Abstract
High levels of genomic and allelic microvariation have been found in major marine planktonic microbial species, including the ubiquitous open ocean cyanobacterium, Prochlorococcus marinus. Crocosphaera watsonii is a unicellular cyanobacterium that has recently been shown to be important in oceanic N2 fixation and has been reported from the Atlantic and Pacific oceans in both hemispheres, and the Arabian Sea. In direct contrast to the current observations of genomic variability in marine non-N2-fixing planktonic cyanobacteria, which can range up to >15% nucleotide sequence divergence, we discovered that the marine planktonic nitrogen-fixing cyanobacterial genus Crocosphaera has remarkably low genomic diversity, with <1% nucleotide sequence divergence in several genes among widely distributed populations and strains. The cultivated C. watsonii WH8501 genome sequence was virtually identical to DNA sequences of large metagenomic fragments cloned from the subtropical North Pacific Ocean with <1% sequence divergence even in intergenic regions. Thus, there appears to be multiple strategies for evolution, adaptation, and diversification in oceanic microbial populations. The C. watsonii genome contains multiple copies of several families of transposases that may be involved in maintaining genetic diversity through genome rearrangements. Although genomic diversity seems to be the rule in many, if not most, marine microbial lineages, different forces may control the evolution and diversification in low abundance microorganisms, such as the nitrogen-fixing cyanobacteria.
Keywords: evolution, genome, marine, nitrogen fixation, Crocosphaera
Recent discoveries in marine microbiology have demonstrated the high degree of diversification of abundant planktonic microorganism lineages examined to date, including Pelagibacter ubique (1), Vibrio splendidus (2), and the ubiquitous open ocean cyanobacterium, Prochlorococcus marinus (3). Comparative genomics of isolates (3, 4), amplification of single genetic loci including rRNA genes or intergenic spacer regions (1, 5), and metagenomic studies (6–9) all have highlighted the extensive genetic diversity in the major planktonic cyanobacterial lineages of Prochlorococcus within single samples and habitats and across ecosystems.
Nitrogen fixation, the reduction of atmospheric dinitrogen (N2) gas to biologically available ammonium, determines the relative availability of oceanic nitrogen (N) and phosphorus (P) over geological time scales (10). Relatively few oceanic microorganisms (e.g., Trichodesmium and Richelia) catalyze this key biogeochemical transformation. Recently, groups of oceanic unicellular cyanobacteria, including Crocosphaera watsonii, have been shown to be important in oceanic N2 fixation in tropical and subtropical waters around the globe (11–14), although they are several orders of magnitude less abundant than the non-N2-fixing unicellular cyanobacterial lineages (15). C. watsonii are several micrometers in diameter, contain the photosynthetic pigment phycoerythrin, and have been observed in several oceans by microscopy, flow cytometry (16, 17), and nitrogenase gene amplification (11, 12, 15, 18, 19).
Given the large degree of sequence and genome divergence that coincides with ecotypes in Prochlorococcus (3), we hypothesized that there would be similar genetic diversity and ecotypic variability within the unicellular diazotrophic cyanobacterial genus Crocosphaera. However, by comparing amplified gene sequences from cultivated strains and complete sequences of two metagenomic fragments, we found that the genomic sequence diversity within this group of microorganisms appears to be extremely low relative to the more abundant sympatric non-N2-fixing cyanobacteria.
Results and Discussion
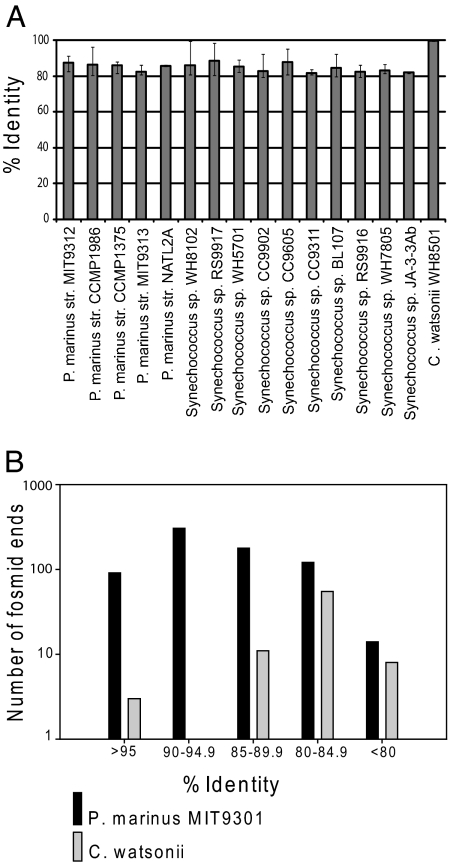
C. watsonii was isolated from the South Atlantic in the 1980s (20), and a draft genome sequence has been completed by the Department of Energy Joint Genome Institute (www.jgi.doe.gov). End sequences of several BAC and fosmid clones from libraries constructed from subtropical North Pacific Ocean picoplankton [collected at Hawaii Ocean Time-series Station ALOHA (22° 45′ N, 158° 00′ W)] were homologous to the C. watsonii WH8501 draft genome sequence (Table 1). Obtaining these metagenomic fragments was initially unexpected, because the abundance of C. watsonii, and N2-fixing microorganisms in general, is several orders of magnitude lower than picoplanktonic prokaryotes in open ocean oligotrophic waters. C. watsonii genomic fragments may have been cloned as a result of a bloom of C. watsonii populations and because of its relatively large genome size. Intriguingly, all end sequences of these metagenomic fragments were generally 97–99% identical to the genome sequence of the cultivated C. watsonii WH8501 (Table 1), even though they were only draft-quality end sequences. In the metagenomic libraries, there were no other end sequences that were >86% identical, showing that there is much less gene and genome sequence variability in C. watsonii than there is in Prochlorococcus (8). This preliminary finding was surprising, because similar analyses of Prochlorococcus, Synechococcus, or heterotrophic bacterial sequences yield groups of sequences that span 78% to 99.5% sequence divergence within a single sample (Fig. 1) and can vary dramatically between individual samples from different depths, habitats, or times (see figure 2 in ref. 9 and Fig. 1).
Table 1.
BAC and fosmid end sequences with homology to the C. watsonii WH8501 genome
| End sequence | GenBank accession no. | Sequence length, bp | Alignment length, bp | % Identity | E value |
|---|---|---|---|---|---|
| BAC sequences | |||||
| HOT0_01C04F | EF089392 | 699 | 688 | 99.6 | 0 |
| HOT0_01C04R | EF089392 | 755 | 753 | 99.1 | 0 |
| HOT0_02H07F | EF089393 | 232 | 232 | 100 | 3.0−130 |
| HOT0_02H07R | EF089393 | 314 | 314 | 99.4 | 3.0−177 |
| HOT0_04C07F | EF089394 | 126 | 126 | 98.4 | 2.0−62 |
| HOT0_04C07R | EF089394 | 568 | 568 | 99.5 | 0 |
| HOT0_07A01F | EF089395 | 397 | 397 | 99.8 | 0 |
| HOT0_07A01R | EF089395 | 541 | 541 | 99.6 | 0 |
| HOT0_10H08F | EF089391 | 778 | 778 | 99.5 | 0 |
| HOT0_10H08R | EF089391 | 677 | 678 | 99.6 | 0 |
| HOT0_04B05R | EF089396 | 526 | 526 | 99.8 | 0 |
| HOT0_02H05 F* | EF089389 | 373 | 373 | 99.7 | 0 |
| HOT0_02H05 R* | EF089389 | 107 | 105 | 98.1 | 5.00−50 |
| HOT0_07D09 F* | EF089390 | 530 | 525 | 100 | 0 |
| HOT0_07D09 R* | EF089390 | 196 | 171 | 96.5 | 2.00−72 |
| Fosmids | |||||
| HF0070_14C03 F* | DU741405.1 | 1,030 | 981 | 99.0 | 0 |
| HF0070_14C03 R* | DU741406.1 | 880 | 776 | 97.1 | 0 |
| HF0010_039H07 F | DU745539.1 | 1,001 | 150 | 93.3 | 4.00−47 |
| HF0010_039H07 R | DU745540.1 | 1,045 | 842 | 97.2 | 0 |
BACs and fosmids that were completely sequenced are indicated by asterisks. F, forward; R, reverse.
Fig. 1.
Diversity of environmental cyanobacteria gene sequences compared with cultivated strains. (A) Range of percent identity of Prochlorococcus and Synechococcus pdxA gene sequences to Global Ocean Sampling (GOS) sequences from Sargasso Sea sites in comparison with C. watsonii gene diversity found in this study (8). Solid bars show the average percent identity to sequences from each species, and error bars show the complete range of sequence identity. Results are shown in comparison to the C. watsonii sequence analysis presented here. (B) Results of a BLASTN (nucleotide) search of the C. watsonii WH8501 and Prochlorococcus MIT9301 genomes against all ALOHA fosmid end sequences (7) binned by percent identity of the top alignment. Only alignments >75 bp were included. Cultivated strains are generally different from environmental populations, there is a range in DNA sequence identity (to 80% or lower) to individual strains of non-N2-fixing cyanobacteria, and the C. watsonii gene sequences are much more highly conserved than those of Prochlorococcus and Synechococcus. Note that within an individual sample or habitat, there are sequences very similar to individual isolates (e.g., Synechococcus sp. WH8102) in the environment, but there is a large range in sequence identity that reflects ecotypes and genetic variants. This variation is also reflected in the lower overall average sequence identity percentages (e.g., <85% for Synechococcus sp. WH8102) in A. In contrast, sequences similar to C. watsonii are either nearly identical, or only distantly related to the WH8501 genome, displaying a pattern that indicates a lack of strain nucleotide sequence variation.
Fig. 2.
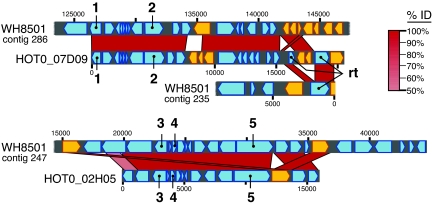
Alignment of BAC and C. watsonii genome sequences showing the extent of sequence similarity. Station ALOHA BACs HOT0_07D09 (GenBank accession no. EF089390) and HOT0_02H05 (GenBank accession no. EF089389) to C. watsonii WH8501 draft genome sequence (GenBank accession no. AADV00000000.2). The degree of nucleotide sequence similarity is indicated by the shade of red cross-hatch between the aligned genome and BAC sequences. Transposase genes are indicated in gold. Numbers indicate the genes that were amplified from cultures (Table 2): 1, cytochrome c oxidase subunit II (Cyt c); 2, peptidase transferase (pt); 3, pyridoxal phosphate biosynthetic protein (pdxA); 4, photosystem II protein W (psbW); 5, DNA polymerase A (DNA pol). The RNA-directed DNA polymerase, which is interrupted by transposases on BAC HOT0_07D09, is also shown (rt).
Before completely sequencing representative BACs, we amplified several additional genes for a multilocus comparison of DNA sequences. We chose genes that we predicted (based on the WH8501 draft genome) would be present on the BACs. These genes were amplified and sequenced from several strains of C. watsonii isolated from the Atlantic and Pacific oceans from different years and different sampling locations to determine whether there was gene sequence diversity among the cultivated strains and the BACs (Table 2). This approach might have also uncovered different genome arrangements, because we anticipated that genomes in different strains might be rearranged and genes present between the BAC end sequences might or might not be the same as those on the homologous genomic region from the isolate C. watsonii WH8501. The nucleotide sequences of genes amplified from seven strains were >99% identical to the homologous C. watsonii WH8501 (GenBank accession no. AADV00000000.2) gene sequences (Table 2). We did not attempt to distinguish between the errors introduced by PCR and draft DNA sequencing, as this degree of similarity is in and of itself distinctive, relative to similar surveys of other open-ocean cyanobacteria and bacteria gene sequence diversity (9) (Fig. 1).
Table 2.
Percent identity of sequences amplified from cultivated strains compared with the C. watsonii WH8501 genome sequence (isolated from South Atlantic Ocean, 1984) homologues
| Strain | Sample source | Date | Latitude/longitude | Cyt c | pt | DNA pol | pdxA | psbW |
|---|---|---|---|---|---|---|---|---|
| WH0005 | North Pacific Offshore Oahu, HI | 2000 | 19°73′ N 155°07′ W | 99 (3) | 99 (3) | 99–100 (3) | 99–100 (3) | ND |
| WH0002 | North PacificStation ALOHA | 2000 | 22°45′ N 158°00′ W | 99–100 (3) | 99 (3) | 99 (2) | 99 (2) | ND |
| WH0003 | North Pacific Station ALOHA | 2000 | 22°45′ N 158°00′ W | 99–100 (3) | 99 (3) | 99–100 (3) | 99 (3) | 99 (3) |
| WH0004 | North Pacific Station ALOHA | 2000 | 22°45′ N 158°00′ W | 99 (1) | ND | ND | ND | ND |
| WH0401 | Atlantic | 2003 | 6°58.78′ N 49°19.70′ W | 99 (3) | 99 (3) | ND | 99–100 (3) | ND |
| WH0403 | North Atlantic | February 1985 | 13°00′ N 59°00′ W | 99–100 (3) | 99 (3) | ND | 99 (3) | 99 (3) |
| WH8502 | Atlantic | March 1984 | 26°00′ S 42°00′ W | 100 (2) | 99 (3) | ND | 99 (3) | 99 (3) |
Genes were amplified and cloned from individual strains. The number of clones sequenced from each strain is shown in parentheses. ND, not determined. Cyt c, cytochrome c; pt, peptide transferase; DNA pol, DNA polymerase; pdxA, pyridoxal phosphate biosynthetic protein; psbW, photosystem II protein W.
Because all amplified sequences from the different strains were virtually identical, we sequenced two complete C. watsonii-like BAC clones (HOT0_07D09 and HOT0_02H05) from Station ALOHA (Fig. 2). We hypothesized that the complete sequences of these BACs would uncover greater sequence divergence from the C. watsonii WH8501 genome sequence, without the potential artifacts of PCR, and may also uncover differences in gene synteny or other genome rearrangements. However, the sequences of these BAC clones, covering a total of 37 kb of DNA, was >99% identical to the C. watsonii WH8501 genome sequence (Fig. 2).
The first 15.6 kb of DNA sequence of BAC HOT0_07D09 is homologous (99.6% identity) to a portion of contig 286 from C. watsonii WH8501, with the exception of a 1.5-kb insertion in HOT0_07D09 (Fig. 2). This short region encodes an apparent chimera of IS66 and IS4 types of transposases, both of which are found in multiple copies throughout the genome. Immediately following this IS66 and IS4 region is a 353-bp segment similar to IS605-type transposases also found in multiple copies throughout the genome. The end (4.9 kb) of BAC HOT0_07D09 encodes a reverse transcriptase (RT) that is 99.6% identical to a RT encoded on contig 235 but is interrupted on the BAC by a transposon insertion containing a chimera of multiply repeated transposases (Fig. 2). After the RT is another transposase that is 99.8% identical to that encoded at the end of C. watsonii WH8501 contig 235, and this transposase type is present in a few, less well conserved (<90% identical) copies in the genome.
BAC HOT0_02H05 is also very similar to the C.watsonii WH8501 genome. The first 12 kb of the 16.2-kb BAC HOT0_02H05 are homologous (99.75% identical) to contig 247 of the genome, including 11 complete ORFs (Fig. 2). There is a 1.7-kb insertion in the BAC relative to the genome, which is followed by a 2.4-kb region of homology (99.6% identity) to contig 247. The 1.7-kb insertion encodes an IS5-type transposase, which is present in multiple copies in the genome. The 2.4-kb region contains two hypothetical proteins that are found in the same order on contig 247 but without the 1.7-kb intervening sequence (Fig. 2). A third Station ALOHA metagenome fragment (fosmid HF0070_14C03), ≈40 kbp in length, which has recently been sequenced, is also >99% identical over its entire length (GenBank accession no. EU125530). The comparison of these BAC sequences to the C. watsonii genome shows that not only are functional gene sequences (e.g., genes sequenced from PCR-amplified samples; Table 2) highly conserved at the DNA level, but that even intergenic DNA sequences and gene synteny over large genomic regions are also highly conserved.
The lack of gene sequence diversity found in the metagenomic sequences (Fig. 2) and amplified sequences from cultivated isolates (Table 2) is consistent with previous observations that nifH and 16S rRNA gene sequences from natural populations of group B cyanobacteria from the North and South Pacific Ocean (12, 21), the Arabian Sea (11), and the North and South Atlantic Ocean (14, 22–24) are nearly identical (>98% at the nucleotide level) to the genes of cultivated strains of C. watsonii (20). Although the nitrogenase (nifH) gene is highly conserved, nifH sequences are more divergent than are the respective 16S rRNA sequences (25), and nifH is used as a taxonomic marker in some studies (26). Evidence from amplification of genes from strains and natural populations, and the analysis of metagenomic fragments, suggests that there is very little gene sequence diversity in C. watsonii strains throughout tropical and subtropical oceans (Fig. 3). There is much more gene sequence variation in strains of Prochlorococcus and Synechococcus. For example, DNA polymerase protein sequences are virtually identical in all C. watsonii strains examined (and the underlying DNA sequences are also 99% identical), but the same protein and encoding DNA sequences are very divergent among other cyanobacterial strains (Fig. 4). The other genetic loci examined had similar patterns, with 99% DNA sequence conservation in C. watsonii strains (Fig. 2 and Table 2), but with high variability among Prochlorococcus and Synechococcus strains (Figs. 1 and 4).
Fig. 3.
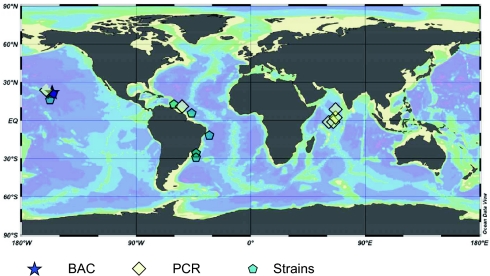
Summary of different types of evidence for similarity of C. watsonii strains from the Atlantic and Pacific oceans and the Arabian Sea. Geographic distribution of gene sequences amplified from C. watsonii isolates (>97% DNA sequence identity; indicated by pentagons), PCR-amplified genes from natural populations including those from previous reports (>97% DNA sequence identity; indicated by diamonds) (11, 12, 18, 22, 24), and sequences from two metagenomic fragments (>99% DNA sequence identity; indicated by a star).
Fig. 4.
Phylogenetic tree of cyanobacterial DNA polymerase I protein sequences showing genetic diversity among Prochlorococcus and Synechococcus strains compared with gene conservation in Crocosphaera strains. Scale bar represents 0.1 amino acid substitutions per site. Nucleotide sequence divergences in Prochlorococcus and Synechococcus are even greater than is exhibited in this amino acid phylogeny, but C. watsonii sequence similarities are 99%, even at the DNA level.
Although there is a high degree of C. watsonii DNA sequence similarity among PCR products, genome sequence, and BAC and fosmid sequences, there undoubtedly exists genetic variants that maintain species diversity. Individual strains of C. watsonii vary in temperature optima, exopolysaccharide production, and other physiological characteristics (J. Waterbury, personal communication). Interestingly, these strains are very closely related, as shown here and by analysis of ITS sequences (J. Waterbury and E. Webb, personal communication). It may ultimately be demonstrated that there are specific allelic variations or SNPs among these strains and natural populations of C. watsonii, but it is remarkable that there is not more diversity in gene sequences as exhibited in the natural populations and cultures of Prochlorococcus. Rusch et al. (9) showed that across environments there were very few DNA fragments showing >85% sequence identity to Prochlorococcus genomes by fragment recruitment analysis. In the case of C. watsonii, with just two random samples, collected from different ocean basins 20 years apart, >40 kb of DNA sequence exhibits >99% sequence identity, including coding and intergenic regions (Fig. 2 and Table 2). It would be impossible to obtain these results from two random C. watsonii samples from different ocean basins if natural variation was similar in magnitude to that of Prochlorococcus (8, 9).
C. watsonii physiological diversity might be maintained by random gene activation/inactivation by transposases, which could foster phenotypic diversity among global populations. Genome rearrangements in the two completely sequenced Pacific Ocean BAC sequences were adjacent to highly repetitive (transposase or hypothetical protein) sequences in the cultivar genome (colored in gold, Fig. 2). There are >400 transposases in the C. watsonii WH8501 genome. Although the rearrangements could be caused by artifacts of the original genome assembly, two pieces of evidence suggest otherwise: (i) the insertions are seen in the BAC fragments, which were not shotgun sequenced and (ii) recent experiments indicate that other cultivated strains of C. watsonii contain the insertion seen in the HOT0_07D09 BAC. Primers designed to the RT, which was interrupted in BAC HOT0_07D09 (Fig. 2), amplified either 2.4- or 0.8-kbp fragments from the C. watsonii strains (S.R.B., unpublished data). Strains C. watsonii WH8501, WH8502, WH0002, WH0003, WH0004, WH0005, and WH0402 had a 0.8-kbp amplification product showing that there was no insertion in the RT gene, but strain C. watsonii WH0401 and BAC HOT0_07D09 had a 2.4-kbp amplification product indicating that the natural populations shared the insertion found in C. watsonii WH0401. Thus, it is likely that genomic rearrangement, rather than gene sequence divergence, is the mechanism that maintains population diversity in C. watsonii. The C. watsonii genome is likely to be an example of the evolutionary strategy of DNA rearrangement, mediated by “evolution genes,” transposases, and other mobile elements, whereas that of the more abundant Prochlorococcus exhibit local sequence change strategies (27). Recently, an analysis of the transposase genes and insertion elements in the C. watsonii genome indicated that the genes are under positive selection and may be involved in increasing host fitness or invasion of new environments (28). Furthermore, in recent analysis of microbial community transcripts (metatranscriptomics) using pyrosequencing (454 Life Sciences, Branford, CT), transcript sequences were obtained that were 100% identical to C. watsonii IS891/IS1136/IS1314 and IS605 elements (GenBank accession nos. EU124865-EU124867).
It is likely that genetic and genomic conservation as reported here is present in other microbial populations in the ocean and elsewhere. The nifH sequences of C. watsonii amplified from the Atlantic Ocean, the Pacific Ocean, and the Arabian Sea are >98% identical at the nucleotide level (Fig. 3) (11, 12, 14, 24). This level of nifH gene conservation is also observed in the open-ocean filamentous cyanobacterial species Trichodesmium (29, 30) and freshwater heterocyst-forming cyanobacteria (Cylindrospermopsis) separated by large geographic distances (26). It is possible that the low genetic diversity observed in C. watsonii is an evolutionary strategy for less abundant microorganisms, such as the diazotrophs. C. watsonii concentrations are several orders of magnitude lower than those of Prochlorococcus (15). Diazotrophs at times can be extremely rare and depend on dispersal of low concentrations of cells that exploit N2-fixing conditions when they occur (23). In contrast, gene space in Prochlorococcus is maintained by large population numbers (31). Abundant microorganisms may benefit from population diversification involving gene loss, genome reduction, and lateral gene transfer mediated by viruses (32, 33). It may be beneficial for rare species, such as planktonic N2-fixing cyanobacteria, to use different evolutionary strategies to adapt to localized marine planktonic niches spanning the global tropical ocean.
In summary, open-ocean populations of C. watsonii have remarkably low genetic diversity, even in temporally and geographically separated samples. The gene sequence diversity among cultivated strains and naturally occurring C. watsonii originating from different ocean basins appears to be much lower than that found among Prochlorococcus cultures (3, 34) and natural populations (7, 8, 35) and suggests that there are multiple strategies of evolution, adaptation, and diversification in oceanic microbial populations.
Methods
Sample Collection.
Water samples were collected at the Hawaii Ocean Time-series Station ALOHA in the North Pacific subtropical gyre by using a conductivity-temperature-depth rosette. Samples for BAC and fosmid cloning were collected in October 2002 from 10- to 500-m depths as described (7). The protocols used for preservation, DNA extraction, and fosmid and BAC library construction are described in refs. 7 and 36.
DNA samples collected from 25-m depth at Station ALOHA (Hawaii Ocean Time-series cruise 129) were used for PCR amplification experiments (37). DNA was extracted from samples, cultures, or enrichments (0.5–2 l) as described (15).
Strains.
Strains of C. watsonii used for PCR amplification were provided by J. Waterbury (Woods Hole Oceanographic Institution, Woods Hole, MA) and E. Webb (University of Southern California, Pasadena). See Table 2.
PCR Amplification.
Several gene fragments were amplified from cultivated isolates. Two BAC clones (HOT0_07D09 and HOT0_02H05) with BAC end sequences HOT0_07D09F and HOT0_07D09R, HOT0_02H05F, and HOT0_02H05R were aligned to the C. watsonii WH8501 genome to identify genes to amplify by PCR. These genes were: cytochrome c oxidase subunit II (Cyt c), peptidase transferase (pt), DNA polymerase A (DNApol), pyridoxal phosphate biosynthetic protein (pdxA), and photosystem II protein W (psbW). The forward (f) and reverse (r) PCR primer sequences are: psbW f, 5′-ACCCCTTCCATCGAGTTCTT-3′; psbW r, 5′-CGATCAATGCTCATCTTT-3′; pdxA f, 5′-GCAAAATCTTGCTGCAAACA-3′; pdxA r, 5′-AGGGACAGCCCTTAGTCCAT-3′; pt f, 5′-CCACAAGGAGGAACCGTTTA-3′, pt r, 5′-AACATCATGGCCTCGTTCTC-3′; DNApol f, 5′-GGCGGAACATTCACAGTTTT-3′, DNApol r, 5′GTGTCCCAAGCAACAGGAAT-3′; Cyt c f, 5′-ATGCAAGCTTCGGCACTATC-3′; and Cyt c r 5′-GTCCTCCCATTGAAGGGAAT-3′. The primers were used in PCRs (50 μl) containing 5–30 ng DNA, 5 μl of 10× buffer without MgCl2 (Promega, Madison, WI), 8 μl MgCl2 (25 mM) (Promega), 1 μl each primer (50 μM each), 1 μl of each nucleotide triphosphate (10 mM), and 0.5 μl of TaqDNA polymerase (5 units/μl) (Promega). Thirty cycles of amplification were performed under the following conditions: 94°C for 30 s, 57°C annealing for 30 s, 72°C extension for 1 min, and one final extension step at 72°C for 7 min. We used special rooms for preparing PCRs to avoid contamination with PCR products or recombinant DNA. We processed PCRs in the DNA clean room where recombinant DNA work is not performed, using separate PCR hoods for preparing reactions, and adding the genomic DNA. Positive controls were added outside this laboratory. No-DNA negative controls were included with each reaction.
BAC and Fosmid Libraries.
The surface water BAC library was collected in December 2001, and BAC libraries were prepared as described (36). Fosmid libraries were collected in October 2002 from 10- to 500-m depths, and fosmid libraries were prepared as described (7).
Sequencing.
PCR products were cloned into pGEM-T vectors and sequenced at the University of California, Berkeley sequencing center. BACs were sequenced by preparing random transposon insertion libraries with Tn5 (EZ-Tn5; Epicentre, Madison WI). Tn5 insertion clones were sequenced in both directions by using KAN-2 FP-1 and KAN-2 RP-1 primers (Epicentre) by BigDye v3.1 cycle sequencing chemistry and an ABI Prism 3700 DNA analyzer (Applied Biosystems, Foster City, CA).
Alignments and Analysis.
PCR-amplified and BAC DNA sequences were compared with the C. watsonii WH8501 genome using BLAST on the National Center for Biotechnology Center server (www.ncbi.nlm.nih.gov) or on an Apple Bioinformatics cluster at the University of California, Santa Cruz. Complete BAC sequences were aligned to the C. watsonii WH8501 genome with BLAST and visualized by using ACT (www.sanger.ac.uk/Software/ACT). DNA polymerase amino acid sequences were retrieved from GenBank and aligned with translations of sequences generated in this study by using HMMR. The resulting alignment was used to construct a neighbor joining tree in ARB (38).
Genome Analysis.
A more recent draft of the C. watsonii WH8501 genome (composed of 120 contigs) was provided before GenBank release by Cliff Han at the Los Alamos National Laboratory, Los Alamos, NM. The most recent draft genome sequence (Joint Genome Institute–Los Alamos National Laboratory) had rearrangements compared with the previous release (comprised of >300 contigs) deposited in GenBank (accession no. AADV00000000.2).
Acknowledgments
We thank D. Karl, M. Church, and the Hawaii Ocean Time-series staff for facilitating sample collection; J. Waterbury and E. Webb for strains of C. watsonii; C. Han for providing access to the updated draft C. watsonii genome sequence; R. Poretsky and I. Hewson for unpublished metatranscriptomics data; and S. W. Chisholm for encouragement and helpful comments on the manuscript. The C. watsonii WH8501 genome sequence data were produced by the U.S. Department of Energy Joint Genome Institute (www.jgi.doe.gov). This research was supported by National Science Foundation Grant OCE-0425363 (to J.P.Z.) and the Gordon and Betty Moore Foundation (J.P.Z. and E.F.D.).
Abbreviation
- RT
reverse transcriptase.
Footnotes
References
- 1.Field KG, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni SJ. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson JR, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt DE, Benoit J, Sarma-Rupavtarm R, Distel DL, Polz MF. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. [DOI] [PubMed] [Google Scholar]
- 3.Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, et al. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- 4.Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Duprat S, Galperin MY, Koonin EV, Le Gall F, et al. Proc Natl Acad Sci USA. 2003;100:10020–10025. doi: 10.1073/pnas.1733211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rocap G, Distel DL, Waterbury JB, Chisholm SW. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Béjà O, Koonin EV, Aravind L, Taylor LT, Seitz H, Stein JL, Bensen DC, Feldman RA, Swanson RV, DeLong EF. Appl Environ Microbiol. 2002;68:335–345. doi: 10.1128/AEM.68.1.335-345.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, Martinez A, Sullivan MB, Edwards R, Brito BR, et al. Science. 2006;311:496–503. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- 8.Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu DY, Paulsen I, Nelson KE, Nelson W, et al. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- 9.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falkowski PG. Nature. 1997;387:272–275. [Google Scholar]
- 11.Mazard SL, Fuller NJ, Orcutt KM, Bridle O, Scanlan DJ. Appl Environ Microbiol. 2004;70:7355–7364. doi: 10.1128/AEM.70.12.7355-7364.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zehr JP, Waterbury JB, Turner PJ, Montoya JP, Omoregie E, Steward GF, Hansen A, Karl DM. Nature. 2001;412:635–638. doi: 10.1038/35088063. [DOI] [PubMed] [Google Scholar]
- 13.Montoya JP, Holl CM, Zehr JP, Hansen A, Villareal TA, Capone DG. Nature. 2004;430:1027–1032. doi: 10.1038/nature02824. [DOI] [PubMed] [Google Scholar]
- 14.Falcón LI, Carpenter EJ, Cipriano F, Bergman B, Capone DG. Appl Environ Microbiol. 2004;70:765–770. doi: 10.1128/AEM.70.2.765-770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Church MJ, Jenkins BD, Karl DM, Zehr JP. Aquat Microb Ecol. 2005;38:3–14. [Google Scholar]
- 16.Neveux J, Lantoine F, Vaulot D, Marie D, Blanchot J. J Geophys Res. 1999;104:3311–3321. [Google Scholar]
- 17.Campbell L, Liu HB, Nolla HA, Vaulot D. Deep-Sea Res I. 1997;44:167–192. [Google Scholar]
- 18.Zehr JP, Mellon MT, Zani S. Appl Environ Microbiol. 1998;64:3444–3450. doi: 10.1128/aem.64.9.3444-3450.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. Appl Environ Microbiol. 2005;71:5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterbury JB, Rippka R. In: Bergey's Manual of Systematic Bacteriology. Staley JT, Bryant MP, Pfenning N, Holt JG, editors. Vol 3. Baltimore: Williams & Wilkins; 1989. pp. 1728–1729. [Google Scholar]
- 21.Hewson I, Moisander PH, Morrison AE, Zehr JP. ISME J. 2007;1:78–91. doi: 10.1038/ismej.2007.5. [DOI] [PubMed] [Google Scholar]
- 22.Falcón LI, Cipriano F, Chistoserdov AY, Carpenter EJ. Appl Environ Microbiol. 2002;68:5760–5764. doi: 10.1128/AEM.68.11.5760-5764.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hewson I, Moisander PH, Achilles KM, Carlson CA, Jenkins BD, Mondragon EA, Morrison AE, Zehr JP. Aquat Microb Ecol. 2007;46:15–30. [Google Scholar]
- 24.Langlois RJ, LaRoche J, Raab PA. Appl Environ Microbiol. 2005;71:7910–7919. doi: 10.1128/AEM.71.12.7910-7919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zehr JP, Jenkins BD, Short SM, Steward GF. Environ Microbiol. 2003;5:539–554. doi: 10.1046/j.1462-2920.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- 26.Dyble J, Paerl HW, Neilan BA. Appl Environ Microbiol. 2002;68:2567–2571. doi: 10.1128/AEM.68.5.2567-2571.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arber W. FEMS Microbiol Rev. 2000;24:1–7. doi: 10.1111/j.1574-6976.2000.tb00529.x. [DOI] [PubMed] [Google Scholar]
- 28.Mes THM, Doeleman M. J Bacteriol. 2006;188:7176–7185. doi: 10.1128/JB.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Porath J, Carpenter EJ, Zehr JP. J Phycol. 1993;29:806–810. [Google Scholar]
- 30.Zehr JP, Limberger RJ, Ohki K, Fujita Y. Appl Environ Microbiol. 1990;56:3527–3531. doi: 10.1128/aem.56.11.3527-3531.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hess WR. Curr Opin Biotechnol. 2004;15:191–198. doi: 10.1016/j.copbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Dufresne A, Garczarek L, Partensky F. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berg OG, Kurland CG. Mol Biol Evol. 2002;19:2265–2276. doi: 10.1093/oxfordjournals.molbev.a004050. [DOI] [PubMed] [Google Scholar]
- 34.Moore LR, Post AF, Rocap G, Chisholm SW. Limnol Oceanogr. 2002;47:989–996. [Google Scholar]
- 35.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 36.de al Torre JR, Christianson LM, Béjà O, Suzuki MT, Karl DM, Heidelberg J, DeLong EF. Proc Natl Acad Sci USA. 2003;100:12830–12835. doi: 10.1073/pnas.2133554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zehr JP, Montoya JP, Jenkins BD, Hewson I, Mondragon E, Short CM, Church MJ, Hansen A, Karl DM. Limnol Oceanogr. 2007;52:169–183. [Google Scholar]
- 38.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]