Abstract
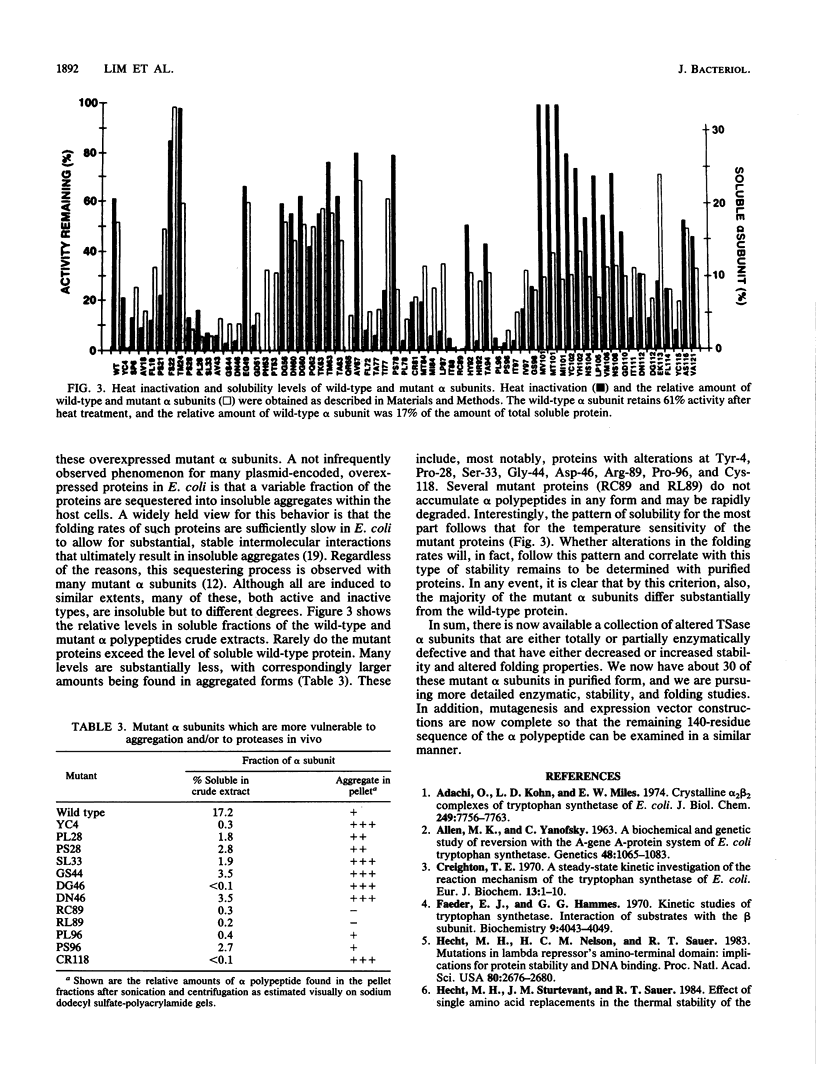
In vitro mutagenesis of the Escherichia coli trpA gene has yielded 66 mutant tryptophan synthase alpha subunits containing single amino acid substitutions at 49 different residue sites and 29 double and triple amino acid substitutions at 16 additional sites, all within the first 121 residues of the protein. The 66 singly altered mutant alpha subunits encoded from overexpression vectors have been examined for their ability to support growth in trpA mutant host strains and for their enzymatic and stability properties in crude extracts. With the exception of mutant alpha subunits altered at catalytic residue sites Glu-49 and Asp-60, all support growth; this includes those (48 of 66) that have no enzymatic defects and those (18 of 66) that do. The majority of the enzymatically defective mutant alpha subunits have decreased capacities for substrate (indole-3-glycerol phosphate) utilization, typical of the early trpA missense mutants isolated by in vivo selection methods. These defects vary in severity from complete loss of activity for mutant alpha subunits altered at residue positions 49 and 60 to those, altered elsewhere, that are partially (up to 40 to 50%) defective. The complete inactivation of the proteins altered at the two catalytic residue sites suggest that, as found via in vitro site-specific mutagenesis of the Salmonella typhimurium tryptophan synthetase alpha subunit, both residues probably also participate in a push-pull general acid-base catalysis of indole-3-glycerol phosphate breakdown for the E. coli enzyme as well. Other classes of mutant alpha subunits include some novel types that are defective in their functional interaction with the other tryptophan synthetase component, the beta 2 subunit. Also among the mutant alpha subunits, 19 were found altered at one or another of the 34 conserved residue sites in this portion of the alpha polypeptide sequence; surprisingly, 10 of these have wild-type enzymatic activity, and 16 of these can satisfy growth requirements of a trpA mutant host. Heat stability and potential folding-rate alterations are found in both enzymatically active and defective mutant alpha subunits. Tyr-4. Pro-28, Ser-33, Gly-44, Asp-46, Arg-89, Pro-96, and Cys-118 may be important for these properties, especially for folding. Two regions, one near Thr-24 and another near Met-101, have been also tentatively identified as important for increasing stability.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALLEN M. K., YANOFSKY C. A BIOCHEMICAL AND GENETIC STUDY OF REVERSION WITH THE A-GENE A-PROTEIN SYSTEM OF ESCHERICHIA COLI TRYPTOPHAN SYNTHETASE. Genetics. 1963 Aug;48:1065–1083. doi: 10.1093/genetics/48.8.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi O., Kohn L. D., Miles E. W. Crystalline alpha2 beta2 complexes of tryptophan synthetase of Escherichia coli. A comparison between the native complex and the reconstituted complex. J Biol Chem. 1974 Dec 25;249(24):7756–7763. [PubMed] [Google Scholar]
- Creighton T. E. A steady-state kinetic investigation of the reaction mechanism of the tryptophan synthetase of Escherichia coli. Eur J Biochem. 1970 Mar 1;13(1):1–10. doi: 10.1111/j.1432-1033.1970.tb00892.x. [DOI] [PubMed] [Google Scholar]
- Faeder E. J., Hammes G. G. Kinetic studies of tryptophan synthetase. Interaction of substrates with the B subunit. Biochemistry. 1970 Oct 13;9(21):4043–4049. doi: 10.1021/bi00823a003. [DOI] [PubMed] [Google Scholar]
- HENNING U., YANOFSKY C. An electrophoretic study of mutationally altered A proteins of the tryptophan synthetase of Escherichia coli. J Mol Biol. 1963 Jan;6:16–21. doi: 10.1016/s0022-2836(63)80077-7. [DOI] [PubMed] [Google Scholar]
- Hecht M. H., Nelson H. C., Sauer R. T. Mutations in lambda repressor's amino-terminal domain: implications for protein stability and DNA binding. Proc Natl Acad Sci U S A. 1983 May;80(9):2676–2680. doi: 10.1073/pnas.80.9.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Kirschner K., Wiskocil R. L., Foehn M., Rezeau L. The tryptophan synthase from Escherichia coli. An improved purification procedure for the alpha-subunit and binding studies with substrate analogues. Eur J Biochem. 1975 Dec 15;60(2):513–523. doi: 10.1111/j.1432-1033.1975.tb21030.x. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lim W. K., Smith-Somerville H. E., Hardman J. K. Solubilization and renaturation of overexpressed aggregates of mutant tryptophan synthase alpha-subunits. Appl Environ Microbiol. 1989 May;55(5):1106–1111. doi: 10.1128/aem.55.5.1106-1111.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luntz T. L., Schejter A., Garber E. A., Margoliash E. Structural significance of an internal water molecule studied by site-directed mutagenesis of tyrosine-67 in rat cytochrome c. Proc Natl Acad Sci U S A. 1989 May;86(10):3524–3528. doi: 10.1073/pnas.86.10.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MALING B. D., YANOFSKY C. The properties of altered proteins from mutants bearing one or two lesions in the same gene. Proc Natl Acad Sci U S A. 1961 Apr 15;47:551–566. doi: 10.1073/pnas.47.4.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matchett W. H. Indole channeling by tryptophan synthase of neurospora. J Biol Chem. 1974 Jul 10;249(13):4041–4049. [PubMed] [Google Scholar]
- Miles E. W., McPhie P., Yutani K. Evidence that glutamic acid 49 of tryptophan synthase alpha subunit is a catalytic residue. Inactive mutant proteins substituted at position 49 bind ligands and transmit ligand-dependent to the beta subunit. J Biol Chem. 1988 Jun 25;263(18):8611–8614. [PubMed] [Google Scholar]
- Miles E. W. Tryptophan synthase: structure, function, and subunit interaction. Adv Enzymol Relat Areas Mol Biol. 1979;49:127–186. doi: 10.1002/9780470122945.ch4. [DOI] [PubMed] [Google Scholar]
- Milton D. L., Napier M. L., Myers R. M., Hardman J. K. In vitro mutagenesis and overexpression of the Escherichia coli trpA gene and the partial characterization of the resultant tryptophan synthase mutant alpha-subunits. J Biol Chem. 1986 Dec 15;261(35):16604–16615. [PubMed] [Google Scholar]
- Murgola E. J., Yanofsky C. Structural interactions between amino acid residues at positions 22 and 211 in the tryptophan synthetase alpha chain of Escherichia coli. J Bacteriol. 1974 Feb;117(2):444–448. doi: 10.1128/jb.117.2.444-448.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers R. M., Lerman L. S., Maniatis T. A general method for saturation mutagenesis of cloned DNA fragments. Science. 1985 Jul 19;229(4710):242–247. doi: 10.1126/science.2990046. [DOI] [PubMed] [Google Scholar]
- Nagata S., Hyde C. C., Miles E. W. The alpha subunit of tryptophan synthase. Evidence that aspartic acid 60 is a catalytic residue and that the double alteration of residues 175 and 211 in a second-site revertant restores the proper geometry of the substrate binding site. J Biol Chem. 1989 Apr 15;264(11):6288–6296. [PubMed] [Google Scholar]
- Shirvanee L., Horn V., Yanofsky C. Escherichia coli mutant trpA34 has an Asp----Asn change at active site residue 60 of the tryptophan synthetase alpha chain. J Biol Chem. 1990 Apr 25;265(12):6624–6625. [PubMed] [Google Scholar]
- YANOFSKY C., HELINSKI D. R., MALING B. D. The effects of mutation on the composition and properties of the A protein of Escherichia coli tryptohan synthetase. Cold Spring Harb Symp Quant Biol. 1961;26:11–24. doi: 10.1101/sqb.1961.026.01.006. [DOI] [PubMed] [Google Scholar]
- YANOFSKY C., RACHMELER M. The exclusion of free indole as an intermediate in the biosynthesis of tryptophan in Neurospora crassa. Biochim Biophys Acta. 1958 Jun;28(3):640–641. doi: 10.1016/0006-3002(58)90533-x. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Horn V. Tryptophan synthetase chain positions affected by mutations near the ends of the genetic map of trpA of Escherichia coli. J Biol Chem. 1972 Jul 25;247(14):4494–4498. [PubMed] [Google Scholar]
- Yanofsky C. Tryptophan synthetase: its charmed history. Bioessays. 1987 Mar;6(3):133–137. doi: 10.1002/bies.950060309. [DOI] [PubMed] [Google Scholar]
- Yutani K., Ogasahara K., Sugino Y., Matsushiro A. Effect of a single amino acid substitution on stability of conformation of a protein. Nature. 1977 May 19;267(5608):274–275. doi: 10.1038/267274a0. [DOI] [PubMed] [Google Scholar]
- Yutani K., Ogasahara K., Tsujita T., Kanemoto K., Matsumoto M., Tanaka S., Miyashita T., Matsushiro A., Sugino Y., Miles E. W. Tryptophan synthase alpha subunit glutamic acid 49 is essential for activity. Studies with 19 mutants at position 49. J Biol Chem. 1987 Oct 5;262(28):13429–13433. [PubMed] [Google Scholar]


