Abstract
We report a multiple-stage ion-trap (IT) mass spectrometric approach with electrospray ionization (ESI) for structural characterization of the [M − 2H + Na]− ion of cardiolipin (CL), a 1,3-bisphosphatidyl-sn-glycerol that consists of four fatty acyl chains and three glycerol backbones designated as A, B, and central glycerol, respectively (see Scheme 1). Following collisionally activated dissociation (CAD), the [M − 2H + Na]− ions of CL yield two prominent fragment ions that arise from the differential losses of the diacylglycerol moieties containing A or B glycerol, respectively. The tentative assignment of the two phosphatidyl moieties attached to the 1′- or 3′-position of the central glycerol is based on the observation that the ions arising from loss of the diacylglycerol moiety containing glycerol B is more abundant than that containing glycerol A. The structures of the above two ions, including the identities of the fatty acyl substituents and the position of fatty acyl substituents on the glycerol backbones (glycerol A and B) are determined by MS3 experiments that give spectra comprising several sets of prominent ions informative for the structural assignment of the fatty acyl substituents on the glycerol A and glycerol B. This method permits the structures of CL in a mixture isolated from Escherichia coli, including species that consist of various isomers, to be unveiled in detail.
ESI with tandem mass spectrometry using tandem quadrupole [1], quadrupole time-of-flight (Q-TOF) [2], and ion-trap [3] instruments have been used for the characterization of cardiolipin and glucosylcardiolipin as the [M − H]− and [M − 2H + Na]− ions in the negative-ion mode and as the [M − 2H + 3Na]+ ion in the positive-ion mode. The subclasses of cardiolipin of glucosyl-, alanyl- and lysocardiolipins, were also previously characterized by FAB with-sector mass spectrometer [4]. These methods are useful for identification of the fatty acyl substituents but are not useful for determination of their positions on the glycerol backbone.
Recently, we described an approach using tandem quadrupole in conjunction with multiple-stage ion-trap mass spectrometric methods for the characterization of cardiolipins as their [M − H]− and [M − 2H]2− ions generated by ESI. The methods demonstrated the potential use of tandem mass spectrometry in the structural characterization of complex CL in mixtures [5]. In this report, we describe an alternative multiple-stage IT mass spectrometric approach that uses the [M − 2H + Na]− ions formed by ESI for structural characterization of CL in a mixture isolated from E. coli. Because the methanolic solution of the disodium salt of CL from biological samples such as from E. coli is readily ionizable and gives prominent [M − 2H + Na]− ion species that yield rich structural information upon collisional activation, characterization of CL as the [M − 2H + Na]− ion can be useful for samples of biological origin.
Materials
Cardiolipin isolated from E. coli was purchased from Avanti Polar Lipid (Alabaster, AL). All chemicals used in the analysis were purchased from Fisher Scientific (Pittsburgh, PA).
Methods
Mass Spectrometry
Low-energy CAD tandem mass spectrometry experiments were conducted on a Finnigan (San Jose, CA) LCQ DECA ion-trap mass spectrometer (ITMS) with the Xcalibur operating system. The methanolic CL mixture from E. coli (10 pmol/μL) was continuously infused (3 μL/min) to the ESI source, where the skimmer was set at ground potential, the electrospray needle was set at 4.5 kV, and temperature of the heated capillary was 260 °C. The automatic gain control of the ion trap was set to 5 × 107, with a maximum injection time of 400 ms. Helium was used as the buffer and collision gas at a pressure of 1 × 10−3 mbar. A zoom scan was used to acquire full scan mass spectra, so that doubly-charged ions from CL can be baseline resolved. The MSn experiments were carried out with a relative collision energy ranged from 30–40% and with an activation q value at 0.25. The activation time was set at 100 ms. Mass spectra were accumulated in the profile mode, typically for 3–5 min for MS2- and MS3-spectra. The mass resolution of the instrument was tuned to 0.6 Da at half peak height.
Nomenclature
To simplify data interpretation, we adopt the rules recommended by IUPAC with modification for designation of CL. Briefly, the three glycerol moieties are designated as A, B, and central glycerol (Scheme 1). The stereospecific numbering (sn) of the C1A and C2A (A glycerol) carbons are designated as sn-1 and sn-2, respectively. The C1B and C2B (B glycerol) carbons are designated as sn-1′ and sn-2′, respectively. The carbon number of the central glycerol is designated as C-1′, C-2′, and C-3′ with the C-1′ attached to the phosphatidic moiety with glycerol A. Abbreviation of cardiolipin, such as (16:0/16:1)(18:0/18:1)-CL signifies that the 16:0-, 16:1-, 18:0-, and 18:1-fatty acyl substituents attach to C1A, C2A, C1B, and C2B, respectively. The production spectra from MSn (n = 2, 3, 4) experiments are denoted as the MSn-spectra (n = 2, 3, 4).
Scheme 1.
Results and Discussion
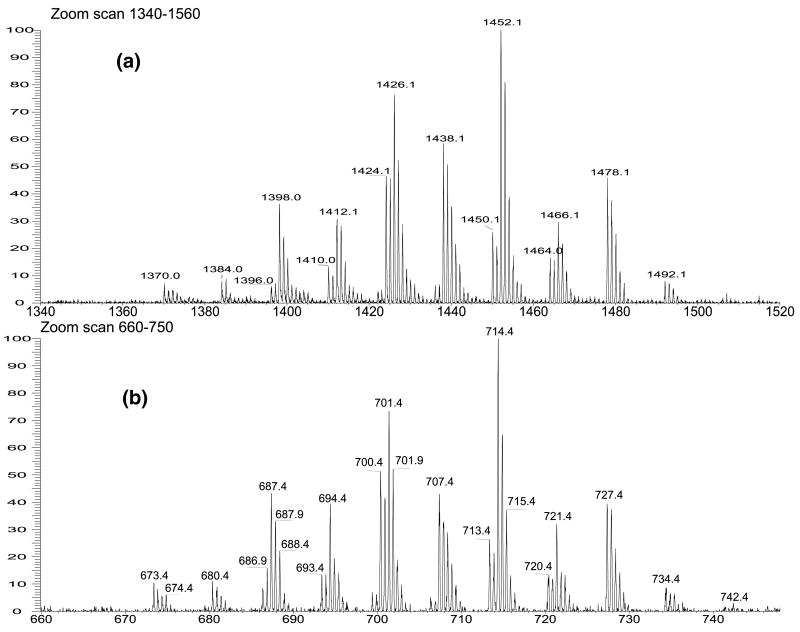
Cardiolipins possess two anionic charge sites [6, 7]; thus they form both [M − H]− and [M − 2H]2− ions, when being subjected to ESI, following a clean-up step to remove Na+ from solution [5]. In this study, we found that both the [M − 2H + Na]− and [M − 2H]2− ions are readily formed when a methanolic solution of the disodium salt of CL was ionized, consistent with that previously reported by Beckedorf et al. [2]. As illustrated in Figure 1, the profiles of the ESI zoom scan mass spectra of the [M − 2H + Na]− (Panel a) and [M − 2H]2− (Panel b) ions species of the CL mixture from E. coli are nearly identical. The profiles are also identical to that observed for [M − H]− ions from the sample in which Na+ has been removed (data not shown). The tandem mass spectrometric methods for characterization of cardiolipin as [M − 2H + Na]− ions from extract of E. coli, including MS4 mass spectrometry are described below.
Figure 1.
The zoom scan ESI/MS spectra of (a) the [M − 2H + Na]− ions and (b) the [M − 2H]2− ions of the cardiolipin mixture isolated from E. coli.
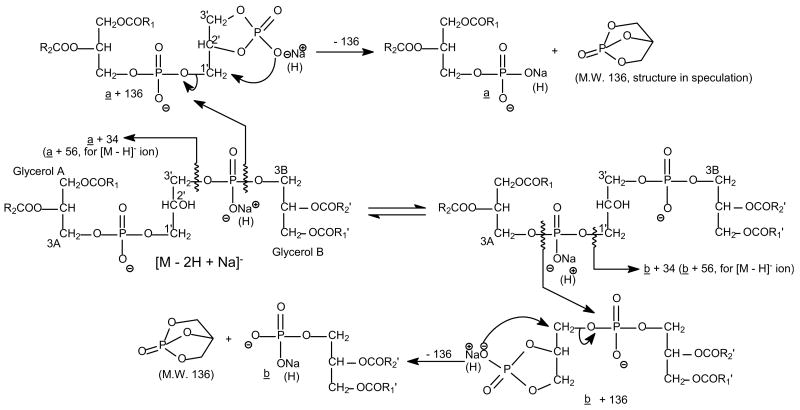
The Fragmentation Pathways of the [M − 2H + Na]− Ions of Cardiolipins Upon Low-Energy Collisional Activation
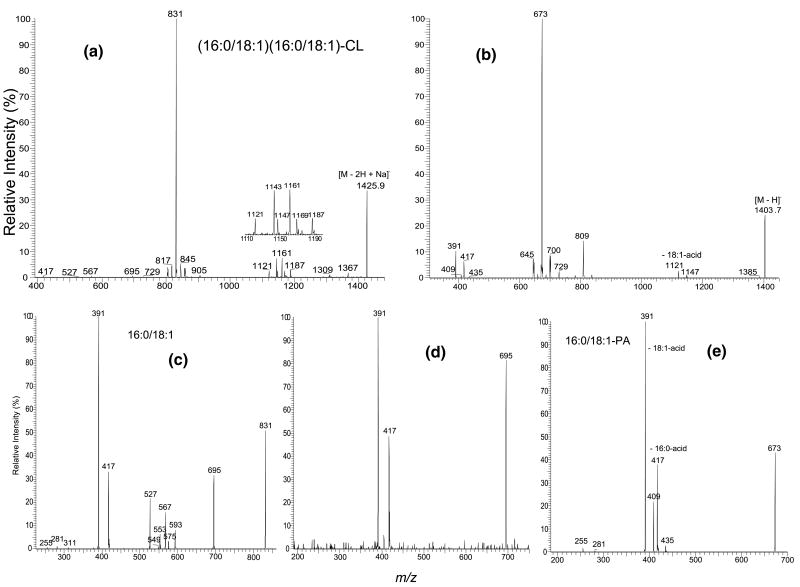
We previously described the fragmentation pathways of the [M − H]− ions of CL under low-energy CAD [5]. Following resonance excitation in ion-trap, the [M − 2H + Na]− ions of CL undergo similar fragmentation processes as those observed for the [M − H]− ions, resulting in the formation of two prominent (a + 136) and (b + 136) ions via cleavages of the C(3A)O–P and the C(3B)O–P bonds, respectively (Scheme 1). As shown in Figure 2, the MS2-spectrum of the [M − 2H + Na]− ions of (16:0/18:1)(16:0/18:1)-CL at m/z 1425.9 (Panel A) from E. coli is dominated by the ion at m/z 831, representing both an (a + 136) and a (b + 136) ions, which yield ions at m/z 695 (a and b ion) by loss of a bicyclic glycerophosphate ester (136 Da) [5, 9]. The pathway is consistent with that previously observed for the [M − H]− ion at m/z 1403.9 (Figure 2b), which contains the ion at m/z 809 and a prominent ion at m/z 673 arising from loss of 136 [5]. The fragmentation pathway is also supported by the MS3-spectrum of m/z 831 (1425→831) (Figure 2c), which gives rise to m/z 695. The MS2-spectrum of the m/z 1425 ion also contains ions at m/z 729 [(a + 34) and (b + 34)] ions via loss of a glycerophosphatidic acid as the sodium salt (Scheme 1). Again, the ion is identical to the m/z 729 ion [(a + 56) and (b +56)] that arises from loss of a glycerophosphatidic acid observed in the MS2-spectrum of the [M − H]− ion (Figure 2b).
Figure 2.
The IT MS2-spectra of (16:0/18:1)(16:0/18:1)-CL of (a) the [M − 2H + Na]− ion at m/z 1425.9, and (b) the [M − H]− ion at m/z 1403.9. The IT MS3-spectra of the m/z 831 ion (1425 → 831) (c), the IT MS4-spectrum of the m/z 695 ion (1425 → 831 → 695) (d), and the IT MS3-spectrum of the m/z 673 ion (1403 → 673) (e) identify the phosphatidyl moieties. Ions at m/z 804, 805, 857, 858 in a, and ions at m/z 645, 646, 700, and 701 in c are from second isotopic contribution of the m/z 1423.9 ([M − 2H + Na]−) and 1401.9 ([M − H]−) ions, respectively, which give the same nominal masses.
Ions at m/z 1161, 1143, and 1121 (Figure 2a) arise from neutral losses of the 18:1-fatty acyl substituent as a ketene, a free acid, and as a sodium salt, respectively, and the ions at m/z 1187, 1169, and 1147 arise from the corresponding losses of the 16:0-fatty acyl substituent. The ions at m/z 1161 and 1143 formed by the various losses of 18:1-fatty acyl substituent are, respectively, more abundant than the ions at m/z 1187 and 1169, reflecting the various losses of 16:0-fatty acyl substituent. The results indicate that the 18:1- and 16:0-fatty acyl substituents are located at sn-2 (or sn-2′) and sn-1 (or sn-1′) of the glycerol A (or B), respectively, and are consistent with the findings that the fragmentation process leading to loss of the fatty acyl substituents at sn-2 (or sn-2′) is more favorable than the corresponding loss at sn-1 (or sn-1′) [8–10].
Further activation of the m/z 695 ion leads to formation of the ions at m/z 417 and 391 via eliminations of 16:0- (loss of C15H31CO2Na, 278 Da) and 18:1-fatty acyl (loss of C17H33CO2Na, 304 Da) moieties as sodium salts, respectively. These fragmentation pathways are supported by the MS4-spectrum of the m/z 695 ion (1425 → 831 → 695) (Figure 2d), which shows the prominent ions at m/z 391 and 417. The m/z 391 ion (695 − C17H33CO2Na) is more abundant than the m/z 417 ion (695 − C15H31CO2Na), consistent with the notion that the 16:0- and 18:1-fatty acyl residues are located at the sn-1 (or sn-1′) and sn-2 (or sn-2′) positions, respectively. The assignment is in agreement with that based on the [M − H]− ion at m/z 1403, which gives a MS3-spectrum of m/z 673 ion (1403 → 673) (Figure 2e) identical to that arising from 16:0/18:1-PA and signifies that the m/z 1403 ion represents a (16:0/18:1)(16:0/18:1)-CL structure [5].
Ions at m/z 593 (831 − C14H29CHC=O), 575 (831 − C15H31CO2H), and 553 (831 − C15H31CO2Na) (Figure 2c) are formed by elimination of the 16:0-fatty acid as a ketene, a free acid, and a sodium salt, respectively; while ions at m/z 567 (831 − C16H31CHC=O), 549 (831 − C17H33CO2H), and 527 (831 − C17H33CO2Na) arise from the analogous losses of the 18:1-fatty acyl substituent. Again, the ions at m/z 567 and 527, reflecting the various losses of the 18:1-fatty acyl substituent at sn-2 are, respectively, more abundant than the ions at m/z 593 and 553 arising from the analogous losses of the 16:0-fatty acyl substituent at sn-1, supporting the idea that loss of the fatty acyl substituent at sn-2 is more favorable than that at sn-1. The ions at m/z 593 and m/z 567, arising from losses of the fatty acyl substituents as ketenes are also, respectively, more abundant than the ions at m/z 575 and 549, arising from losses of the similar fatty acyl substituents as acids. The preferential losses of the fatty acyl substituents as ketenes over the losses as free fatty acids are similar to those observed for the [M − H]− ions of phosphatidylethanolamine (PE), suggesting that the gas-phase basicities of the [(b + 136) and (a + 136)] ions at m/z 831 may be similar to that of the [M − H]− ion of PE [10].
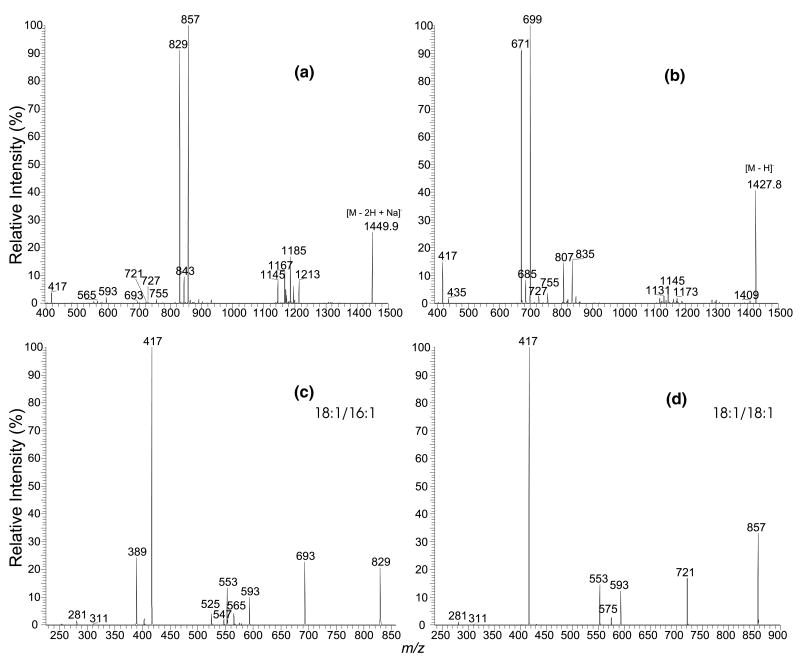
The formation of two distinct (a + 136) and (b + 136) ions by the cleavages of the C(3B)O–P and the C(3A)O–P bonds, respectively, is seen in the MS2-spectra of the [M − 2H + Na]− ions of cardiolipins that consist of two various phosphatidyl moieties. As illustrated in Figure 3a, the MS2-spectrum of the [M − 2H + Na]− ions at m/z 1449.9 is dominated by the ion at m/z 857 (a + 136), which is more abundant than another prominent ion at m/z 829 (b + 136). The results are also similar to those observed in the MS2-spectrum of the [M − H]− ion at m/z 1427.9. This spectrum shows a higher abundance of the m/z 699 (a ion) ion than of the m/z 671 (b ion) ion (Figure 3b). The above results are consistent with the notion that CL contains two chemically distinct phosphatidyl moieties [7, 11, 12] that, when liberated by the differential cleavage of the C(3A)O–P and C(3B)O–P bonds upon CAD [5], result in the preferential formation of the m/z 857 ions over the m/z 829 ion. This result provides information for assignment of the phosphatidyl moieties residing at glycerol A and at glycerol B, respectively.
Figure 3.
The IT MS2-spectra of (a) the [M − 2H + Na]− ion at m/z 1449.9, and (b) the [M − H]− ion at m/z 1427.9. The IT MS3-spectra of the m/z 829 (1449 → 829) (c) and the m/z 857 (1449 → 857) (d) ions provide information to determine the 18:1/16:1- and 18:1/18:1-structures, respectively.
The m/z 417 ion (829 − 136 − C15H29CO2Na) arising from further loss of the 16:1-fatty acyl substituent as a sodium salt is more abundant than the m/z 389 ion (829 − 136 − C17H33CO2Na) arising from the analogous loss of the 18:1-fatty acyl substituent in the MS3-spectrum of the m/z 829 ion (1449 → 829) (Figure 3c), indicating that the 16:1- and 18:1-fatty acyl moieties reside at sn-2′ and sn-1′ of glycerol B, respectively. This assignment is also consistent with the observation of a higher abundance of the m/z 593 ion (829 − 236) arising from loss of 16:1-fatty acyl moiety at sn-2′ as ketene than the m/z 565 ion (829 − 264), arising from the analogous loss of the 18:1-fatty acyl moiety at sn-1′. The MS3-spectrum of the m/z 857 ion (1449 → 857) (Figure 3d) contains a major ion at m/z 417 (721 − 304), arising from secondary fragmentation of the m/z 721 ion (857 − 136) via loss of the 18:1-fatty acyl substituent as sodium salt, and ions at m/z 593, 575, and 553, arising from losses of the 18:1-fatty acyl substituent as a ketene, a free acid, and sodium salt, respectively, suggesting that the m/z 857 ion represent a 18:1/18:1-structure. The above results demonstrate that the m/z 1449 ion consists of a (18:1/18:1)(18:1/16:1)-CL structure. This structural assignment is consistent with that obtained from the corresponding [M − H]− ion (m/z 1427), which gives MS2-spectrum contains ions at m/z 699 and 671, representing an 18:1/18:1-PA and 18:1/16:1-PA, respectively (data not shown).
In the MS2-spectrum of m/z 1449, an ion at m/z 843, representing both a (b + 136) and a (a + 136) ion is also observed. The MS3-spectrum of m/z 843 (1449 → 843) (shown later in Figure 4b) contains ions that are typical of an 18:1/17:1-structure. The results indicate that the m/z 1449 ion may also consist of a (18:1/17:1)(18:1/17: 1)-CL structure. Structural characterizations of cardiolipin species consisting of multiple isomeric structures are described below.
Figure 4.
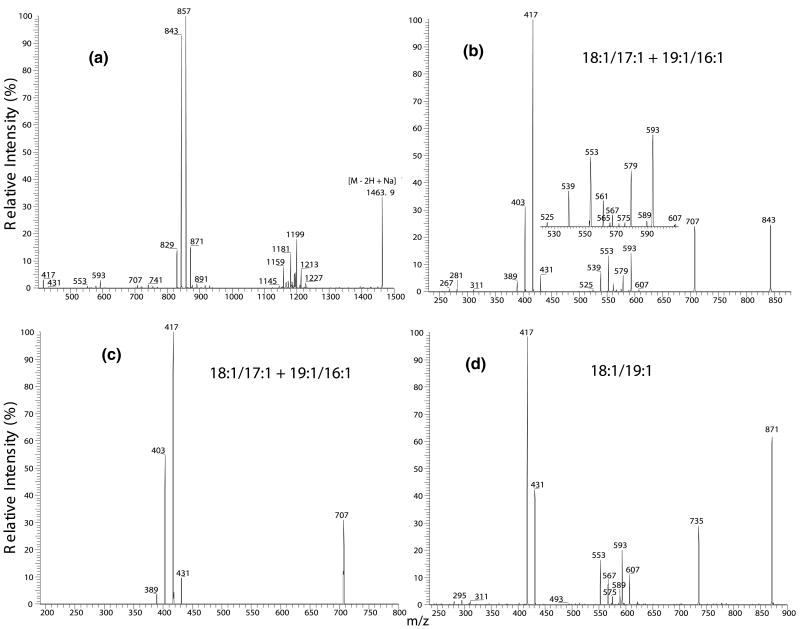
The IT MS2-spectrum of (a) the [M − 2H + Na]− ion at m/z 1463.9, (b) the MS3-spectrum of the m/z 843 ion (1463 → 843), (c) the MS4-spectrum of the m/z 707 ion (1463 →843 →707), and (d) the MS3-spectrum of the m/z 871 ion (1463 → 871). The combined information from the above spectra identifies the (18:1/18:1)(18:1/17:1)-CL, (18:1/18:1)(19:1/16:1)-CL, and (18:1/19:1)(18:1/16:1)-CL species for the m/z 1463 ion.
Structural Characterization of Cardiolipin Molecules Consisting of Multiple Isomers
At least two isomers were identified in each of the CL molecular species in a mixture isolated from E. coli. (Table 1). The approach to structural characterization is exemplified by that of the m/z 1463.9 ion, which yields a MS2-spectrum containing two prominent ions at m/z 857 (a + 136) and 843 (b + 136) along with ions at m/z 721 (a), 707 (b), 755 (a + 34), and 741 (b + 34) that are of low-abundance (Figure 4a). The MS3-spectrum of the m/z 857 ion (1463 → 857) is identical to that shown in Figure 3d, which represents an 18:1/18:1-structure. The MS3-spectrum of the m/z 843 ion (1463 → 843) (Figure 4b) contains ions at m/z 417 and 403, arising from the ion at m/z 707 (843 − 136) by neutral loss of 136, followed by further losses of the 17:1- and 18:1-fatty acyl substituents as sodium salts, respectively, and is a typical spectrum of a 18:1/17:1-structure. However, the spectrum also contains a greater abundance of the ion at m/z 431, arising from loss of 16:1-fatty acyl substituent as sodium salt, than the ion at m/z 389, arising from the analogous loss of the 19:1-fatty acyl substituent. These results indicate that the m/z 843 ion also consists of an isomer with 19:1/16:1-structure. The presence of 19:1/16:1-isomer is further supported by the observation of the set of ions at m/z 607, 589, and 567 (Figure 4b, inset) arising from losses of a 16:1-fatty acyl substituent as a ketene, free acid and a sodium salt, respectively, as well as the set of ions at m/z 565, 547 (not seen) and 525, arising from the analogous losses of the 19:1-fatty acyl substituent. The ions in the former set are each more abundant than the analogous ions in the latter set, consistent with the assignment that the 19:1- and 16:1-fatty acyl substituents reside at sn-1 and sn-2, respectively. The presence of the 19:1/16:1-structure is also consistent with the observation of a higher abundance of the m/z 431 ion than the m/z 389 ion in the MS4-spectrum of the m/z 707 ion (1463 → 843 → 707) (Figure 4c). The combined information indicates that the m/z 857 and 843 ion-pair in the MS2-spectra arise from both the (18:1/18:1)(18:1/17:1)-CL and (18:1/18:1)(19:1/16: 1)-CL structures.
Table 1.
Cardiolipin species identified in Escherichia coli by tandem mass spectrometry
| Phosphatidyl moieties | Fatty acids | |||||
|---|---|---|---|---|---|---|
| [M − 2H + Na] m/z | a + 136 | b + 136 | A | B | Isomer % amount | Relative abundance (%) |
| 1369.916 | 803 | 803 | 16:0/16:1 | 16:0/16:1 | 83% | 8 |
| 775 | 831 | 14:0/16:1 | 16:0/18:1 | 10% | ||
| 829 | 777 | 18:1/16:1 | 16:0/14:0 | 7% | ||
| 1371.931 | 805 | 803 | 16:0/16:0 | 16:0/16:1 | 50% | 2 |
| 777 | 831 | 16:0/14:0 | 16:0/18:1 | 50% | ||
| 1383.931 | 817 | 803 | 16:0/17:1 | 16:0/16:1 | 55% | 9 |
| 803 | 817 | 16:0/16:1 | 16:0/17:1 | 43% | ||
| 789 | 831 | 14:0/17:1 | 16:0/18:1 | 2% | ||
| 1395.932 | 803 | 829 | 16:0/16:1 | 18:1/16:1 | 78% | 7 |
| 801 | 831 | 16:1/16:1 | 16:0/18:1 | 10% | ||
| 857 | 775 | 18:1/18:1 | 14:0/16:1 | 7% | ||
| 815 | 817 | 17:1/16:1 | 16:0/17:1 | 5% | ||
| 1397.946 | 803 | 831 | 16:0/16:1 | 16:0/18:1 | 78% | 38 |
| 817 | 817 | 16:0/17:1 | 16:0/17:1 | 17% | ||
| 857 | 777 | 18:1/18:1 | 16:0/14:0 | 4% | ||
| 1409.962 | 817 | 829 | 16:0/17:1 | 18:1/16:1 | 65% | 14 |
| 829 | 817 | 18:1/16:1 | 16:0/17:1 | |||
| 843 | 803 | 18:1/17:1 | 16:0/16:1 | 26% | ||
| 815 | 831 | 16:1/17:1 | 16:0/18:1 | 8% | ||
| 789 | 857 | 14:0/17:1 | 18:1/18:1 | 2% | ||
| 801 | 845 | 16:1/16:1 | 16:0/19:1 | 1% | ||
| 775 | 871 | 14:0/16:1 | 18:1/19:1 | 1% | ||
| 1411.962 | 817 | 831 | 16:0/17:1 | 16:0/18:1 | 90% | 28 |
| 803 | 845 | 16:0/16:1 | 16:0/19:1 | 10% | ||
| 1421.946 | 829 | 829 | 18:1/16:1 | 18:1/16:1 | 78% | 5 |
| 857 | 801 | 18:1/18:1 | 16:1/16:1 | 18% | ||
| 815 | 843 | 17:1/16:1 | 18:1/17:1 | 4% | ||
| 1423.962 | 829 | 831 | 18:1/16:1 | 16:0/18:1 | 48% | 49 |
| 857 | 803 | 18:1/18:1 | 16:0/16:1 | 42% | ||
| 843 | 817 | 18:1/17:1 | 16:0/17:1 | 10% | ||
| 1425.978 | 831 | 831 | 16:0/18:1 | 16:0/18:1 | 100% | 65 |
| 1435.962 | 843 | 829 | 18:1/17:1 | 18:1/16:1 | 76% | 7 |
| 815 | 857 | 17:1/16:1 | 18:1/18:1 | 23% | ||
| 871 | 801 | 18:1/19:1 | 16:1/16:1 | 1% | ||
| 1437.978 | 857 | 817 | 18:1/18:1 | 16:0/17:1 | 60% | 61 |
| 843 | 831 | 18:1/17:1 | 16:0/18:1 | 26% | ||
| 829 | 845 | 18:1/16:1 | 16:0/19:1 | 26% | ||
| 871 | 803 | 18:1/19:1 | 16:0/16:1 | 4% | ||
| 1449.978 | 857 | 829 | 18:1/18:1 | 18:1/16:1 | 95% | 28 |
| 843 | 843 | 18:1/17:1 | 18:1/17:1 | 5% | ||
| 1451.994 | 857 | 831 | 18:1/18:1 | 16:0/18:1 | 90% | 100 |
| 871 | 817 | 18:1/19:1 | 16:0/17:1 | 5% | ||
| 843 | 845 | 18:1/17:1 | 16:0/19:1 | 5% | ||
| 1463.994 | 857 | 843 | 18:1/18:1 | 18:1/17:1 | 85% | 18 |
| 857 | 843 | 18:1/18:1 | 19:1/16:1 | |||
| 871 | 829 | 18:1/19:1 | 18:1/16:1 | 15% | ||
| 1466.009 | 857 | 845 | 18:1/18:1 | 16:0/19:1 | 70% | 25 |
| 871 | 831 | 18:1/19:1 | 16:0/18:1 | 30% | ||
| 1478.009 | 857 | 857 | 18:1/18:1 | 18:1/18:1 | 95% | 49 |
| 843 | 871 | 18:1/17:1 | 18:1/19:1 | 5% | ||
| 1492.025 | 871 | 857 | 18:1/19:1 | 18:1/18:1 | 54% | 9 |
| 857 | 871 | 18:1/18:1 | 18:1/19:1 | 44% | ||
| 885 | 843 | 18:1/20:1 | 18:1/17:1 | 2% | ||
In Figure 4a, the MS2-spectrum of the m/z 1463.9 ion also contains another set of ions at m/z 871 (a +136) and 829 (b +136), indicating the presence of an additional isomer. The MS3-spectrum of m/z 829 (1463 → 829) is identical to that shown in Figure 3c, which represents the 18:1/16:1-structure and the MS3-spectrum of m/z 871 (1463 → 871) (Figure 4d) contains ions reflecting an 18:1/19:1-configuration. Because the m/z 871 ion is more abundant than the m/z 829 ion, the CL with a (18:1/19: 1)(18:1/16:1)-CL structure can also be assigned for the m/z 1463 ion.
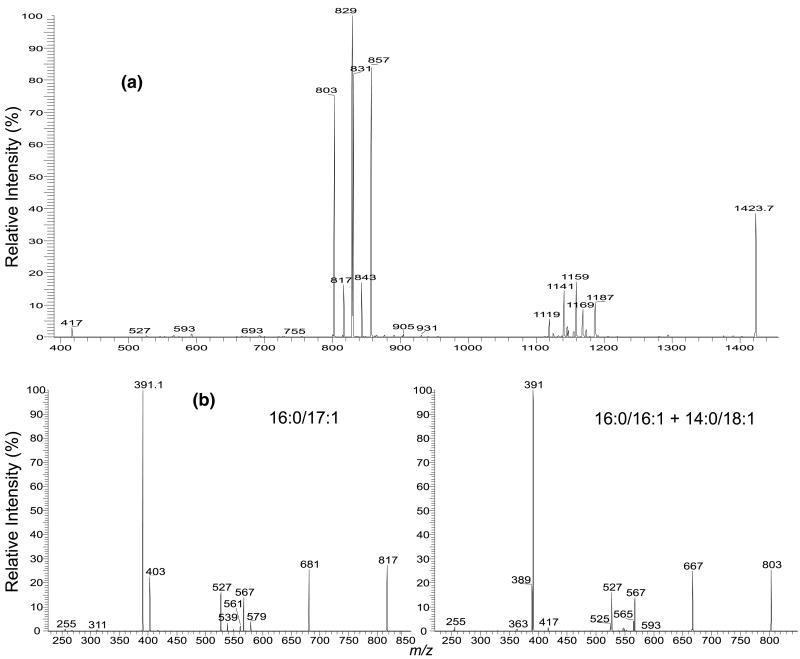
The MS2-spectrum of the m/z 1423.9 ion (Figure 5a) contains at least three sets of (a +136)/(b +136) ion pairs that are observed at m/z 829/831, 857/803, and 843/817, respectively. Again, the identities and the position of the fatty acyl substituents can be easily determined by the profiles of the MS3-spectra of the individual ions in the pairs. The MS3-spectra of m/z 829 (1423 → 829), 831 (1423 → 831), 843 (1423 → 843), and 857 (1423 → 857) are identical to those shown earlier (Figure 2–4), indicating 18:1/16:1-, 16:0/18:1-, 18:1/17: 1-, 18:1/18:1- structures, respectively; while the MS3-spectra of m/z 817 (1423 → 817) (Figure 5b) and 803 (1423 → 803) (Figure 5c) represent 16:0/17:1- and 16:0/16:1-structures, respectively. The combined information suggests that ion at m/z 1423 consists of an (18:1/16:1)(16:0/18:1)-CL, an (18:1/18:1)(16:0/16:1)-CL, and an (18:1/17:1)(16:0/17:1)-CL. The percent amount of these isomers are 48, 42, and 10%, respectively (Table 1), based on the calculation using the following formula: Percent amount of , Where H (a + 136) and H (b + 136) denote the peak height of (a + 136) and (b + 136) ions, respectively.
Figure 5.
The IT MS2-spectrum of (a) the [M − 2H +Na]− ions at m/z 1423, and the MS3-spectra of the m/z 843 ion (1423 → 817) (b), and the m/z 803 ion (1423 → 803) (c). The MS2- and MS3-spectra identify the (18:1/16:1)(16:0/18:1)-CL, (18:1/18:1)(16:0/16:1)-CL, (18:1/17:1)(16:0/17:1)-CL, and (18: 1/18:1)(14:0/18:1)-CL molecules for the m/z 1423.9 ion. Because the m/z 843 ion also represent a 19:1/16:1 structure (Figure 4b), a minor species with a (19:1/16:1)(16:0/17:1)-CL [or (16:0/17:1)(19:1/16:1)-CL] structure can also be assigned.
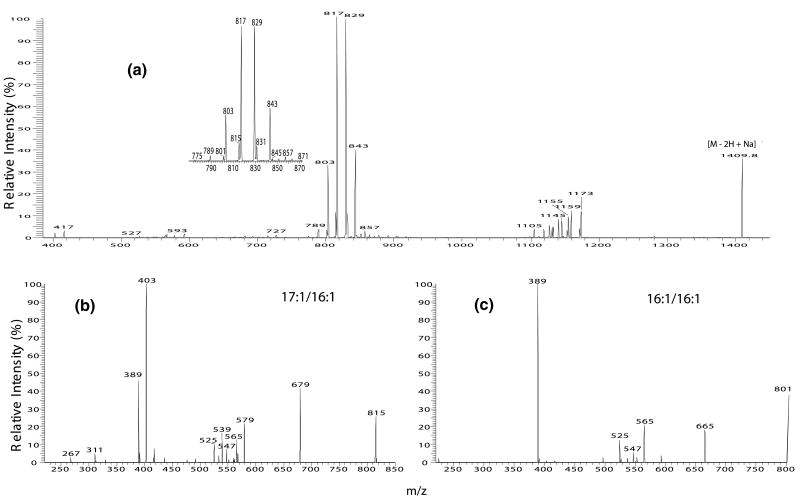
There are six sets of the (a + 136)/(b + 136) ion pairs in the MS2-spectrum of the m/z 1409.9 ion (Figure 6a). These ions are observed at m/z 817/829, 843/803, 815/831, 801/845, 789/857, and 775/871, respectively (Figure 6a, inset). The former ion is more abundant than the latter in each set, and thus, the position of the phosphatidyl group attached to 1′- or 3′-position of the central glycerol can be determined. The MS3-spectra of the m/z 803, 829, 831, 843, 817, 857, and m/z 871 are identical to those observed earlier and represent 16:0/16:1-, 18:1/16:1-, 16:0/18:1-, 18:1/17:1-, 16:0/17:1-, 18:1/18:1-, and 18:1/19:1-structures, respectively; while the MS3-spectra of m/z 815 (1409 → 815) (Figure 6b), 801 (1409 → 801) (Figure 6c), 845 (1409 → 845) (not shown), 789 (1409 → 789) (not shown), and 775 (not shown) reveal the 17:1/16:1-, 16:1/16:1-, 16:0/19:1-, 14:0/17:1-, and 14:0/16:1-structures, respectively. These results clearly demonstrate that the m/z 1409 ion is a mixture consisting of (16:0/17:1)(18:1/16:1)-CL, (18:1/17:1)(16:0/16:1)-CL, (16:1/17:1)(16:0/18:1)-CL, (17:1/14:0)(18:1/18:1)-CL, (16:1/16:1)(16:0/19:1)-CL, and (14:0/16:1)(18:1/19:1)-CL isomers.
Figure 6.
The IT MS2-spectrum of (a) the [M − 2H + Na]− ions at m/z 1409, and the MS3-spectra of the m/z 815 ion (1409 →815) (b), and the m/z 801 ion (1409 → 801) (c). The m/z 1409 ion consists of a major (16:0/17:1)(18:1/16:1)-CL and (18:1/16:1) (16:0/17:1)-CL species (from 817/829 pair), along with (18:1/17:1)(16:0/16:1)-CL (from 843/803 pair), (17:1/16:1)(16:0/18:1)-CL (from 815/831 pair), (14:0/17:1)(18:1/18:1)-CL (from 789/857 pair), (16:1/16:1)(16:0/19:1)-CL (from 801/845 pair), and (14:0/16:1)(18:1/19:1)-CL (from 775/871 pair) molecular species. To simplify, only the major structure in each pair is shown. For example, the m/z 843 ion consists of a major 18:1/17:1 structure and a minor 19:1/16:1 structure, and the m/z 803 ion consists of a major 16:0/16:1- structure and a minor structure 14:0-18:1-, but only the (18:1/17:1)(16:0/16:1)-CL structure was assigned to the 843/803 ion pair.
Structural Characterization of Cardiolipin Isomers Varied by the Positions of the Phosphatidyl Moieties
As described earlier, the determination of the phosphatidyl moieties attached to 1′- or 3′-position of the central glycerol is based on the findings that the (a + 136) ion is more abundant than the (b + 136) ion. The smallest abundance ratio of (b + 136) ion to (a + 136) ion, for example, for the m/z 829/831 and 857/803 ion pairs that reflect the (18:1/16:1)(16:0/18:1)-CL and (18:1/18:1)(16:0/16:1)-CL, respectively, observed in the MS2-spectrum of the m/z 1423 ion (Figure 5a), is ∼0.85. This ratio was also observed for the m/z 843/803 ion-pair (Figure 6a) in the MS2-spectra of m/z 1409.8, and for the major ion-pair observed in the MS2-spectra from other CL species examined. However, the abundance ratio of the m/z 817 (a + 136) ion to the m/z 829 (b + 136) ion in Figure 6a, for instance, is 0.95. The m/z 817 ion, which reflects a 16:0/17:1-structure, is slightly more abundant than the m/z 829 ion, reflecting an 18:1/16:1-structure, indicating that the m/z 817/829 ion pair arises mainly from a (16:0/17:1)(18:1/16:1)-CL. However, the abundance ratio of 0.95, which is higher than the expected ratio of 0.85 for the two ions indicate that the 18:1/16:1- and 16:0/17:1-phosphatidyl residues may also reside at 1′- and 3′-position of the central glycerol, respectively, to form a (18:1/16:1)(16:0/17:1)-CL isomer. The (18:1/16:1)(16:0/17:1)-CL gives a MS2-spectrum similar to that of (16:0/17:1)(18:1/16:1)-CL but preferentially yields a higher abundance of the m/z 829 ion than the m/z 817 ion upon CAD, resulting in the rise of the abundance ratio of these two ions. Thus, ions at m/z 817 and 829 observed in the MS2-spectrum of m/z 1423.9 may derive from both a (16:0/17:1)(18:1/16:1)-CL and a (18:1/16:1)(16:0/17:1)-CL isomers rather than from a single isomer. The percent amount of the former isomer is 55%, whereas that of the latter isomer is 45%, based on a calculation that the ratio for a single isomer is 0.85. The observation of the positional isomers of CL was previously reported [13, 14].
Conclusions
One important finding from this study is that the (a + 136) or (b + 136) ions with same m/z values represent identical phosphatidyl structures, regardless of the various CL species that give rise to the same ions. For examples, all the m/z 829 ions observed in the MS2-spectra of the ions at m/z 1395, 1409, 1421, 1423, 1435, 1437, 1449, and 1463 (Table 1) consist of a 18:1/16:1-structure. The m/z 831 ion, which reflects a 16:0/18:1-structure, is produced for the ions at m/z 1379, 1411, 1423, 1425, 1437, 1451, and 1465. The finding is in accord with the facts that the PG molecular species found in the E. coli from which the CL was isolated contain 16:0/16:1-PG (35%), 16:0/17:1-PG (60%), 18:1/16:1-PG (23%), 16:0/18:1-PG (100%), 16:0/19:1-PG (25%), and 18:1/18:1-PG (45%) as the major PG species, based on our tandem mass spectrometric analysis. [Unpublished results based on our tandem mass spectrometric analysis of phosphatidylglycerol from E. coli (E. coli B-ATCC 11303) from Avanti Polar Lipid Co., Alabaster, AL.] The findings are also consistent with the notion that in the prokaryotic biosynthesis pathway, two molecules of phosphatidylglycerol are involved in the synthesis of CL [15].
The structural assignments of various CLs from the [M − 2H + Na]− ions are also in accord with those obtained from the [M − H] − ions.
The advantage of characterizing CLs in the [M − 2H + Na]− form over that in the [M − H]− form is that the [M − 2H + Na]− ions are readily formed when CL samples of biological origin are subjected to ionization by ESI, attributable to the fact that Na+ ions are ubiquitous in biological samples. In contrast, to form principally [M − H]− ions requires further desalting steps. In addition, the MS3-spectra of the (a + 136) and (b + 136) ions derived from the [M − 2H + Na]− ions contain multiple sets of secondary fragment ions that are informative for identifying and locating the fatty acyl substituents. This feature is useful for confirming a structural assignment, in particular, for the ions that consist of several isomers. This was shown in the structural determination of, for example, the m/z 843 ion (Figures 4b), which consists of two isomeric structures. The multiple-stage IT mass spectrometric methods described above readily provide a simple means leading to complete structural characterization of CL. Nevertheless, investigations of standard cardiolipins with four distinct fatty acyl substituents are desired to confirm the structural assignments.
Acknowledgments
This research was supported by U.S. Public Health Service grants P41-RR-00954, R37-DK-34388, P60-DK-20579, P01-HL-57278, P30-DK-56341, and a grant (996003) from the Juvenile Diabetes Foundation.
References
- 1.Valianpour F, Wanders RJA, Barth PG, Overmars H, van Gennip AH. Quantitative and Compositional Study of Cardiolipin in Platelets by Electrospray Ionization Mass Spectrometry: Application for the Identification of Barth Syndrome Patients. Clin Chem. 2002;48:1390–1397. [PubMed] [Google Scholar]
- 2.Beckedorf AI, Schaffer C, Messner P, Peter-Katalinic J. Mapping and Sequencing of Cardiolipins from Geobacillus stearothermophilus NRS 2004/3a by Positive and Negative Ion Nano ESI-QTOF-MS and MS/MS. J Mass Spectrom. 2002;37:1086–1094. doi: 10.1002/jms.369. [DOI] [PubMed] [Google Scholar]
- 3.Lesnefsky ES, Stoll MSK, Minkler PE, Hoppel CL. Separation and Quantitation of Phospholipids and Lysophospholipids by High-Performance Liquid Chromatography. Anal Biochem. 2000;285:246–254. doi: 10.1006/abio.2000.4783. [DOI] [PubMed] [Google Scholar]
- 4.Peter-Katalinic J, Fischer W. α-D-Glucopyranosyl-, D-alanyl-, and L-Lysylcardiolipin from Gram-Positive Bacteria: Analysis by Fast Bombardment Mass Spectrometry. J Lipid Res. 1998;39:2286–2292. [PubMed] [Google Scholar]
- 5.Hsu FF, Turk J, Rhoades ER, Russell DG, Shi Y, Groisman EA. Structural Characterization of Cardiolipin by Tandem Quadrupole and Multiple-Stage Quadrupole Ion-Trap Mass Spectrometry with Electrospray Ionization. J Am Soc Mass Spectrom. 2005;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 6.LeCocq J, Ballou CE. On the Structure of Cardiolipin. Biochemistry. 1964;155:976–80. doi: 10.1021/bi00895a023. [DOI] [PubMed] [Google Scholar]
- 7.Powell GL, Jacobus J. The Nonequivalence of the Phosphorus Atoms in Cardiolipin. Biochemistry. 1974;13:4024–4026. doi: 10.1021/bi00716a032. [DOI] [PubMed] [Google Scholar]
- 8.Hsu FF, Turk J. Characterization of Phosphatidylinositol, Phosphatidylinositol-4-Phosphate, and Phosphatidylinositol-4,5-Bisphosphate by Electrospray Tandem Mass Spectrometry: A Mechanistic Study. J Am Soc Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 9.Hsu FF, Turk J. Charge-Driven Fragmentation Processes in Diacyl Glycerophosphatidic Acids Upon Low-Energy Collisional Activation. A Mechanistic Proposal. J Am Soc Mass Spectrom. 2000;11:797–803. doi: 10.1016/S1044-0305(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 10.Hsu FF, Turk J. Charge-Driven and Charge-Remote Fragmentation Processes in Diacyl Glycerophosphoethanolamine Upon Low-Energy Collisional Activation. A Mechanistic Proposal. J Am Soc Mass Spectrom. 2000;11:892–899. doi: 10.1016/S1044-0305(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 11.Henderson TO, Glonek T, Myers TC. Phosphorus-31 Nuclear Magnetic Resonance Spectroscopy of Phospholipids. Biochemistry. 1974;13:623–628. doi: 10.1021/bi00700a034. [DOI] [PubMed] [Google Scholar]
- 12.Kates M, Syz JY, Gosser D, Haines TH. pH-Dissociation Characteristics of Cardiolipin and its 2′-deoxy Analog. Lipids. 1993;28:877–882. doi: 10.1007/BF02537494. [DOI] [PubMed] [Google Scholar]
- 13.Schlame M, Brody S, Hostetler KY. Mitochondrial Cardiolipin in Diverse Eukaryotes. Comparison of Biosynthetic Reactions and Molecular Acyl Species. Eur J Biochem. 1993;212:727. doi: 10.1111/j.1432-1033.1993.tb17711.x. [DOI] [PubMed] [Google Scholar]
- 14.Schlame M, Otten D. Analysis of Cardiolipin Molecular Species by High-Performance Liquid Chromatography of Its Derivative 1,3-Bisphosphatidyl-2-Benzoyl-sn-Glycerol Dimethyl Ester. Anal Biochem. 1991;195:290–295. doi: 10.1016/0003-2697(91)90332-n. [DOI] [PubMed] [Google Scholar]
- 15.Hirschberg CB, Kennedy EP. Mechanism of the Enzymatic Synthesis of Cardiolipin in Escherichia coli. Proc Natl Acad Sci U S A. 1972;69:648–651. doi: 10.1073/pnas.69.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]