Abstract
MicroRNAs (miRNAs) are important genetic regulators of development, differentiation, growth, and metabolism. The mammalian genome encodes ≈500 known miRNA genes. Approximately 50% are expressed from non-protein-coding transcripts, whereas the rest are located mostly in the introns of coding genes. Intronic miRNAs are generally transcribed coincidentally with their host genes. However, the nature of the primary transcript of intergenic miRNAs is largely unknown. We have performed a large-scale analysis of transcription start sites, polyadenylation signals, CpG islands, EST data, transcription factor-binding sites, and expression ditag data surrounding intergenic miRNAs in the human genome to improve our understanding of the structure of their primary transcripts. We show that a significant fraction of primary transcripts of intergenic miRNAs are 3–4 kb in length, with clearly defined 5′ and 3′ boundaries. We provide strong evidence for the complete transcript structure of a small number of human miRNAs.
MicroRNAs (miRNAs) are short (21- to 23-nt) RNAs that bind to the 3′ untranslated regions (3′ UTRs) of target genes (1, 2). This binding event causes translational repression of the target gene (3) or stimulates rapid degradation of the target transcript (4). miRNAs control the expression of large numbers of genes (5, 6) and are involved in crucial biological processes, including development, differentiation, apoptosis, cell proliferation, and disease (7). Indeed, recent studies have implicated miRNAs in numerous human diseases such as colorectal cancer, chronic lymphocytic leukemia, and Fragile X syndrome (8–12). Expression analyses show that miRNAs are highly and differentially expressed, with specific miRNAs active in specific tissues at specific times (13). In many cancers, miRNA expression is significantly altered, which has been shown to be a useful diagnostic tool (14).
Although our knowledge of miRNA biology has advanced rapidly, attention has been focused on processing and targeting. A miRNA is processed from a longer transcript, referred to as the primary transcript (pri-miRNA) (1, 15, 16). The pri-miRNA includes a precursor miRNA hairpin (pre-miRNA), or sometimes more than one cotranscribed pre-miRNA (16–18). The processed pre-miRNA hairpin is exported to the cytoplasm, where the final mature miRNA is excised (1, 15). Although we know the genomic coordinates of the mature and precursor sequences on the genome, we often know very little about the primary transcript. Approximately 50% of human miRNAs appear to be expressed from introns of protein-coding transcripts (19). However, only a handful of intergenic pri-miRNAs have been characterized. These studies indicate that they can be up to several kilobases long and contain a 5′ 7-methylguanosine cap and a 3′ polyadenylated [poly(A)] tail (17, 18, 20), implying transcription by RNA polymerase II. Past studies have also identified canonical TATA box motifs upstream of miRNA genes (21). In contrast, a recent study provided evidence for transcription of a few miRNAs by polymerase III, where these miRNAs were found to be interspersed by Alu repeats (22). Full understanding of miRNA transcription requires a complete description of the location and extent of pri-miRNAs, including transcription start sites, promoters, and transcription factor (TF)-binding sites.
Annotation of the primary transcripts of miRNAs is extremely important to our understanding of the biology of miRNAs and their regulatory targets. This information allows (i) the prediction of upstream regulatory regions such as TF-binding sites; (ii) the detection of other sequence and structural motifs around the miRNA that may be recognized by the processing machinery; (iii) the mapping of SNPs and mutations that may be outside the mature miRNA but affect its transcription or processing; and it (iv) provides information required to build targeting constructs for miRNA knockout.
Although previous studies (23, 24) have looked at transcriptional features associated with miRNAs, they focused on individual features in isolation [expressed sequence tags (ESTs) and TF-binding sites]. In this work, we seek to combine multiple sets of features from large-scale genomic data with experimentally validated miRNA data to predict and describe features of pri-miRNA transcripts and to delineate their genomic boundaries. Similar analyses have proved invaluable for coding gene prediction (25). We located intergenic miRNAs on the human genome and mapped the following features to the surrounding regions: transcription start site (TSS) predictions, EST matches, CpG island predictions, gene identification signature–paired-end ditags (GIS-PET) matches, TF-binding site predictions, and poly(A) signal predictions. We found consistent 5′ and 3′ sequence features outside pre-miRNAs that clearly delineate the boundaries of miRNA transcripts. For many intergenic miRNAs, we predict the extent of their primary transcripts by using these data. Our results indicate that the majority of primary transcripts of intergenic miRNAs are shorter than protein-coding transcripts, with TSSs located within 2,000 bp upstream and poly(A) signals located within 2,000 bp downstream of the pre-miRNA. Additional features such as ESTs, conserved TF-binding sites, and expression ditags provide further insight into the structure of primary transcripts and the transcription of clustered miRNAs as polycistronic transcripts.
Results and Discussion
We analyzed six sets of data: TSSs, CpG islands, ESTs, TF-binding sites, expression ditags, and poly(A) signal predictions. Each of these features is discussed in detail below.
TSSs.
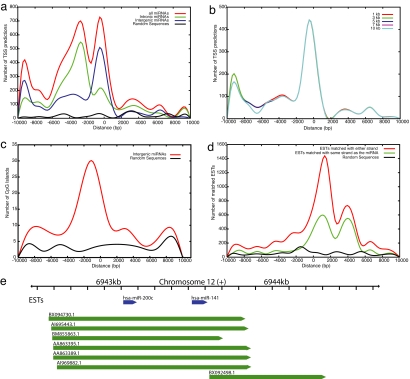
We searched 10 kb upstream and downstream of all 474 human miRNAs for TSSs by using the Eponine method (26). At a threshold of 0.990, Eponine predicted TSSs for 203 (43%) miRNAs. The total number of TSS predictions was 5,190; 172 miRNAs had multiple predicted TSSs, whereas 31 miRNAs were found to have only one predicted TSS. The distribution of distances of predicted TSSs from the pre-miRNA is shown (Fig. 1a). As expected, TSS predictions are located almost exclusively in the upstream region. The majority of TSSs occur within 6 kb upstream of the pre-miRNA start. Moreover, this distribution can be divided into three distinct regions: the first lying close to the pre-miRNA within 2 kb upstream and containing 22% of the total TSS predictions; the second broad region from −2 kb to −6 kb, with the highest peak centered on −4 kb comprising 39% of TSSs; and the third region from −9 kb to −10 kb, containing 7% of TSSs. We speculate that the three different regions correspond to different sets of miRNAs. Indeed, intronic and intergenic miRNAs have distinct distributions (Fig. 1a). There are 2,875 and 2,315 TSS predictions for 104 intronic and 99 intergenic miRNAs, respectively. TSS predictions for intronic miRNAs lie predominantly in the region from −2 kb to −6 kb (Fig. 1a). The other two regions contain higher peaks for intergenic miRNAs than the intronic ones. Overall, 36% and 64% of TSS predictions in these regions belong to intronic and intergenic miRNAs, respectively.
Fig. 1.
Distribution of TSS, CpG, and EST distances from the 5′ end of the pre-miRNAs (positioned at zero) and in random intergenic sequences. (a) TSS predictions. (b) TSS predictions for clustered intergenic pre-miRNAs. (c) CpG predictions. (d) EST predictions. (e) The miRNA cluster hsa-mir-200c∼141 overlapped by six ESTs aligned completely in the upstream and downstream regions.
It is accepted that intronic miRNAs are generally transcribed along with their host genes (19). Eponine predicted TSSs of intronic miRNAs should therefore correspond with the annotated TSSs of the host transcript. Indeed, we find that the majority of predictions lie within 1 kb of the annotated TSS, and the distribution of distances between predicted and annotated TSSs is peaked ≈0 [see supporting information (SI) Fig. 3]. The 1-kb variation in position corresponds well with the published positional accuracy of the Eponine method (26).
Intergenic miRNAs are independent transcription units, with their own transcriptional regulatory elements. For such miRNAs, there are two separate and prominent peaks containing predicted TSSs (Fig. 1a). The first lies close to the pre-miRNA (within 2 kb) and contains 31% of the total TSS predictions, whereas the second (approximately −10 kb) lies further from the pre-miRNA and contains 11% of the TSS predictions. We find 15 miRNAs that possess TSSs in the region from −9 kb to −10 kb. Interestingly, all of these miRNAs have more than one predicted TSS, with the majority of predictions lying between −8 kb and −10 kb. No other annotated transcripts are identified in these regions. We conclude that a minority of intergenic pri-miRNAs are long with TSSs centered on −10 kb. As a control, we used Eponine to predict TSSs in randomly selected intergenic sequences of length 20 kb (Fig. 1a). The distribution of predictions in random intergenic sequences differs greatly from that surrounding miRNAs, indicating that the distribution of TSSs at −2 kb and −10 kb of intergenic miRNAs is highly nonrandom.
Many intergenic miRNAs are clustered in the genome, suggesting that more than one pre-miRNA may be processed from the same primary transcript (27). We clustered miRNAs at different distance cutoffs (1–10 kb) and tested whether TSS predictions support their cotranscription. With an inter-miRNA distance of <1 kb, there are 38 clusters formed by 97 miRNAs. Of 38 clusters, 17 are found to have predicted TSSs within 10 kb upstream. The TSS distribution remains largely unchanged, even at large clustering distances, which suggests that there are very few TSS predictions in the regions between pre-miRNAs within a single cluster. For example, of a total of 17 clusters with predicted TSSs, only 6 clusters have TSS predictions between the clustered miRNAs. We conclude that some miRNAs separated by up to 10 kb are transcribed together in a single primary transcript.
CpG Islands.
CpG islands (28) are useful aids for promoter prediction because they are known to colocalize with TSSs. We looked for CpG islands within 10 kb upstream and downstream of intergenic miRNAs. In total, 111 CpG islands were identified for 82 intergenic miRNAs. Twenty-one miRNAs were found to possess more than one predicted CpG island, with most of these associated with two CpG islands. One exception to this finding was hsa-mir-9-3, which had five CpG islands. The mean length of predicted CpG islands was 1.4 kb. Furthermore, 25 pre-miRNAs were found to be completely embedded in CpG islands. In a recent study, it was shown that the expression of CpG-embedded miRNAs is regulated by hypo/hypermethylation of their associated CpG islands (29), which can further contribute to their distinct expression profiles in normal and cancerous cells (29, 30). Although CpG islands are frequently associated with constitutively expressed housekeeping genes, their role in regulation of expression of miRNAs is still poorly understood.
The distribution of distances of CpG islands from the start of the pre-miRNA and in random intergenic sequences is shown in Fig. 1c. There is a considerable difference in the distribution plots for CpG islands of miRNAs and random intergenic sequences. CpG islands are identified both upstream and downstream of miRNA sequences, with a significant proportion (40%) overlapping with predicted TSS sites within 4 kb upstream. This colocalization provides compelling evidence that promoters of intergenic miRNAs are often located within 4 kb upstream of the pre-miRNA. Treating clusters of miRNAs (defined at different inter-miRNA distances) as single units does not significantly alter the distribution (see SI Fig. 4).
ESTs.
We searched for ESTs mapped to within 10 kb of all 249 intergenic miRNAs. A total of 222 miRNAs had 7,014 matching ESTs. Only 55 pre-miRNA sequences were overlapped by 336 ESTs, whereas the remaining 167 had EST matches in their flanking sequences. More ESTs (5,021) matched downstream of the pre-miRNA than upstream (1, 657). Among the miRNAs with matched ESTs, 19 miRNAs had only one EST match, 117 had 2–10 matched ESTs, 20 had 11–20 matched ESTs, and 66 had >20 EST matches. Furthermore, the multiple ESTs matching the flanking sequences of a pre-miRNA exhibited significant overlap, providing excellent evidence for the boundaries of the pri-miRNAs.
The distribution of matched ESTs to the flanking sequences of intergenic miRNAs is shown in Fig. 1d. Most of the EST matches are located between −2 kb and +4.5 kb. There are more ESTs matched in the downstream region, and a small excess match the antisense strand. We note that the strand specificity of ESTs is often poorly determined, but the excess of antisense ESTs matching immediately downstream of pre-miRNAs warrants further investigation. These findings are consistent with a previous study by Gu et al. (23) who also reported that ≈76% of the investigated miRNAs have matched ESTs in their upstream 2.5 kb and downstream 4 kb flanking sequences and cover mostly the downstream region of pre-miRNAs.
We also investigated the distribution of matched 5′ and 3′ ESTs separately. We find that: (i) there are more matches to 3′ ESTs (40% of matched ESTs are 3′, 31% are 5′ ESTs, and the rest are unannotated); (ii) the majority of 3′ ESTs matched in the downstream region, whereas 5′ ESTs matched both in the downstream and upstream regions; and (iii) most of the 5′ ESTs are matched to the same strand as the pre-miRNAs. ESTs contained within 2 kb upstream of pre-miRNA overlap with the distribution of predicted TSSs and CpG islands (Fig. 1d).
We examined EST support for clusters of miRNAs expressed from a single primary transcript. Different clustering distances do not significantly alter the distribution of EST overlap (see SI Fig. 5), with matched ESTs spanning from −2 kb to +4.5 kb. At the 10-kb clustering distance, there are 34 clusters, of which 30 have matched ESTs. Only five clusters have ESTs that map across the whole cluster (SI Table 3), providing strong evidence that each cluster is transcribed as a single transcript. For instance, the cluster formed by the miRNAs hsa-mir-374∼545 on chromosome X is entirely overlapped by 6 ESTs, where the majority are 5′ ESTs in the same orientation as the cluster (SI Table 3). Similarly, the cluster on chromosome 12 formed by miRNAs hsa-mir-200c∼141 is found to be completely overlapped by 6 ESTs with significant overlap in the upstream and downstream regions (Fig. 1e).
The remaining 25 clusters are found to have ESTs matched only to their flanking sequences or partially overlapping the cluster. We observed that in such instances the flanking ESTs are often perfectly overlapped with each other, defining either the 5′ or 3′ end of a cluster. Aligned flanking ESTs are useful in demarcating the transcript boundaries and thus estimating the length of the pri-miRNA. For instance, the cluster formed by miRNAs hsa-mir-23a∼27a∼24-2 has 10 ESTs in its downstream region with a perfect overlap at 1,771 bp from the 3′ end of hsa-mir-24-2, defining its 3′ boundary, which is in agreement with reported experimental data regarding the 3′ end structure of hsa-mir-23a∼27a∼24-2 (18). Another cluster formed by miRNAs hsa-mir-29b-2 and hsa-mir-29c is found to be overlapped by >20 ESTs aligned with perfect 3′ overlap, 333 bp downstream of hsa-mir-29c. Similarly, the cluster formed by hsa-let-7a-1∼7f-1∼7d has perfect overlap of 3′ ends of flanking ESTs at 637 bp downstream of hsa-let-7d.
It is also possible to derive hypotheses regarding the expression profile of miRNAs based on tissue expression data of mapped ESTs. For example, hsa-mir-122a is found to have 26 ESTs matching with its flanking regions, 24 of which are expressed in liver. Previous reports have determined the liver specificity of hsa-mir-122a by detailed and laborious cloning studies (31). Similarly, we are able to derive the tissue expression of skeletal muscle-enriched miRNAs (133b, 206, 143) (32) and brain-specific miRNAs (124a-1, 124a-2, 124a-3) (32) based solely on the flanking matched ESTs.
TF-Binding Sites.
Generally, TF-binding sites are 8–15 bp long and located in upstream regions near the TSS. We searched for potential TF-binding sites in the flanking regions of clustered and unclustered miRNAs to define their positional preferences and their class representation. TF-binding sites can also be used to locate the core promoters of miRNA genes. To date, only the promoters of two miRNA genes (hsa-mir-23a∼27a∼24-2 and hsa-mir-371∼372∼373) are known experimentally (18, 21).
In total, 163 miRNAs were found to contain a total of 731 predicted TF-binding sites for 161 TFs. Of these miRNAs, 29 were found to possess only one predicted TF-binding site, most of which were located in the upstream regions. The maximum number of TF-binding site predictions was obtained for hsa-mir-9-2, with 26 TF-binding sites followed by hsa-mir-125b-1 with 16 sites.
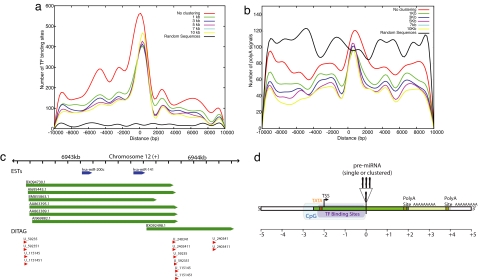
The distribution of TF-binding site distances from pre-miRNAs is shown in Fig. 2a. The flanking regions of miRNAs contain significantly more binding sites than random sequences, and the distribution is highly nonrandom. The distribution of TF-binding sites around miRNAs heavily overlaps with that of predicted TSSs and CpG islands. Several other observations follow from the mapping of TF-binding sites. (i) TF-binding site predictions are enriched in upstream regions (69.6% and 30.4% of TF-binding sites lie in upstream and downstream regions, respectively). The presence of few downstream binding sites may represent some distal regulatory elements (enhancers or silencers). (ii) A large fraction of TF-binding sites lie close to the pre-miRNA: 37% of the total upstream TF-binding sites lie within 2 kb upstream. (iii) Thirty-six miRNAs have TF sites overlapping with their CpG islands. (iv) Many of the TF sites are clustered and may act cooperatively: 60% of miRNAs have more than half of their TF sites within 1 kb. Similar distribution profiles are also obtained after clustering of miRNAs (Fig. 2a).
Fig. 2.
Distribution of TF-binding site distances in intergenic miRNAs and random sequences. (a) TF-binding site predictions. (b) Poly(A) signal predictions. (c) The miRNA cluster hsa-mir-200c∼141 with six matched ESTs and the 5′/3′ ends supported by mapped ditags (U_59235 and U_115145). (d) Structure of a canonical human intergenic pri-miRNA.
Recently, Zhou et al. (24) predicted putative promoter regions of miRNAs and analyzed their positional distributions with respect to the corresponding miRNA precursors. They reported that 100 (93.5%) of the 107 human miRNA genes they analyzed have putative promoters within 500 bp upstream. Our findings also suggest that the TF-binding sites are close to the pre-miRNA. In addition, Lee et al. (18) provided the first direct evidence of transcription of hsa-mir-23a∼27a∼24-2 and identified the promoter region within ≈600 bp upstream of the cluster. Our analysis shows the 400-bp upstream region of cluster hsa-mir-23a∼27a∼24-2 to be highly enriched in TF-binding sites.
We further wished to determine whether some of these putative TFs are more abundant in miRNAs. We searched for those factors that are enriched and overrepresented in miRNAs, focusing on TFs found within 2 kb upstream of a pre-miRNA. Z scores and P values were then calculated for each of the TFs identified. We compared them with a control consisting of 2 kb upstream of the annotated transcription start sites of RefSeq genes (33) obtained from the University of California, Santa Cruz (UCSC) Genome Browser (34). The mapping of conserved TF-binding sites and the TFs in RefSeq upstream sequences were obtained from the TF-binding site track of UCSC Genome Browser.
We found binding sites for 72 TFs within 2 kb upstream of pre-miRNAs. Among these TFs, the top five statistically significant ones with P values (≤ 0.05) are listed in Table 1. For each TF, we show the TransFac ID, TransFac class, factor name, the associated Gene Ontology function, and the P value. Interestingly, most of these TFs are related to growth and developmental processes. The top two TFs, MSX1_01 and NCX_01, are homeobox proteins, which are known to be involved in developmental and regulatory processes. These TFs are also predicted as TFs of miRNAs in a previous study (35). In addition, we also found a correspondence between the presence of a particular TF with the tissue expression of miRNAs. The upstream regions of brain-specific miRNAs hsa-mir-124a-1, 124a-2, 125b-1, 219-1, and 9-3 contain many binding sites for homeobox TFs. For example, hsa-mir-124a-2 is associated with sites for TFs MSX, ZIC1, MEIS1BHOXA9_01, and CREB1CJUN_01, which are known to regulate brain and nervous developmental processes. A T cell-specific miRNA, hsa-mir-142, has a binding site for TF SEF1_C, which is known to be important for T cell-specific gene expression. Moreover, let-7 family miRNAs, which are known to be involved in differentiation processes, are found to be enriched in the TFs CDP, EVI1, and NKX, involved in cell growth and development. It is important to note that there may be a large number of miRNA-specific TF-binding sites that are not annotated in the databases. The data presented here provide a platform for subsequent detection of novel TF motifs important for regulation of miRNA expression.
Table 1.
Top 5 of 72 TFs identified within 2 kb upstream of human intergenic miRNAs
| TransFac ID | TransFac class | Factor name | Gene ontology | P value |
|---|---|---|---|---|
| MSX1_01 | Homeobox | MSX1, HOX7: Homeobox protein MSX-1 | Skeletal development | 0.00004 |
| NCX_01 | Homeobox | TLX2, HOX11L1, NCX: T cell leukemia homeobox protein 2 | Development | 0.0047 |
| CDC5_01 | Tryptophan clusters | Cell division control protein 5 | Regulation of transcription | 0.0047 |
| SRF_01 | Mads | Serum response factor | RNA polymerase II transcription factor activity; signal transduction | 0.0264 |
| RP58_01 | Unannotated | Zinc finger protein 238 | Negative regulation of transcription from RNA polymerase II | 0.0264 |
In contrast, the five significant motifs in upstream regions of RefSeq genes are NRF2, SREBP1_01, CREBP1CJUN_01, E2F_02, and RFX1_02 (SI Table 4). NRF2 is the mitochondrial respiratory chain regulator and was also detected as one of the strongest motifs by a previous study (36). Overall, 11 TFs are obtained with significant P values and among them, 6 are consistent with the TFs identified by a previous study (36).
Poly(A) Signals.
Pri-miRNAs have been shown to be poly(A) (17, 18, 20). To obtain an estimate of the 3′ boundary of transcripts, we searched for four different poly(A) hexamers in the flanking regions of miRNAs by using the dnafsminer tool (37). The distribution of poly(A) signals is shown in Fig. 2b. We identified a total of 1,560 instances of these poly(A) hexamers in 249 intergenic miRNAs. A total of 28 miRNAs had one putative poly(A) signal, 41 miRNAs had two poly(A)s, 22 miRNAs had three poly(A)s, and the remainder had more than four signals. The most frequently occurring hexamers were AATAAA and ATTAAA, comprising 31.7% and 25.3% of the total poly(A) hexamers identified, respectively. Treating clustered miRNAs as single transcriptional units results in a greater proportion of poly(A) predictions falling <1.8 kb downstream of the last miRNA. The mean distance of the first AATAAA was 830 bp from the 3′ end of the pre-miRNA. The distribution of poly(A) site predictions in randomly chosen intergenic sequences is also shown (Fig. 2b). The number of predictions in random intergenic sequences is higher in regions flanking miRNAs, demonstrating that the specificity of the prediction is unsurprisingly poor. A poly(A) hexanucleotide signal alone is therefore insufficient evidence for a 3′ end. However, the peak of poly(A) predictions between 0 and 2 kb downstream of pre-miRNAs remains clearly above that in surrounding regions.
Very few miRNAs have experimentally verified poly(A) tails. Lee et al. (18) experimentally characterized the presence of a poly(A) tail at 1,752 bp downstream of hsa-mir-24-2. We also identified a putative poly(A) signal AATAAA with a significant dnafsminer score (0.939) at 1,752 bp downstream of the 3′ end of hsa-mir-24-2 in a cluster formed by hsa-mir-23a∼27a∼24-2. As discussed above, we also identified >10 ESTs in the downstream region of this cluster, perfectly aligned at 1,771 bp from the 3′ end of hsa-mir-24-2. However, no single EST covers the entire cluster. In addition, we found other examples of putative poly(A) signals supported by proximally aligned ESTs. For instance, a cluster formed by miRNAs hsa-mir-29b-2∼29c possesses a poly(A) signal (dnafsminer score = 0.952) at ≈311 bp downstream of the 3′ end of hsa-mir-29c, close to the ESTs aligned at ≈333 bp.
GIS-PET.
GIS analysis covalently links the 5′ and 3′ signatures of each full-length transcript into a paired-end ditag (PET) (38). Ditags provide unique identifiers for transcript 5′ and 3′ ends and are useful in defining their boundaries (38, 39). The prediction of TSS sites/CpG islands can be supported by mapping the 5′ end of ditags or 5′ CAGE tags in the upstream region. Similarly, mapping of the 3′ end of ditags can confirm the location of poly(A) tails. We identified a few instances where the ditag 5′ ends are mapped in the upstream region close to the predicted TSSs or CpG islands (within 100 bp). For example, the cluster of miRNAs hsa-mir-181c∼181d is found to contain the 5′ end of ditags U_443297 and U_166195 at −1,607 bp from the 5′ end of hsa-mir-181c overlapping with predicted CpG islands (present at −1,916 bp upstream of hsa-mir-181c). Similarly, many 3′ ditags overlap 3′ ends of ESTs. For example, the cluster formed by miRNAs, hsa-mir-200c and hsa-mir-141 is found to contain ditags U_59235 and U_115145 at 473 bp from the 5′ end of hsa-mir-200c and their 3′ counterparts present at 230 bp from the 3′ end of hsa-mir-141. The 3′ ditags support the putative poly(A) signal identified at ≈211 bp and overlapping ESTs ending at 250 bp from the 3′ end of hsa-mir-141 (Fig. 2c). Taken together, these data strongly suggest that the putative 5′ and 3′ boundaries of cluster hsa-mir-200c∼141 are approximately −400 bp and +250 bp, respectively, with primary transcript length of ≈1,150 bp.
Prediction of Putative Boundaries of Pri-miRNAs.
The data presented in this work provide strong evidence for the length and boundaries of pri-miRNAs and the transcription of clustered polycistronic miRNAs. Although each feature represents only a small piece of evidence for a transcript end, the combined weight of the diversity and number of analyzed features delineates many 5′ and 3′ boundaries of pri-miRNAs with high confidence. The 15 best supported pri-miRNAs [by TSS, CpG, EST, ditag, and poly(A) data] are given (Table 2). Detailed graphical views of these and other examples are provided in SI Table 5. It is clear that the length of the primary transcript varies greatly, from 0.5 kb to 7 kb in the 15 examples. In addition, a small set of pri-miRNAs (hsa-mir-374∼545, hsa-mir-9-2, hsa-mir-193b∼365-1, hsa-mir-181c∼181d) appear to have length >10 kb (see SI Table 6). Furthermore, we are able to predict only the 5′ or 3′ end of the primary transcript of a much larger number of miRNAs (SI Table 7). Based on these results, we derive a canonical structure of the mammalian miRNA primary transcript (Fig. 2d) showing the TSS, CpG islands, and overlapping TF sites within 2 kb upstream and poly(A) signal 2 kb downstream of the precursor miRNA.
Table 2.
High-confidence predictions of boundaries and lengths of primary transcripts of 15 intergenic miRNAs
| miRNA/cluster | Chromosome (strand) | Predicted 5′ end of primary transcript | Predicted 3′ end of primary transcript | Predicted length of primary transcript, bp | Supporting evidence |
|---|---|---|---|---|---|
| hsa-mir-200c∼141 | 12 (+) | 6942737 | 6943865 | 1,128 | TSS, CpG, ESTs, ditags, poly(A) |
| hsa-mir-497–195 | 17 (−) | 6861211 | 6863698 | 2,487 | ESTs, 5′ CAGE |
| hsa-mir-34b∼34c | 11 (+) | 110888630 | 110889820 | 1,190 | TSS, CpG, 5′ CAGE, ESTs, poly(A) |
| hsa-mir-29b-2∼29c | 1 (−) | 206041489 | 206046102 | 4,613 | ESTs, 5′ CAGE, ditags, poly(A) |
| hsa-mir-572 | 4 (+) | 10979348 | 10984460 | 5,112 | TSS, CpG, 5′ CAGE, ditags, ESTs, poly(A) |
| hsa-mir-124a-1 | 8 (−) | 9794986 | 9800634 | 5,648 | TSS, CpG, 5′ CAGE, ESTs, poly(A) |
| hsa-mir-99b∼let-7e∼125a | 19 (+) | 56885133 | 56888521 | 3,388 | TSS, CpG, 5′ CAGE, ESTs |
| hsa-mir-424-503 | X (−) | 133508008 | 133511322 | 3,314 | TSS, CpG, 5′ CAGE, ESTs |
| hsa-mir-200b∼200a∼429 | 1 (+) | 1088033 | 1094500 | 6,467 | TSS, CpG, 5′ CAGE, ditags, ESTs |
| hsa-mir-223 | X (+) | 65152025 | 65156338 | 4,313 | ESTs, 5′ CAGE, poly(A) |
| hsa-mir-23a∼27a∼24–2 | 19 (−) | 13804510 | 13808884 | 4,374 | ESTs, ditags |
| hsa-mir-219-2 | 9 (−) | 130193109 | 130195318 | 2,209 | TSS, CpG, 5′ CAGE, ditags, ESTs |
| hsa-mir-210 | 11 (−) | 555660 | 558587 | 2,927 | TSS, CpG, 5′ CAGE, ESTs, poly(A) |
| hsa-let-7i | 12 (+) | 61283559 | 61284132 | 573 | TSS, CpG, ESTs, ditags, poly(A) |
| hsa-mir-92b | 1 (+) | 153431072 | 153432003 | 931 | TSS, CpG, 5′ CAGE, ESTs, ditags |
Conclusions
Little is known about the structure of pri-miRNA transcripts. Previous studies have focused on individual features (23, 24). In contrast, we present a survey of multiple genomic features of approximately hundreds of annotated miRNAs in the human genome, the most comprehensive analysis of miRNA transcriptional features to date. This analysis has allowed us to delineate the boundaries of a significant proportion of intergenic human miRNAs.
Our results demonstrate that transcriptional features in flanking sequences of miRNA precursors provide strong evidence for the boundaries of pri-miRNAs and for the cotranscription of clustered miRNAs. TSS and CpG island predictions demarcate the 5′ ends of many intergenic miRNA transcripts. The data analyzed indicate that a significant fraction of human intergenic miRNAs possess TSS sites within 2 kb of the pre-miRNA. Most CpG islands are found in close proximity to these TSS sites. Poly(A) sites define and predict 3′ boundaries. We show that the distribution of poly(A) signals peaks at 2 kb downstream of the pre-miRNA.
Combining these signal prediction results with EST and expression ditag data provides strong evidence that many human intergenic miRNAs are encoded by primary transcripts 3–4 kb long, with a small fraction of longer transcripts up to 6 kb (Fig. 2d). Previous detailed experimental studies of small numbers of pri-miRNAs have also shown transcript lengths of 1–4 kb (17, 18, 20).
Obviously, experimental work is required to determine unambiguously exact pri-miRNA transcript lengths. In contrast, the computational results presented here provide bounds on pri-miRNA transcript lengths, together with a set of high confidence, strongly supported pri-miRNA predictions in an efficient and timely manner. These predictions provide a large-scale look at features surrounding intergenic miRNAs and as such represent a significant step in our understanding of their transcription.
Materials and Methods
Obtaining Human Pre-miRNAs.
We obtained genomic coordinates of 474 human pre-miRNAs from the miRBase miRNA sequence database (version 9.0) (40). The human genome sequences and annotations were obtained from Ensembl release 42 (Wellcome Trust Sanger Institute, Cambridge, U.K.) (41) and are clustered according to their genomic distance. For full details, see SI Materials and Methods.
Obtaining Genomic Features.
Flanking sequence data, TSSs (SI Fig. 5), and GIS-PET data were obtained from Ensemble (41, 42). TF-binding sites were obtained using the UCSC browser (34, 43, 44). Poly(A) signals were predicted using the dnafsmimer method. For details, please see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank members of Team101 at the Wellcome Trust Sanger Institute for useful discussion and advice. H.K.S. was supported by a Glaxo-SmithKline postdoctoral fellowship. S.G.-J. was supported by the Wellcome Trust and the University of Manchester, and A.J.E. was supported by the Wellcome Trust.
Abbreviations
- GIS-PET
gene identification signature-paired end ditags
- miRNA
microRNA
- poly(A)
polyadenylated
- pre-miRNA
precursor miRNA
- pri-miRNA
primary transcript of microRNA
- TF
transcription factor
- TSS
transcription start site
- UCSC
University of California Santa Cruz.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703890104/DC1.
References
- 1.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Pasquinelli AE, Hunter S, Bracht J. Curr Opin Genet Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 3.Wightman B, Ha I, Ruvkun G. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 4.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 6.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez-Garcia I, Miska EA. Development (Cambridge, UK) 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caudy AA, Myers M, Hannon GJ, Hammond SM. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McManus MT. Semin Cancer Biol. 2003;13:253–258. doi: 10.1016/s1044-579x(03)00038-5. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 13.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 15.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cullen BR. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Cai X, Hagedorn CH, Cullen BR. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bracht J, Hunter S, Eachus R, Weeks P, Pasquinelli AE. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houbaviy HB, Dennis L, Jaenisch R, Sharp PA. RNA. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borchert GM, Lanier W, Davidson BL. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 23.Gu J, He T, Pei Y, Li F, Wang X, Zhang J, Zhang X, Li Y. Mamm Genome. 2006;17:1033–1041. doi: 10.1007/s00335-006-0007-9. [DOI] [PubMed] [Google Scholar]
- 24.Zhou X, Ruan J, Wang G, Zhang W. PLoS Comput Biol. 2007;3:e37. doi: 10.1371/journal.pcbi.0030037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stormo GD. Genome Res. 2000;10:394–397. doi: 10.1101/gr.10.4.394. [DOI] [PubMed] [Google Scholar]
- 26.Down TA, Hubbard TJ. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bird AP. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 29.Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A, Sanchez-Cespedes M, Gitt A, et al. Cancer Res. 2007;67:1424–1429. doi: 10.1158/0008-5472.CAN-06-4218. [DOI] [PubMed] [Google Scholar]
- 30.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 31.Barad O, Meiri E, Avniel A, Aharonov R, Barzilai A, Bentwich I, Einav U, Gilad S, Hurban P, Karov Y, et al. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pruitt KD, Tatusova T, Maglott DR. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karolchik D, Baertsch R, Diekhans M, Furey TS, Hinrichs A, Lu YT, Roskin KM, Schwartz M, Sugnet CW, Thomas DJ, et al. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sethupathy P, Megraw M, Barrasa MI, Hatzifeorgiou AG. Lecture Notes Comput Sci. 2005;3746:457–468. [Google Scholar]
- 36.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Han H, Li J, Wong L. Bioinformatics. 2005;21:671–673. doi: 10.1093/bioinformatics/bth437. [DOI] [PubMed] [Google Scholar]
- 38.Ng P, Wei CL, Sung WK, Chiu KP, Lipovich L, Ang CC, Gupta S, Shahab A, Ridwan A, Wong CH, et al. Nat Methods. 2005;2:105–111. doi: 10.1038/nmeth733. [DOI] [PubMed] [Google Scholar]
- 39.Peters BA, Velculescu VE. Nat Methods. 2005;2:93–94. doi: 10.1038/nmeth0205-93. [DOI] [PubMed] [Google Scholar]
- 40.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Birney E, Andrews D, Caccamo M, Chen Y, Clarke L, Coates G, Cox T, Cunningham F, Curwen V, Cutts T, et al. Nucleic Acids Res. 2006;34:D556–D561. doi: 10.1093/nar/gkj133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stabenau A, McVicker G, Melsopp C, Proctor G, Clamp M, Birney E. Genome Res. 2004;14:929–933. doi: 10.1101/gr.1857204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.