Abstract
Fundamental building blocks of life have been detected extraterrestrially, even in interstellar space, and are known to form nonenzymatically. Thus, the HCN pentamer, adenine (a base present in DNA and RNA), was first isolated in abiogenic experiments from an aqueous solution of ammonia and HCN in 1960. Although many variations of the reaction conditions giving adenine have been reported since then, the mechanistic details remain unexplored. Our predictions are based on extensive computations of sequences of reaction steps along several possible mechanistic routes. H2O- or NH3-catalyzed pathways are more favorable than uncatalyzed neutral or anionic alternatives, and they may well have been the major source of adenine on primitive earth. Our report provides a more detailed understanding of some of the chemical processes involved in chemical evolution, and a partial answer to the fundamental question of molecular biogenesis. Our investigation should trigger similar explorations of the detailed mechanisms of the abiotic formation of the remaining nucleic acid bases and other biologically relevant molecules.
Keywords: reaction mechanism, DNA, RNA, density functional theory, gas phase model
How did life on earth begin? The presence of biomolecules was a prerequisite, but the origin of even the simplest of these remains a fascinating but unsolved puzzle (1, 2). Numerous experiments have demonstrated that amino acids, nucleotides, carbohydrates, and other essential compounds form under simulated primitive earth conditions from simple starting materials, hydrocarbons, HCN, cyano compounds, aldehydes, and ketones (3, 4). HCN, a high-energy prebiotic precursor, is produced in appreciable amounts, for example, by the action of electric discharges on simulated primitive atmospheres (5). The HCN pentamer adenine (a constituent of DNA, RNA, and many coenzymes) is one of the most abundant biochemical molecules. The abiotic synthesis of adenine from a solution of HCN and ammonia was first reported by Oró and colleagues in 1960 (6–9). Although many experimentalists have contributed to our understanding of possible prebiotic processes, low yields and mechanistic complexities rule out detailed investigations (7, 10–15). The key riddle remains: how do five HCN molecules combine to form adenine under prebiotic conditions? We now propose a detailed, thermochemically favorable mechanism based on density functional theory computations.
In Oró's first experiment (1960), adenine was formed in 0.5% yield by heating solutions of ammonium cyanide (>1.0 M) at 70°C for several days. Since then, the abiotic synthesis of adenine from the polymerization of HCN under various conditions has been achieved many times (7, 10–15). The highest yield of adenine (20%) resulted from the sealed-tube reaction of HCN with liquid ammonia (16). In 1978, Ferris et al. (17) detected 0.04% adenine from 0.1 M NH4CN kept in the dark at room temperature for 4–12 months. In a particularly striking experiment, Levy et al. (18) detected 0.035–0.04% adenine from dilute solution of ammoniacal HCN, frozen for 25 years at −20°C and −78°C (simulated prebiotic synthetic process in the Jovian moon Europa).
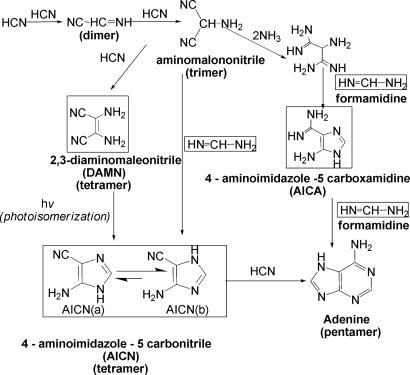
Experimental findings have suggested the outlines of possible prebiotic pathways for the formation of adenine in prebiotic earth conditions (6, 10–15) (Fig. 1). However, the mechanistic details have not been explored (apart from an ab initio mechanistic study of the HCN dimerization and adenine protonation) (19, 20). Experimental investigations would be very difficult because adenine is not formed cleanly, yields are small, and many steps are involved. However, clues are provided by four putative intermediates detected in the product mixtures (Fig. 1): formamidine, 2,3-diaminomaleonitrile (DAMN), 4-amino-5-cyanoimidazole (4-aminoimidazole-5-carbonitrile; AICN) (10, 11, 13–15), and 4-aminoimidazole-5-carboxamidine (9, 11).
Fig. 1.
Proposed steps for the formation of adenine in aqueous ammonium cyanide solution (7, 10–15). Experimentally detected putative intermediates in the abiotic formation of adenine are enclosed in boxes. Two tautomers of AICN can exist; AICN(b) is the more stable. Note that a photoisomerization step is proposed for the formation of AICN from DAMN. DAMN has not been demonstrated to be an adenine intermediate in a nonphotolytic reaction.
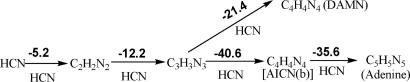
The pentamerization of HCN to give adenine is very exothermic overall (ΔG298 = −53.7 kcal/mol; Fig. 2). Likewise, each successive step, HCN dimerization and the sequential HCN additions to give trimer, tetramer, and pentamer, also is quite exothermic. But a favorable thermochemistry does not ensure that reactions actually will proceed, because they may be prevented by insurmountable kinetic impediments.
Fig. 2.
Thermochemistry of pentamerization of HCN. The relative energies in gas phase are in kilocalories per mole computed at B3LYP/6–311+G**+ZPVE. Entropy is unfavorable but is not included in each step. Overall energy for pentamerization of adenine (5 HCN → C5H5N5) is −93.8 kcal/mol (ΔG298 = −53.7 kcal/mol). Note that the last crucial step for formation of pentamer (adenine) from tetramer [AICN(b)] is highly exothermic.
Reaction mechanisms involving several intermediate and transition states (which are difficult to detect and identify experimentally) can be studied effectively computationally. These allow selection among various possibilities. Our investigations have identified a plausible detailed step-by-step mechanism for the formation of adenine based on density functional theory computations (21) (described in Methods).
We focus on the last, crucial stage, the reaction of the HCN tetramer, AICN, with a fifth HCN to give adenine. This is the key, rate-limiting step in the general adenine formation pathway starting from HCN (see Fig. 1). Although DAMN and AICN are both C4H4N4 isomers, AICN was selected as the adenine precursor because of its much lower energy (19.3 kcal/mol according to our computations) and greater structural similarity to adenine (Fig. 2). Like adenine, AICN has a C–C–C–N sequence, whereas DAMN has a C–C–C–C backbone not present in adenine (Fig. 1). DAMN may form experimentally as a kinetically controlled side product. Although DAMN cannot be involved directly in the pathway, it may serve as a “material reservoir.” Indeed, many details of photoisomerization of DAMN to AICN in water are established (13, 15). In contrast, the nonphotolytic conversion of DAMN to AICN or to adenine has not been observed (11). Here, we describe our investigation of the mechanism for adenine formation from AICN under nonphotochemical conditions (reactions in the dark); consideration of DAMN as adenine precursor will be reported subsequently.
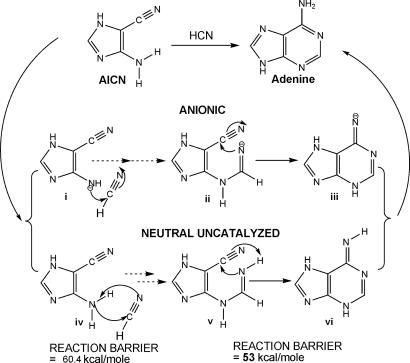
An anionic mechanism for the abiotic formation of adenine seems plausible from a physical organic chemist's point of view. HCN is a weak acid and adenine forms in ammoniacal solution, but mechanisms involving unsolvated anionic species proved not to be feasible computationally. Ring closure (Fig. 3ii → iii) is precluded because the putative anionic intermediates ii and iii are not minima, and iii reverts to the ring-opened isomers on optimization. Mechanisms involving free radical intermediates also were unpromising. In contrast, optimizations of the reactant, product, and transition states for the neutral closed-shell mechanism were successful.
Fig. 3.
Anionic mechanisms are unfeasible in isolation. On optimization, both ii and iii revert back to i (reactants). Free radical and neutral uncatalyzed mechanisms are also not viable because of the very large reaction barriers for the two steps shown.
We first point out explicitly that two AICN tautomeric forms, AICN(a) and AICN(b) (Fig. 1), can exist and that AICN(b) is favored strongly at equilibrium (by ΔG298 = 3.73 kcal/mol in the gas phase and 1.73 kcal/mol with bulk solvation, according to our computations). [Bulk solvation (22) is described in Methods.] The pathway to adenine from the less stable AICN(a) is precluded by the second high reaction barrier associated with the six-membered ring closure step. The reaction profile is depicted in Fig. 4.
Fig. 4.
Gas-phase reaction profile [B3LYP/6-31G*] for adenine formation from the less stable AICN(a) isomer, when one explicit H2O molecule is included as catalyst. This pathway is precluded by a second high reaction barrier.
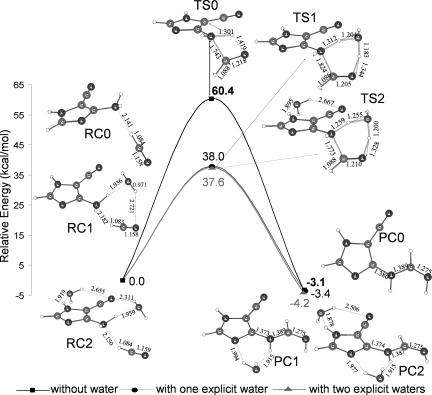
We report the favorable mechanism of adenine formation starting from the thermodynamically more stable AICN(b) tautomer. However, the first step from AICN(b), the addition of HCN to the NH2 group, cannot proceed directly through a four-center transition state: the reaction barrier (60.4 kcal/mol in the gas phase relative to the energy of the 1:1 HCN–AICN complex; Figs. 3 and 5) is much too high. Moreover, a prohibitive reaction barrier (53 kcal/mol) also precludes a subsequent step in the neutral uncatalyzed pathway (involving the concerted six-membered ring closure and H transfer; Fig. 3). Therefore, the neutral, uncatalyzed mechanism for adenine formation also is unlikely. The involvement of additional molecules serving as catalysts is required to lower the high barriers.
Fig. 5.
Gas-phase potential energy profiles (in kilocalories per mole) for the first key addition step of HCN to AICN(b) (B3LYP/6-311+G** + ZPVE). The uncatalyzed reaction, shown by the dashed line at the top, has a prohibitively high 60.4 kcal/mol barrier. Optimized geometries (in angstroms) are given for reactant complexes (RC), transition structures (TS), and product complexes (PC) having one or two explicit catalytic H2O molecules. Note that these are active participants and reduce the barrier to 38.0 and 37.6 kcal/mol, respectively (entropy was not considered).
The unfavorable four-center transition structure (TS0) associated with the first step from AICN(b) is shown in Fig. 5. The catalytic participation of a water molecule in the six-center alternative (TS1) reduces the barrier considerably (ammonia or similar molecules can function similarly). The importance of water-assisted proton transfer is well known in keto–enol tautomerization (23, 24). The water bridge connects the donor and acceptor sites and stabilizes the transition structure. The classical proton transfer barrier is lowered substantially. The energetically most favorable six-center cyclic transition structure leading to the HCN–AICN(b) addition product (TS1) (Fig. 5) is very different from TS0 for the neutral uncatalyzed mechanism. The water molecule in TS1 transfers its hydrogen-bonded proton to the HCN nitrogen concertedly with the formation of the new bond between the AICN amine nitrogen and the electron-deficient HCN carbon.
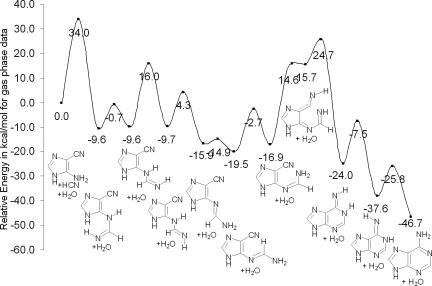
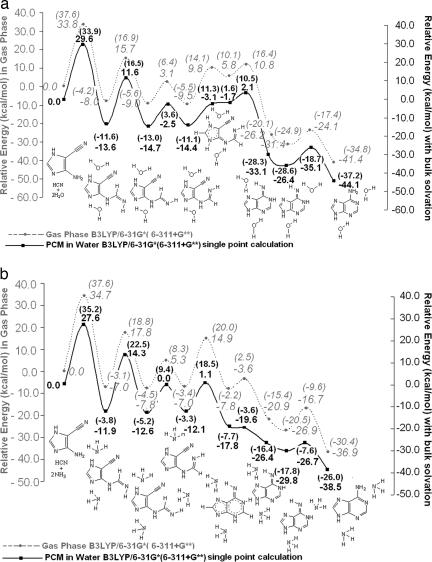
The inclusion of one specific water molecule decreases the reaction barrier drastically, from 60.4 kcal/mol (without water) to 38.0 kcal/mol (with a single H2O) (Fig. 4). Consequently, we also explored the effect of more than one solvent molecule in the mechanism. A second H2O does not participate in the proton relay effectively, but can form relatively strong hydrogen bonds stabilizing the reactant, product, and transition state complexes. The energetically most favorable transition structure with two water molecules (TS2) also is depicted in Fig. 5. However, inclusion of two explicit H2O molecules decreases the reaction barrier by only an additional 0.4 kcal/mol (to 37.6 kcal/mol). It does not seem likely that additional explicit H2O molecules would have much of a further effect. However, a complete solvation shell does have a significant influence (see below). Almost all of the reported abiotic syntheses of adenine were carried out with HCN dissolved in water–ammonia solutions. Besides maintaining the pH of the medium, ammonia might also participate mechanistically. Indeed, an explicit NH3 molecule is as good a catalyst as an explicit H2O molecule. The reaction barrier for the first step, the NH3-catalyzed addition of HCN to AICN(b) (with one explicit NH3), is 37.1 kcal/mol, as compared with 38.0 kcal/mol with an explicit H2O. Although computations based on species in isolation (gas phase) may simulate extraterrestrial conditions, they do not suffice to predict prebiotic processes on the primitive earth, where reactions might have taken place in solution. In addition to the explicit solvent modeling (see above), we simulated the bulk solvation effect by employing the polarizable continuum model (PCM) (22), which considers the solvent as a macroscopic continuum of dielectric constant. Bulk solvation stabilizes structures involving greater charge separation (e.g., transition states) preferentially. [This is a simple alternative to explicit solvation modeling (involving a box of water molecules) by using computationally demanding ab initio molecular dynamics methods (25).] Indeed, bulk solvation reduces the reaction barriers for the rate-determining step to 33.9 kcal/mol (from 37.6 kcal/mol) and to 35.2 kcal/mol (from 37.6 kcal/mol) for specific H2O/NH3-catalyzed mechanisms, respectively. These barriers are low enough to be consistent with the experimental observations as well as conjectures regarding the abiotic genesis of adenine. The subsequent steps from the HCN–AICN adduct are depicted in Fig. 6a (for catalysis by two H2O molecules) and Fig. 6b (for catalysis by two NH3 molecules). The effects of bulk solvation are included in both plots. Except for the syn–anti hydrogen transfer and the C–N bond rotation in the second and third steps, all of the stages require the catalytic participation of at least one H2O or NH3 molecule. The proton relays across six- and five-membered rings are interesting mechanistic features, because two H2O or NH3 molecules are needed to complete the H-bonded “circuit.” One molecule participates in the proton transfer directly as a catalyst, while the second H2O or NH3 assists by hydrogen bonding. The detailed step-by-step mechanism for the formation of [adenine·(H2O)2] starting from AICN(b) + HCN + 2 H2O is depicted in Fig. 6a. This reaction profile includes the relative energies of each stationary point (with respect to AICN + HCN + 2 H2O 1:1:2 reactant complex) without and with bulk solvation. The first step is rate-determining (as stated in our earlier discussion). The subsequent steps have lower reaction barriers.
Fig. 6.
Reaction profiles for the formation of adenine starting from AICN(b) and HCN in the gas phase and with simulated bulk water solvation by means of explicit solvent-catalyzed mechanisms (two solvent molecules). (a) Reaction profile with water as explicit catalytic molecule (gas phase vs. simulated bulk water solvation). (b) Reaction profile with ammonia as explicit catalytic molecule (gas phase vs. simulated bulk water solvation). The two H2O or NH3 molecules facilitate a “proton relay” by forming an H-bonded “circuit” for the proton transfer in a six-membered transition state. All species shown are stable minima. (Dotted lines depict partial bonds in complexes or transition states.) The comparisons with the gas-phase profiles show the large extent to which simulated water solvation reduces the barrier electrostatically. The first step is rate-determining in all cases. The basis set dependency of the barrier heights is shown by the comparison data at 6-31G* and at 6-311+G** (in parentheses).
All of the mechanistic features of the NH3- (Fig. 6b) and the H2O-catalyzed (Fig. 6a) pathways are not alike, because ammonia and water have different Lewis base properties. Although the first three steps, that is, addition of HCN to AICN catalyzed by ammonia, syn–anti hydrogen transfer, and rotation around C–N, are similar, concerted six-membered ring closure and 1,4 H transfer take place in the NH3-catalyzed mechanism before 1,3 H transfer.
Our computed reaction profile with low energy barriers reveals the feasibility of adenine formation from AICN under abiotic conditions. But are the computed reaction barriers reliable? The B3LYP density functional theory method we used is known to give relatively accurate structures and spectroscopic properties of first-row molecules, but activation barriers may be more problematical (26, 27). Our computed reaction barriers at B3LYP/6–31G* are ≈3–4 kcal/mol lower than at B3LYP/6-311+G**. The key rate-determining step was recomputed at the MP2/6-311+G** and the gas-phase reaction barrier with zero-point vibrational energy (ZPVE) correction is 43.9 kcal/mol, which is higher than that at B3LYP/6-311+G**. It has been reported that although MP2 in general gives accurate geometries for reaction complexes, it also overestimates barrier heights (28, 29). Thus for calibration, we performed higher-level CCSD(T)/cc-pVTZ single-point computations on the simpler HCN + NH3 + H2O system on MP2 geometries optimized at 6-311+G** level. This models the key AICN(b) + HCN + H2O rate-determining step, because it also involves the addition of a NH bond to HCN. (The geometries of the reactants, transition structures, products, and barriers, computed at various levels of theory, are given in supporting information (SI) Fig. 7). The CCSD(T)/aug-cc-pVTZ single-point reaction barrier is only 3 kcal/mol higher than the B3LYP/6–311+G** result.
Conclusion
Our detailed computational investigations of step-by-step formation pathways from AICN(b) show how adenine can arise abiotically. Although the formation of adenine by the pentamerization of HCN is very exothermic, this process is quite unlikely in isolation (gas phase). Not only must five HCN molecules come together, but also the reaction barriers are very high. The intimate participation of an additional molecule, such as H2O or NH3 (or perhaps HCN) is needed to lower the barriers considerably to realistic energies. Moreover, an aqueous medium facilitates the reaction, because both specific and bulk solvation lowers the barrier of the rate-determining step further. The reaction energetics are mostly governed by the enthalpy change (see Fig. 2 legend) especially at the low temperatures of some of the experiments.
Finding a viable, thermodynamically feasible, step-by-step mechanism that can account for the formation of adenine did not prove to be easy. Our approach has a model character to it. The “first model” is without water or ammonia, the second model is with explicit participation of ammonia (along with water) in gas phase, and the third model adds bulk solvation. Because there are no quantitative experimental data to match, a highly refined study, for example, by using the approach described by Jorgensen and his coworkers (30) to model the medium effects, is not called for at this initial stage. The computed activation energy is reasonable considering the long time scale of chemical evolution toward complex organic systems on primitive earth. An alternate pathway based on the experimental isolation of 2- and 8-cyanoadenine or adenine 8-carboxamide as adenine precursor suggests a further complex mechanism involving hexamer and heptamers of HCN (31). The feasibility of that alternate pathway requires further investigation. The subject is of continuing interest, and detailed calculations on reactions of this kind are needed before a full picture emerges. Our investigation should trigger similar explorations of the detailed mechanisms of the abiotic formation of the remaining nucleic acid bases and other biologically relevant molecules.
Methods
The computations were performed at the B3LYP density functional theory level with the 6-31G* and the 6-311+G** basis sets. In addition, CCSD(T)/aug-ccpVTZ ab initio computations simulated the first HCN + RNH2 addition step with the highest barrier. All computed harmonic frequencies of fully optimized minima were real, whereas transition structures (states) had a single imaginary frequency. Intrinsic reaction coordinate analyses of the minimum-energy pathways confirmed that the transition states led to the reactants and products shown in the figures. All of the relative energies are corrected by the ZPVE and correspond to the classical reaction barriers (i.e., without proton tunneling). The reaction barriers are defined as the difference in sum of electronic and zero-point energies (ε0 + εZPE) of the reactant complex and transition state. ΔG is defined as difference between the sum of electronic and thermal free energies (ε0 + Gcorr) of reactant and product.
Instead of modeling bulk solvation by an explicit shell involving many solvent molecules surrounding the solute (25), we used the PCM implemented in the Gaussian 98 program (21). The PCM “bulk solvent medium” is simulated as a continuum of dielectric constant ε. This surrounds a solute cavity, which is defined by the union of a series of interlocking spheres centered on the atoms. Our computed single-point PCM bulk solvent simulations used the optimized equilibrium geometries. We have added an estimated ≈2.5 kcal/mol ZPVE correction to the single-point PCM energy. The derivation of this corrected energy value is discussed in detail in the SI Dataset. More sophisticated solvation treatments are not called for in the absence of experimental data.
During the review of this article, Glaser et al. (32) proposed and analyzed mechanisms of pyrimidine-ring formation of isomeric monocyclic HCN pentamers. Pyrimidine-ring formation from a monocyclic HCN pentamer, either by proton-catalyzed or by uncatalyzed cyclization, was found not to be viable thermochemically. However, they propose that photoactivation of either of these cyclization paths might lead eventually to the imino form of adenine.
Supplementary Material
Acknowledgments
We thank Prof. T. T. Tidwell for calling our attention to this problem and for his encouragement and the referees who made constructive suggestions, which led to improvements in the article. This work was supported by National Science Foundation Grants CHE-0209857 and CHE-0716718 and the University of Georgia.
Abbreviations
- AICN
4-amino-5-cyanoimidazole
- DAMN
2,3-diaminomaleonitrile
- PCM
polarizable continuum model
- ZPVE
zero-point vibrational energy.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708434104/DC1.
References
- 1.Miller SL, Orgel LE, editors. The Origins of Life on Earth. Englewood Cliffs, NJ: Prentice-Hall; 1974. [Google Scholar]
- 2.Schopf JW, editor. Earth's Earliest Biosphere: Its Origin and Evolution. Princeton: Princeton Univ Press; 1983. [Google Scholar]
- 3.Eschenmoser A, Loewenthal E. Chem Soc Rev. 1992;21:1–16. [Google Scholar]
- 4.Orgel LE. Crit Rev Biochem Mol Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- 5.Abelson PH. Proc Natl Acad Sci USA. 1966;55:1365–1372. doi: 10.1073/pnas.55.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oró J. Biochem Biophys Res Commun. 1960;2:407–412. doi: 10.1016/j.bbrc.2011.07.075. [DOI] [PubMed] [Google Scholar]
- 7.Oró J. Nature. 1961;191:1193–1194. doi: 10.1038/1911193a0. [DOI] [PubMed] [Google Scholar]
- 8.Oró J., Kimball AP. Arch Biochem Biophys. 1961;94:217–227. doi: 10.1016/0003-9861(61)90033-9. [DOI] [PubMed] [Google Scholar]
- 9.Oró J., Kimball AP. Arch Biochem Biophys. 1962;96:293–313. doi: 10.1016/0003-9861(62)90412-5. [DOI] [PubMed] [Google Scholar]
- 10.Ferris JP, Sanchez RA, Orgel LE. J Mol Biol. 1968;33:693–704. doi: 10.1016/0022-2836(68)90314-8. [DOI] [PubMed] [Google Scholar]
- 11.Shuman RF, Shearin WE, Tull RJ. J Org Chem. 1979;44:4532–4536. [Google Scholar]
- 12.Voet AB, Schwartz AW. Bioorg Chem. 1983;12:8–17. [Google Scholar]
- 13.Ferris JP, Orgel LE. J Am Chem Soc. 1966;88:3829–3831. doi: 10.1021/ja00968a028. [DOI] [PubMed] [Google Scholar]
- 14.Ferris JP, Orgel LE. J Am Chem Soc. 1965;87:4976–4977. doi: 10.1021/ja00949a068. [DOI] [PubMed] [Google Scholar]
- 15.Ferris JP, Orgel LE. J Am Chem Soc. 1966;88:1074–1074. doi: 10.1021/ja00968a028. [DOI] [PubMed] [Google Scholar]
- 16.Wakamatsu H, Yamada Y, Saito T, Kumashiro I, Takenishi T. J Org Chem. 1966;31:2035–2036. [Google Scholar]
- 17.Ferris JP, Joshi PC, Edelson EH, Lawless JG. J Mol Evol. 1978;11:293–311. doi: 10.1007/BF01733839. [DOI] [PubMed] [Google Scholar]
- 18.Levy M, Miller SL, Brinton K, Bada JL. Icarus. 2000;145:609–613. doi: 10.1006/icar.2000.6365. [DOI] [PubMed] [Google Scholar]
- 19.Kikuchi O, Watanabe T, Satoh Y, Inadomi Y. J Mol Struct. 2000;507:53–62. [Google Scholar]
- 20.Tureček F, Chen X. J Am Soc Mass Spectrom. 2005;16:1713–1726. doi: 10.1016/j.jasms.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery AJ, Jr, Stratmann RE, Burant JC, et al. GAUSSIAN 98. Pittsburgh: Gaussian; 1998. Revision A.9. [Google Scholar]
- 22.Miertus S, Scrocco E, Tomasi J. Chem Phys. 1981;55:117–129. [Google Scholar]
- 23.Yamabe S, Tsuchida N, Miyajima K. J Phys Chem A. 2004;108:2750–2757. [Google Scholar]
- 24.Kiruba GSM, Wong MW. J Org Chem. 2003;68:2874–2881. doi: 10.1021/jo0266792. [DOI] [PubMed] [Google Scholar]
- 25.Aida M, Yamataka H, Dupuis M. Chem Phys Lett. 1998;292:474–480. [Google Scholar]
- 26.Barone V. Chem Phys Lett. 1994;226:392–398. [Google Scholar]
- 27.Martell JM, Goddard JD, Eriksson LA. J Phys Chem A. 1997;101:1927–1934. [Google Scholar]
- 28.Wiberg KB, Ochterski JW. J Comput Chem. 1997;18:108–114. [Google Scholar]
- 29.Gonzales-Garcia N, Gonzales-Lafont À, Lluch JM. J Comput Chem. 2005;26:569–583. doi: 10.1002/jcc.20190. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen WL, Alexandrova AN. J Phys Chem B. 2007;111:720–730. doi: 10.1021/jp066478s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borquez E, Cleaves HJ, Lazcano A, Miller SL. Origins Life. 2005;35:79–90. doi: 10.1007/s11084-005-5945-9. [DOI] [PubMed] [Google Scholar]
- 32.Glaser R, Hodgen B, Farrelly D, McKee E. Astrobiology. 2007;7:455–470. doi: 10.1089/ast.2006.0112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.