Abstract
In both vertebrates and invertebrates, ion channels of the TRP superfamily are known to be influenced by a variety of accessory factors, but the list of interacting proteins is acknowledged to be incomplete. Although previous work showed that Drosophila TRP function is disrupted by mutations in the inaF locus, the mechanism of this effect has remained obscure. Here we show that a previously overlooked small protein, INAF-B, is encoded by the locus and fulfills its critical role in retinal physiology. The 81-aa INAF-B gene product is an integral membrane protein that colocalizes to rhabdomeres along with TRP channels. Immunoprecipitation experiments demonstrate that the two proteins participate in a complex, and blotting experiments show that neither protein survives in the absence of the other. Both proteins are normally part of a large supramolecular assembly, the signalplex, but their interaction persists even in the absence of the scaffold for this structure. The inaF locus encodes three other proteins, each of which has diverged from INAF-B except for a 32-aa block of residues that encompasses a transmembrane domain. This conserved sequence defines an inaF motif, representatives of which are found in proteins from organisms as diverse as nematodes, fish, and humans. Given the role of INAF-B, these proteins are good candidates for interacting partners of other members of the TRP superfamily.
Keywords: accessory protein, complex formation, ion channel, transmembrane interaction
Members of the superfamily of TRP channels play important roles in the biology of many cell types, largely through their capacity to serve as conduits for the entry of calcium ions (1). An important theme that has emerged from research on the superfamily is that these channels are often found in association with other proteins (2, 3). Such accessory factors serve to target the channels to appropriate locations within the cell, control their receptivity to extracellular and intracellular signals, and relay the resulting local calcium signals to other cellular components and compartments. In these studies, a large role has been played by the Drosophila TRP protein, the principal channel for light-induced calcium entry in photoreceptors of the fly (2). TRP was not only the founding member of the superfamily, studies of this channel were instrumental in developing the concept that a multiprotein array (called the signalplex) is critical for its function. Core components of this array include NORPA, a phospholipase C, INAC, a protein kinase, and INAD, a PDZ-type scaffolding protein (2, 4). In this work we identify an additional signalplex component, one encoded by the gene that is the locus of inaF mutations (5).
The hallmark of inaF mutants is their inability to sustain a photoreceptor potential in response to a long pulse of light, a defect that is also typical of trp mutants (5). Further evidence for a connection between the two genes is that levels of TRP protein are greatly reduced in inaF mutants (5). The original study documented that a 3.1-kb inaF transcript (Fig. 1A) is normally expressed at high levels in the retina. The authors also pointed out that the second exon of this message contains a 241-aa ORF; subsequent annotations of inaF (also known as CG2457) have assumed that this ORF is the gene product (6). However, the 241-aa polypeptide starts >500 nt from the 5′-end of the message, many of its codons are uncommon, and close matches to its sequence are not evident in a wide variety of organisms. Spurred by our interest in inaF as a gene that affects the response of Drosophila to volatile anesthetics (J. L. Campbell and H.A.N., unpublished data), we reexamined the coding potential of the transcript. Doing so revealed that the first exon contains an 81-aa ORF that is much nearer the 5′ end of the message than the proposed CG2457 product and includes a lower proportion of uncommon codons. In this work we identify this small polypeptide (Fig. 1B) as the critical gene product, demonstrate its presence in photoreceptors, and show that it associates with TRP. Moreover, we have used the sequence of this inaF polypeptide as a seed to identify a 32-aa motif that is found in small proteins from diverse invertebrates and vertebrates.
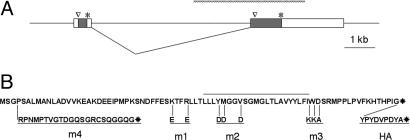
Fig. 1.
The inaF transcript and its coding potential. (A) Diagram of genomic DNA from the region of the inaF message. Two exons are shown that are spliced to make an abundant eye-enriched 3.1-kb transcript. The second exon contains an ORF whose first methionine and stop codon are, respectively, indicated by a caret and asterisk; this 241-aa polypeptide has been proposed to be the CG2457 gene product (5, 6). However, the first exon contains an 81-aa ORF that we show herein to be necessary and sufficient for inaF function. The hatched bar above the exon/intron diagram shows the extent of the P106x deletion (5) that inactivates the gene. (B) Sequence of the 81-aa protein. The overlined region marks a predicted membrane-spanning domain that runs from Leu-42 to Ile-63. Below the sequence are given the amino acid changes for three substitution mutants (m1–m3), a frameshift mutant (m4), and the additional amino acids that are appended to the C terminus to generate a tag (HA).
Results and Discussion
A Polypeptide from the inaF Locus That Is Needed for Proper TRP Function.
To assess the relative importance of the ORFs encoded by the inaF transcript, we performed genomic rescue experiments in a strain in which the endogenous message was inactivated by a deletion allele, inaFP106x (5). This X-linked deletion, which removes the splice acceptor site for the second exon as well as its coding potential (Fig. 1A), confers a strong inaF phenotype that is highlighted by a major depression in levels of TRP protein (Fig. 2A, lane b). Against this background, nearly complete rescue was provided by an autosomally inserted transgene (described in Materials and Methods) that includes both exons and the intervening intron of the inaF locus. Comparable rescue was seen regardless of whether the transgene used the wild-type sequence or a version in which the ORF from the first exon carried a C-terminal tag (Fig. 2A, lanes c and d). In contrast, there was little or no rescue with versions of the transgene in which the 81-aa ORF was engineered to suffer nonconservative substitutions in and around its putative transmembrane domain. In three of three such cases, each with a different multiple substitution (Fig. 1B), little or no TRP could be detected (Fig. 2A, lane e, and data not shown). Similarly, a transgene in which a frameshift was introduced just downstream of the predicted start of this ORF (Fig. 1B) failed to provide rescue (Fig. 2A, lane f). On the other hand, a transgene with a similarly frameshifted version of the 241-aa polypeptide was able to provide strong rescue of the inaF phenotype (Fig. 2A, lane g). It thus appears that the 81-aa polypeptide but not the 241-aa polypeptide is necessary for TRP channels to accumulate to proper levels in photoreceptors. A corollary of this conclusion is that the inaFP106x mutation disrupts function of the native (X-linked) gene not by deleting the 241-aa ORF but by damaging an essential 3′ UTR for its 81-aa ORF.
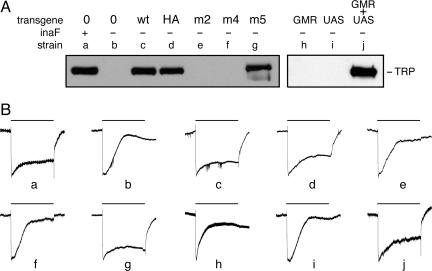
Fig. 2.
Transgenic rescue of the inaF phenotype. (A) Total extracts of head proteins from various strains were probed with anti-TRP. All strains (except for a) carried the X-linked inaFP106x deletion allele. Strains labeled c–g also carried a 13-kb segment of genomic DNA from the inaF region on an autosome. In strain c, this transgene carried the wild-type sequence; in strain d, the wild-type sequence was kept intact, but an HA tag was appended to the C terminus of the 81-aa ORF; in strains e and f, this ORF was modified by introduction of a triple substitution and a frameshift, respectively; in strain g, the 241-aa ORF on the transgene was modified by introduction of a frameshift. Strains labeled h–j carried, as indicated, a transgene bearing the eye-specific GMR-GAL4 driver (23), a UAS transgene bearing the 3.1-kb inaF cDNA, or both transgenes. Construction of the inaF transgenes is described in Materials and Methods; the sequence alteration of the transgene in strains d–f is given in Fig. 1B. (B) Electroretinograms recorded as described (24) from adults of the strains whose genotype is given in A. The bar above each trace shows the 20-s period during which these dark-adapted flies were exposed to white light (88,000 lux). Each trace was normalized to the potential difference that developed 0.25–0.50 s after the onset of light pulse. In traces a–g and h–j, the absolute value of this potential ranged from 16 to 22 mV and from 12 to 14 mV, respectively. In unrescued or poorly rescued flies (b, e, f, h, and i), by the end of the light pulse the photoreceptor potential decayed to the baseline or close to it. Conversely, in wild-type and well rescued flies (a, c, d, g, and j), the photoreceptor potential is still substantial at the end of the light pulse.
To test whether the TRP protein rescued with the help of the various transgenes is functional, we recorded electroretinograms from the strains described above, which demonstrated that normal visual function accompanied restoration of protein level by wild-type and tagged transgenes (Fig. 2B, traces c and d). It also proved that if the inaF locus plays a role in targeting TRP channels to the rhabdomere, the 241-aa polypeptide is not necessary to fulfill it (Fig. 2B, trace g).
The INAF Protein Colocalizes with and Interacts with TRP Channels.
Because trp message levels are unaffected by loss of inaF function (Y.C., unpublished observation), we infer that the 81-aa protein (named here as INAF-B, for reasons we explain below) is a critical element for either the translation or stability of TRP channels. The subcellular location of INAF-B should help to distinguish between these alternatives. If this gene product is involved in TRP synthesis and/or posttranslational modification, it should be found in the photoreceptor cell body along with the extensive endoplasmic reticulum and Golgi apparatus (4). On the other hand, if INAF-B is needed to stabilize mature channels, it should be found in the highly invaginated membranes of the photoreceptor rhabdomere, where TRP channels are concentrated (4). To decide between these alternatives, after unsuccessful attempts at raising antibodies to INAF-B, we constructed a tagged version. As described above, this modification does not interfere with inaF function in vivo, so the subcellular location of the tag should faithfully reflect that of the native protein. Strikingly, this strategy reveals that INAF-B is almost exclusively found in the rhabdomere, completely overlapping the distribution of TRP (Fig. 3A). Western blots [supporting information (SI) Fig. 5A] not only confirmed the specificity of antibody staining but demonstrated that the tag was associated with a polypeptide of the predicted size (10.1 kDa). Blotting experiments also provided evidence for a tight relationship between TRP and INAF-B, in that the two proteins shared the same developmental profile, showing little or no accumulation during embryonic and larval stages and being expressed almost exclusively in late pupae and adult heads (Y.C., unpublished observation).
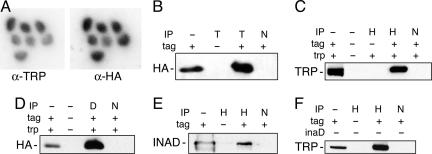
Fig. 3.
Association of HA-tagged INAF-PB with TRP channels. (A) Immunolocalization. A representative confocal section of retina, costained with anti-TRP and anti-HA, from adult flies bearing the tagged version of inaF-PB, is shown. To eliminate interference by eye pigment, the transgenic line also carried bw and st mutations. Within the ommatidium shown, signal from INAF-B is detected in rhabdomeres (and not their surrounding cell bodies) and colocalizes with signal from TRP. (B–F) Immunoprecipitation. The top line of the key for each panel shows the antibody used to precipitate dodecyl-β-maltoside extracts of fly heads: anti-HA (H), anti-TRP (T), anti-INAD (D), or a nonspecific control antibody (N). The second line of the key indicates the presence (+) or absence (−) of an HA tag on a transgenic copy of INAF-B. Where appropriate, the key also indicates the wild-type (+) or mutant (−) status of the locus whose genotype varies in the samples of the panel. Immunoprecipitates were electrophoresed and then probed with the antibody indicated at the left of each panel. The lanes with IP status marked (−) contain a sample (≈3%) of material used as input for the corresponding immunoprecipitations.
The INAF-B polypeptide is predicted to have a single transmembrane domain that is likely to act as a type II signal anchor (www.cbs.dtu.dk/services/SignalP/). To test this prediction, we subjected the tagged protein from fly heads to differential centrifugation and incremental solubilization, which demonstrated that, like TRP, INAF-B is an integral membrane protein (SI Fig. 5B). The intimate colocalization of INAF-B and TRP raises the possibility that these proteins interact. Indeed, immunoprecipitation of detergent extracts of fly heads with anti-TRP followed by probing of Western blots with antibody to the HA tag provides clear evidence of such interaction (Fig. 3B). The converse experiment using anti-HA as the precipitant and anti-TRP as the probe (Fig. 3C) confirms this interpretation. The interaction between these two proteins appears to be mutually vital: not only are TRP levels low in the absence of inaF (Fig. 2A, lane b), extracts of trp mutant heads have undetectable levels of INAF-B (SI Fig. 5C). Moreover, the interaction is specific. Coprecipitation experiments (data not shown) detect little or no association between the tagged version of INAF-B and either TRPγ or TRPL, close relatives of TRP that are also present in the rhabdomere (2). This specificity mimics the mutant phenotype, in that TRPL levels and function are unaffected by inaF mutations (5).
Similar experiments were performed to determine whether INAF-B takes part in the signalplex. When an antibody to the key scaffold for this structure, INAD, is applied to detergent extracts from fly heads, strong signals are seen when immunoprecipitates are probed for the HA tag of INAF-B (Fig. 3D). Conversely, a clear signal is seen when anti-HA precipitates are probed with anti-INAD (Fig. 3E). Thus, INAF-B is indeed an element of the signalplex, even though the sequence of the 81-aa ORF lacks an obvious match to known INAD-binding sites (7). Importantly, INAF-B and TRP continue to coimmunoprecipitate even when the inaD gene has been inactivated by a null mutation (Fig. 3F), showing that the association of these two proteins is not merely a consequence of their incorporation in the signalplex. This finding raises the possibility that TRP and INAF-B interact directly, but we cannot rule out the possibility that another protein acts as a bridge that holds the channel and the 81-aa ORF together during immunoprecipitation experiments. Regardless of whether the interaction is direct or indirect, our observations make it clear that INAF-B should be added to the list of small proteins with a single transmembrane domain, such as MinK (8, 9), that complex with and influence the function of ion channels.
The inaF Family and the inaF Motif.
Flanking the exon that encodes INAF-B there are three stretches of the Drosophila genome that also encode short ORFs (Fig. 4A). Remarkably, each of these (whose full sequence is given in SI Fig. 6) includes a segment with a recognizable match to a segment of the 81-aa polypeptide (Fig. 4B). None of these additional ORFs appears to be included in the standard database of Drosophila genes (6), although they have been noted by at least one gene-finding program (10). To our knowledge, none is represented in EST databases, but, using RT-PCR, we find that each of them is transcribed and spliced to the same downstream exon used by inaF-B. Because the coding potential of each ORF is distinct, the four alternatively spliced isoforms can be considered as separate but related genes encoded by an “inaF locus.” We propose that they be given letter designations according to their genomic order, e.g., inaF-A, inaF-B (hence the name assigned to the 81-aa ORF of Fig. 1B), inaF-C, and inaF-D. The coding potential of inaF-A and inaF-C, like that of inaF-B, is entirely contained in the upstream exon. However, the upstream exon of inaF-D does not contain a stop codon; its ORF continues into the downstream exon and thus includes the sequence annotated as CG2457 plus the coding sequence that lies upstream of its purported initial methionine. Semiquantitative RT-PCR on heads from wild-type and eyes-absent (eya) adults indicates that inaF-C is nearly eye-specific, albeit expressed at much lower levels than the inaF-B transcript. The other isoforms may also be expressed in the eye, but the significant expression outside the eye (i.e., in heads from eya mutants) confounds the analysis. Nevertheless, the fact that there is eye expression of multiple isoforms raises the possibility that TRP function requires the participation of more than one inaF gene. To address this issue, we used the GAL4/UAS system (11) to express a cDNA for the inaF-B isoform in a strain bearing the inaFP106x mutation. Driving this cDNA in photoreceptors restored TRP levels to nearly normal levels (Fig. 2A, lane j), with a consequent restoration of the ERG (Fig. 2B, trace j). Because removal of their common 3′ UTR should inactivate all four inaF genes, it appears that the 81-aa INAF-B product of the locus is necessary and sufficient for conferring normal TRP function.
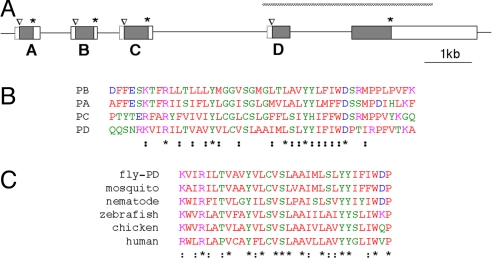
Fig. 4.
The inaF motif. (A) Diagram of the Drosophila genome from the inaF region showing additional exons that flank the exon for INAF-B. Shaded boxes delineate ORFs in the exons; the initial methionine and stop codon for each of them are marked with a caret and an asterisk, respectively. Open boxes enclosed by solid lines indicate UTRs deduced from the sequence of cDNAs. These cDNAs reveal that exons A–D are each spliced to the acceptor site of the rightmost exon; as a result, the methionine in this exon that was marked with a caret in Fig. 1A does not appear to function as the start of an ORF (it is an internal methionine of the inaF-D gene) and is now left unmarked. Open boxes enclosed by dashed lines indicate 5′ UTRs whose extent is uncertain. As in Fig. 1A, the extent of the P106x deletion is diagrammed with a hatched bar. (B) Alignment of a segment of the four ORFs diagramed in A. Residues are colored according to the Clustal (www.ebi.ac.uk/clustalw) convention. Asterisks and semicolons below the last sequence mark positions of identity or conservative substitution, respectively. (C) Alignment of the inaF motif found in INAF-D with similar motifs found in from ORFs from the indicated organisms. Residues are colored as in B, as are positions of conservation.
One might dispute the assumption that the three ORFs that flank inaF-B represent real genes and suggest instead that these DNA segments, despite detectable levels of transcription, are merely the incidental products of a recent set of duplication events. Arguing against this point of view is the presence of all four ORFs in other drosophilids. Even the genome of Drosophila mojavensis, which diverged from melanogaster more than 40 million years ago, contains a cluster of four ORFs whose sequences (SI Fig. 6) align in proper order and from end-to-end with those of melanogaster. Importantly, comparison of the nucleotide regions that encompass these genes in the two organisms reveals that there is little or no sequence conservation outside the coding regions (data not shown). Thus, the four ORFs have specifically resisted genetic drift, implying functional significance.
The portion of the protein that is largely conserved among inaF isoforms (Fig. 4B) is likely to be of special importance. Because the sequence pattern of this segment has not been previously recognized, we propose that it be named the inaF motif. The preponderance of nonpolar residues in a long stretch of this motif implies that it forms a transmembrane domain, and this hypothesis is supported by analysis of each ORF with a topology prediction program (12). Membrane-spanning segments generally can accommodate any of several hydrophobic amino acids at each position. Similarly, the charge but not the identity of residues that flank a transmembrane domain often controls its orientation within the membrane (12). However, in the four examples of the inaF motif found in drosophilids, there are five positions (three within the putative membrane-spanning segment and two flanking it) that are absolutely conserved (Fig. 4B). This sequence conservation implies that the residues of the inaF motif constitute more than just a generic transmembrane domain but are involved in specific interactions. In the case of INAF-B, which may interact directly with TRP, we propose that the residues of the motif contact the intramembrane and perimembrane portions of the channel, whereas the residues that flank the motif might contact its intracellular and extracellular domains. This proposal leads to the hypothesis that the other drosophilid proteins with this motif interact with other members of the TRP superfamily, with specificity for particular channels being derived from small variations in the motif as well as the distinctive flanking residues.
We have searched databases to see the extent to which the inaF motif is found in other organisms. In this exercise, the D isoform proved to be the most useful, as if it were the ancestral form from which the others diverged. Remarkably, in every organism that we scrutinized, we could find at least one predicted protein that incorporated a recognizable version of the inaF motif. Fig. 4C shows an alignment of the conserved segment of these proteins; full sequences are given in SI Fig. 6. Several features are noteworthy. First, 4 of the 5 residues that are perfectly conserved among the drosophilid isoforms (Fig. 4B) are found in all other instances of this motif; they must be of particular functional importance. Second, the variant of the motif found in the drosophilid D isoform must be under very strong selective pressure because 8 additional residues found in this isoform are perfectly conserved during evolution. Third, although some of these ORFs have not been included in curated lists of genes (e.g., the vertebrate members shown in Fig. 4C are not in the Uniprot database), EST databases indicate that all are expressed. Fourth, outside the insect species, we find no evidence for a tandem array of related inaF genes. In some organisms (Caenorhabditis elegans, Gallus gallus, and Danio rerio) the ORF shown is the only one found. In Homo sapiens, two ORFs with the inaF motif are found on separate chromosomes. One of them (shown in Fig. 4C) has a distinct variant of the inaF motif, whereas the other (shown only in SI Fig. 6) is quite closely related to the chicken and zebrafish members. Orthologs of both human ORFs are found in mouse genome, again on separate chromosomes, as if a gene duplication/translocation event occurred early in the mammalian lineage. Finally, despite the impressive conservation of residues within the inaF motif, the amino acid sequences that flank it show considerable evolutionary divergence. As noted above, this divergence is obvious when one compares the four Drosophila tandem genes with each other. The same is true if one compares the sequences of the two human proteins that contain an inaF motif with each other or if one compares the sequence of any of the vertebrate members with any of the invertebrate members. In each of these cases, outside the inaF motif there is virtually no recognizable homology. The variability in flanking amino acid sequence within and between species implies that, if proteins with the inaF motif function as interactors, the partners they contact either covary with them or engage them in ways that are satisfied by nonspecific contacts.
Concluding Remarks.
In this work we have uncovered a protein that interacts with and influences the function of Drosophila TRP channels. In addition to explaining the phenotype described for inaF mutations almost 10 years ago (5), our analysis of the locus opens several promising avenues for future work. For example, it is widely acknowledged that expression of Drosophila TRP in heterologous systems fails to recapitulate the native properties of the channel and may even not yield active channels at all (13, 14). Because the INAF-B protein has an intimate and physiologically significant interaction with these channels, it could be the missing ingredient in such experiments. Furthermore, because the inaF locus encodes three other proteins with a motif found in INAF-B, experiments can now explore whether any of these proteins is partnered with one of the 12 additional members of the TRP superfamily that are encoded in the Drosophila genome so as to enable physiological functions such as hearing, thermosensation, nociception, etc. Finally, our observation that vertebrate genomes contain ORFs with the inaF motif not only empowers a study of these putative proteins but suggests the direction that a search for their interacting partners should take.
Materials and Methods
Fly Strains.
A strain containing the inaFP106x mutation was obtained from William Pak (Purdue University, West Lafayette, IN). The extent of the deletion in this line had previously been determined by EcoRI restriction mapping (5); we refined this estimate by generating a PCR product with primers designed to flank the deletion. The resulting sequence showed that the deletion extends from ≈400 bp upstream of the ORF for inaF-D to the middle of the downstream exon. Strains with the trpP301 and inaD1 mutations were obtained from William Pak and Chales Zuker (University of California at San Diego, La Jolla, CA), respectively. Transgenes were introduced into a w1118 strain by Genetic Services, Inc. (Sudbury, MA), by using coinjection of one of the DNA constructs described below and a helper plasmid as the source of transposase protein. Mutations and transgenes were combined by using standard genetic techniques with balancer chromosomes (15).
DNA Constructs. pP[inaFG].
A genomic DNA clone (dr060) containing the inaF locus in λJ1vector (16) was kindly provided by F. Rob Jackson (Tufts University, Boston, MA). A 2.6-kb EcoRI–BamHI fragment from dr060 was cloned into pBluescript II KS, and the KpnI–EcoRI fragment of this plasmid was replaced by a 1.4-kb PCR fragment of the inaF upstream sequence, obtained by amplification of genomic DNA (using primers 5′-cggggtaccgcggccgcaatgacactgtctcatag-3′ and 5′-ttcgtggtgatctacctgatc-3′) followed by digestion with KpnI and EcoRI. To the resultant plasmid was added a 9.6-kb BamHI fragment of inaF from dr060, yielding the pBS[inaFG] construct. A 13.6-kb NotI fragment from pBS-inaFG, which comprises genomic DNA from 3 kb DNA upstream of the inaF-A gene (Fig. 4A) to 1.3 kb DNA downstream of the final exon of the locus, was cloned into pCaSpeR-4 to yield pP[inaFG].
pP[inaFG-B_HA].
Plasmid pP[inaFG] was modified with recombineering techniques (17, 18) so as to place a HA tag in frame at the C terminus of the presumptive inaF-B ORF. An aliquot (50 μl) of competent cells from strain DY330, kindly provided by Donald Court (National Cancer Institute, Frederick, MD), was transformed by electroporation with 0.8 ng of pP[inaFG] and 7.5 pmol of a PCR product designed to flank an HA tag (YPYDVPDYA) with homology to the appropriate segment of the inaF locus. The latter was generated by amplification (Vent polymerase) of two overlapping primers (5′-gccgccgctgcccgtgttcaagcacacgcatccgattggctacccctacgacgtccccgattacgcc3-′ and 5′-ggagatggatggctaactaaatgatagccatccgatcctaggcgtaatcggggacgtcgtaggggta-3′). Transformant colonies were screened by colony hybridization with a probe to the HA tag (5′-ggcgtaatcggggacgtcgt-3′). The structure of the resulting plasmid, named pP[inaFG-B_HA], was verified by sequencing.
The construction of plasmids in which pP[inaFG] or its HA derivative was modified by substitution (m1–m3) or frameshift (m4 and m5) mutations is described in SI Materials and Methods.
pP[UAS::inaF-B_HA].
A full-length cDNA (GH09956) corresponding to the eye-specific 3.1-kb message from the inaF locus was obtained from the Drosophila Genomic Resource Center (Bloomington, IN). The cDNA was modified by insertion of a HA tag at the C-terminal residue of the inaF-B gene. To do so, RT-PCR was carried out on total RNA isolated from inaFp106X flies bearing a [PinaFG-B_HA] transgene, using primers 5′-ggcagatctaggcggttacggttgcgatt-3′ and 5′-cgattgtttgcctccagctg-3′. The RT-PCR product was used to replace the BglII–SacII fragment of GH09956; a 3.1-kb BglII–XhoI fragment from the resulting chimera was isolated and cloned into the pUAST vector (11). The structure of the resulting plasmid, named pP[UAS::inaF-B_HA], was verified by sequencing.
Western Blots and Immunohistochemistry.
Extracts were made by homogenizing 20–50 fly heads in 50–100 μl of 0.1 M Hepes (pH 7.5) containing 0.1 M KCl, 50 mM EDTA, 5% (vol/vol) glycerol, 1% Triton X-100, and a mixture of protease inhibitors (Roche, Indianapolis, IN). The homogenate was centrifuged for 10 min at 10,000 × g, the supernatant was centrifuged again, and ≈30 μg of protein from the final supernatant was mixed with Novex Tris–glycine SDS sample buffer or NuPage LDS sample buffer (Invitrogen, Carlsbad, CA), heated at 75–85°C for 5 min, and subjected to electrophoresis. After transfer to nitrocellulose membrane, blots were probed with one of the following primary antibodies: TRP [mouse monoclonal MAb83F6 (19), obtained from Seymour Benzar, California Institute of Technology, Pasadena, CA], TRPL and TRPγ [rabbit polyclonals (20), obtained from Craig Montell, The Johns Hopkins University, Baltimore], INAD [rabbit polyclonal (21), obtained from Craig Montell], HA tag [mouse monoclonal HA.11, obtained from Covance Research Products (Berkeley, CA)]. All primary antibodies were used at 1:1,000 dilution. Secondary antibodies were peroxidase-linked donkey anti-IgG, obtained from Jackson ImmunoResearch Laboratories (West Grove, PA) and used according to the supplier's instructions. Blots were developed with the ECL detection system (GE HealthCare, Piscataway, NJ).
Immunohistochemistry on dissected retinas was performed essentially as described (22), except that fixation was with 2% paraformaldehyde. Primary antibodies were rabbit HA-probe Y-11 (Santa Cruz Biotechnology, Santa Cruz, CA), used at 1:100 dilution and mouse TRP mAb83F6, used at 1:600 dilution. Secondary antibodies were Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 568 goat anti-rabbit IgG (Molecular Probes, Eugene, OR).
Immunoprecipitation.
Heads from ≈150 flies (typically 3–7 days old but 1 day old for experiments with inaD1 flies) were homogenized with 400 μl of ice-cold immunoprecipitation buffer (20) and centrifuged at 100,000 × g for 30 min. To the resulting detergent-soluble supernatant, an aliquot of undiluted primary antibody (2 μl for anti-HA, 30 μl for anti-TRP, 5 μl for anti-INAD, 10 μl for control antibody) and 50 μl of protein G–agarose beads (Roche) were added; the mixture was then rotated at 4°C for 2.5 h. The beads were collected by centrifugation at 10,000 × g for 1 min at 4°C and subjected five washes, each with 400 μl of ice-cold immunoprecipitation buffer. The immune complexes remaining in the beads were recovered by heating at 85°C for 10 min with 50 μl of SDS sample buffer followed by 2 min of centrifugation at 10,000 × g. An aliquot (25 μl) of the supernatant was loaded on either an 8% Novex acrylamide/Tris–glycine gel or a 10% NuPage Novex 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)-1,3-propanediol gel (Invitrogen) for electrophoresis and subsequent Western blotting.
Supplementary Material
Acknowledgments
Joy Qun Gu provided expert and efficient technical help with every aspect of this work. We also thank Robert Scott for instruction in electroretinography and for performing immunohistochemical staining. We acknowledge Michael Perry (Accelrys, San Diego, CA) for advice about assembling databases in the GCG package. This work was supported by the Intramural Research Program of the National Institute of Mental Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. EU146932 (inaF-RA), EU146933 (inaF-RD), EU146934 (inaF-RC), and EU153100 (inaF[P106x] deletion allele)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0708368104/DC1.
References
- 1.Ramsey IS, Delling M, Clapham DE. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 2.Montell C. Sci STKE. 2005;2005:re3. doi: 10.1126/stke.2722005re3. [DOI] [PubMed] [Google Scholar]
- 3.Kiselyov K, Shin DM, Kim JY, Yuan JP, Muallem S. Handb Exp Pharmacol. 2007;179:559–574. doi: 10.1007/978-3-540-34891-7_33. [DOI] [PubMed] [Google Scholar]
- 4.Raghu P. Semin Cell Dev Biol. 2006;17:646–653. doi: 10.1016/j.semcdb.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Geng C, Leung HT, Hong YS, Strong LL, Schneuwly S, Pak WL. Proc Natl Acad Sci USA. 1999;96:13474–13479. doi: 10.1073/pnas.96.23.13474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drysdale RA, Crosby MA. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaccaro P, Dente L. FEBS Lett. 2002;512:345–349. doi: 10.1016/s0014-5793(02)02220-2. [DOI] [PubMed] [Google Scholar]
- 8.McCrossan ZA, Abbott GW. Neuropharmacology. 2004;47:787–821. doi: 10.1016/j.neuropharm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Panaghie G, Tai KK, Abbott GW. J Physiol (London) 2006;570:455–467. doi: 10.1113/jphysiol.2005.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hild M, Beckmann B, Haas SA, Koch B, Solovyev V, Busold C, Fellenberg K, Boutros M, Vingron M, Sauer F, et al. Genome Biol. 2003;5:R3. doi: 10.1186/gb-2003-5-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand AH, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 12.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 13.Hardie RC. Annu Rev Physiol. 2003;65:735–759. doi: 10.1146/annurev.physiol.65.092101.142505. [DOI] [PubMed] [Google Scholar]
- 14.Minke B, Parnas M. Annu Rev Physiol. 2006;68:649–684. doi: 10.1146/annurev.physiol.68.040204.100939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greenspan RJ. Fly Pushing: The Theory and Practice of Drosophila Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2004. [Google Scholar]
- 16.DiBartolomeis SM, Akten B, Genova G, Roberts MA, Jackson FR. Mol Genet Genomics. 2002;267:564–576. doi: 10.1007/s00438-002-0700-7. [DOI] [PubMed] [Google Scholar]
- 17.Yu D, Sawitzke JA, Ellis H, Court DL. Proc Natl Acad Sci USA. 2003;100:7207–7212. doi: 10.1073/pnas.1232375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. Proc Natl Acad Sci USA. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pollock JA, Assaf A, Peretz A, Nichols CD, Mojet MH, Hardie RC, Minke B. J Neurosci. 1995;15:3747–3760. doi: 10.1523/JNEUROSCI.15-05-03747.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu XZ, Chien F, Butler A, Salkoff L, Montell C. Neuron. 2000;26:647–657. doi: 10.1016/s0896-6273(00)81201-5. [DOI] [PubMed] [Google Scholar]
- 21.Wes PD, Xu XZ, Li HS, Chien F, Doberstein SK, Montell C. Nat Neurosci. 1999;2:447–453. doi: 10.1038/8116. [DOI] [PubMed] [Google Scholar]
- 22.Wu JS, Luo L. Nat Protoc. 2006;1:2110–2115. doi: 10.1038/nprot.2006.336. [DOI] [PubMed] [Google Scholar]
- 23.Freeman M. Cell. 1996;87:651–660. doi: 10.1016/s0092-8674(00)81385-9. [DOI] [PubMed] [Google Scholar]
- 24.Rajaram S, Scott RL, Nash HA. Proc Natl Acad Sci USA. 2005;102:17840–17845. doi: 10.1073/pnas.0508858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.