun N, Gil A, Huq A, Colwell RR. Appl Environ Microbiol. 2003;5:599–606. doi: 10.1046/j.1462-2920.2003.00443.x. [DOI] [PubMed] [Google Scholar]
- 7.Colwell RR, Huq A. In: Vibrio cholerae and Cholera: Molecular to Global Perspectives. Wachsmuth IK, Blake PA, Olsvik O, editors. Washington, DC: Am Soc Microbiol; 1994. pp. 117–133. [Google Scholar]
- 8.Rollins DM, Colwell RR. Appl Environ Microbiol. 1986;52:531–538. doi: 10.1128/aem.52.3.531-538.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nilsson L, Oliver JD, Kjelleberg S. J Bacteriol. 1991;173:5054–5059. doi: 10.1128/jb.173.16.5054-5059.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. Mol Microbiol. 2001;39:223–235. doi: 10.1046/j.1365-2958.2001.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grimes DJ, Atwell RW, Brayton PR, Palmer LM, Rollins DM, Roszak DB, Singleton FL, Tamplin ML, Colwell RR. Microbiol Sci. 1986;3:324–329. [PubMed] [Google Scholar]
- 12.Kogure K, Simidu U, Taga N. Can J Microbiol. 1979;25:415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- 13.Colwell RR, Brayton PR, Roszak DB, Huq SA, Palmer LM. Biotechnology. 1985;3:817–820. [Google Scholar]
- 14.Rahman I, Shahamat M, Chowdhury MAR, Colwell RR. Appl Environ Microbiol. 1996;62:115–120. doi: 10.1128/aem.62.1.115-120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colwell RR, Brayton P, Herrington D, Tall B, Huq A, Levine MM. World J Microbiol Biotechnol. 1996;12:28–31. doi: 10.1007/BF00327795. [DOI] [PubMed] [Google Scholar]
- 16.Alam M, Hasan NA, Sadique A, Bhuiyan NA, Ahmed KU, Nusrin S, Nair GB, Siddique AK, Sack RB, Sack DA, et al. Appl Environ Microbiol. 2006;72:4096–4104. doi: 10.1128/AEM.00066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruque SM, Islam MJ, Ahmed QS, Faruque ASG, Sack DA, Nair GB, Mekalanos JJ. Proc Natl Acad Sci USA. 2005;102:6119–6124. doi: 10.1073/pnas.0502069102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De SN, Chatterje DN. J Pathol Bacteriol. 1953;66:559–562. doi: 10.1002/path.1700660228. [DOI] [PubMed] [Google Scholar]
- 19.Oliver JD. J Microbiol. 2005;43:93–100. [PubMed] [Google Scholar]
- 20.Gupte AR, De Rezende CL, Joseph SW. Appl Environ Microbiol. 2003;69:6669–6675. doi: 10.1128/AEM.69.11.6669-6675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Islam MS, Goldar MM, Morshed MG, Khan MN, Islam MR, Sack RB. Can J Microbiol. 2002;48:793–800. doi: 10.1139/w02-073. [DOI] [PubMed] [Google Scholar]
- 22.Miller CJ, Feacham RG, Drasar BS. Lancet. 1985;i:261–263. doi: 10.1016/s0140-6736(85)91036-0. [DOI] [PubMed] [Google Scholar]
- 23.Pickar JH, Sochard MR, Bellanti JA, Colwell RR. Dev Ind Microbiol. 1973;14:337–345. [Google Scholar]
- 24.Kierec K, Watnick PI. Appl Environ Microbiol. 2003;69:5079–5088. doi: 10.1128/AEM.69.9.5079-5088.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davey ME, O'Toole GA. Microbiol Mol Biol Rev. 2000;64:847–867. doi: 10.1128/mmbr.64.4.847-867.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flemming HC. Water Sci Technol. 1993;27:1–10. [Google Scholar]
- 27.Waldor MK, Mekalanos JJ. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 28.Pearson GD, Woods A, Chiang SL, Mekalanos JJ. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faruque SM, Kamruzzaman M, Asadulghani, Sack DA, Mekalanos JJ, Nair GB. Proc Natl Acad Sci USA. 2003;100:1280–1285. doi: 10.1073/pnas.0237385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faruque SM, Siddique AK, Saha MN, Asadulghani, Rahman MM, Zaman K, Albert MJ, Sack DA, Sack RB. J Clin Microbiol. 1999;37:1313–1318. doi: 10.1128/jcm.37.5.1313-1318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.World Health Organization. Wkly Epidemiol Rec. 1984;59:1–17. [Google Scholar]
- 32.Roszak DB, Colwell RR. Microbiol Rev. 1987;51:365–379. doi: 10.1128/mr.51.3.365-379.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duncan S, Grover LA, Killham K, Prosser JI. Appl Environ Microbiol. 1994;60:1308–1316. doi: 10.1128/aem.60.4.1308-1316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramaiah N, Ravel J, Straube WL, Hill RT, Colwell RR. J Appl Microbiol. 2002;93:108–116. doi: 10.1046/j.1365-2672.2002.01666.x. [DOI] [PubMed] [Google Scholar]
- 35.Reissbrodt R, Rienaecker I, Romanova JM, Freestone PP, Haigh RD, Lyte M, Tschape H, Williams PH. Appl Environ Microbiol. 2002;68:4788–4794. doi: 10.1128/AEM.68.10.4788-4794.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weichart D, Oliver JD, Kjelleberg S. FEMS Microbiol Lett. 1992;79:205–210. doi: 10.1111/j.1574-6968.1992.tb14041.x. [DOI] [PubMed] [Google Scholar]
- 37.Bogosian G, Aardema ND, Bourneuf EV, Morris PJ, O'Neil JP. J Bacteriol. 2000;182:5070–5075. doi: 10.1128/jb.182.18.5070-5075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Politzer R. Cholera. No 43. Geneva: World Health Organization; 1959. WHO Monograph Series. [Google Scholar]
- 39.Faruque SM, Biswas K, Udden SM, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. Proc Natl Acad Sci USA. 2006;103:6350–6355. doi: 10.1073/pnas.0601277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brayton PR, Colwell RR. J Microbiol Methods. 1987;6:309–314. [Google Scholar]
- 41.Hoshino K, Yamasaki S, Mukhopadhyay AK, Chakraborty S, Basu A, Bhattacharya SK, Nair GB, Shimada T, Takeda Y. FEMS Immunol Med Microbiol. 1998;20:201–207. doi: 10.1111/j.1574-695X.1998.tb01128.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.