Abstract
The structure of intrinsic factor (IF) in complex with cobalamin (Cbl) was determined at 2.6-Å resolution. The overall fold of the molecule is that of an α6/α6 barrel. It is a two-domain protein, and the Cbl is bound at the interface of the domains in a base-on conformation. Surprisingly, two full-length molecules, each comprising an α- and a β-domain and one Cbl, and two truncated molecules with only an α- domain are present in the same asymmetric unit. The environment around Cbl is dominated by uncharged residues, and the sixth coordinate position of Co2+ is empty. A detailed comparison between the IF-B12 complex and another Cbl transport protein complex, trans-Cbl-B12, has been made. The pH effect on the binding of Cbl analogues in transport proteins is analyzed. A possible basis for the lack of interchangeability of human and rat IF receptors is presented.
Keywords: incomplete dimer, x-ray, cobalt, transport protein, glycoprotein
Vitamin B12 [cobalamin (Cbl)] [supporting information (SI) Fig. 5] is a water-soluble vitamin and is essential for the growth and development of all mammals, including humans. Cbl is a member of the corrinoid series of cobalt-containing compounds and is distinguished from some other members of this group by possessing a nucleotide side chain terminating in dimethylbenzimidazole (1, 2). It serves as a key enzymatic cofactor for a number of methyltransferase and mutase reactions occurring in nature (3). In mammals, it contributes the prosthetic group for two important enzymes, methionine synthase (a methyltransferase) and methylmalonyl-CoA mutase (1, 2, 4).
In mammals, three proteins are involved in the uptake, transport, and storage of Cbl. These are gastric intrinsic factor (IF), trans-Cbl II (TC), and haptocorrin (HC). These are immunologically distinct proteins with a protein core of ≈46 kDa. IF and HC are heavily glycosylated, but TC is not (5, 6). In humans, the genes for IF and HC are located in chromosome 22 (7, 8), whereas the gene for TC is located in chromosome 11 (9). Human IF contains 399-aa residues plus ≈15% carbohydrate, giving it a molecular mass of ≈60 kDa, as observed by gel filtration (10–13). All three proteins promote Cbl entry through endocytosis involving distinct cell surface receptors (5, 14–20).
Dietary Cbl is bound first to HC in the gastric lumen but is transferred to IF in the duodenum after degradation of HC by pancreatic proteases. IF is responsible for transit of Cbl through the small intestine and delivery of it to the endothelial cells that line the ileum. The IF-Cbl complex is then recognized by the IF-Cbl receptor, cubilin (CUB), located on the luminal side of the intestinal mucosal cells, and mediates its internalization. Lysosomal degradation of the internalized IF-Cbl complex releases Cbl, which is then able to bind to TC, probably within the enterocyte. The TC-Cbl complex is then translocated across the basolateral membrane and released into the circulation, to be taken up by TC-Cbl receptor-mediated process by all cells in the body. Lysosomal dissociation of the complex then occurs along with transfer of Cbl to the coenzyme methyl- and Ado-Cbl in the cytoplasm and in mitochondria, respectively.
In view of the above factors, the x-ray structure of IF in complex with Cbl will provide insight into the mechanism behind Cbl uptake and release. Recently, the crystal structure of TC was reported (18). In this paper, we present the crystal structure of human IF at 2.6-Å resolution and discuss the implications of the structure on its biological function.
Results
Overall Structure.
The crystals of the IF-Cbl complex contain four molecules in the asymmetric unit, the first six residues of which were absent in the electron density. Two of the molecules are nearly full length, each consisting of residues 7–399 plus one Cbl molecule bound to it. The other two molecules in the asymmetric unit are truncated, each consisting only of residues 7–273 and containing no bound Cbl.
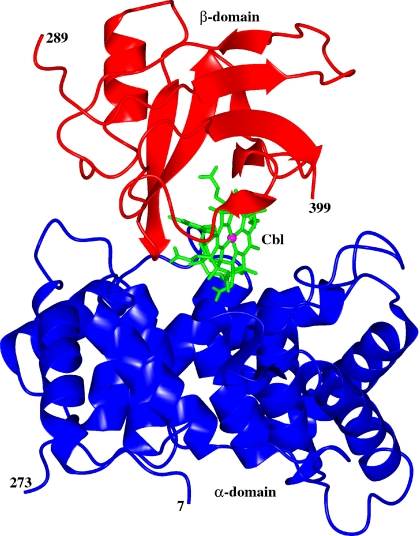
The full-length molecule comprises two domains, an α-domain containing ≈270 residues (7–273) and a β-domain containing ≈110 residues (289–399) (Fig. 1). The Cbl molecule is bound at the interface of the two domains and is mostly buried and shielded from solvent. No electron density is visible for residues 274–288, and this segment appears to be absent or disordered. For the following segment, 289–312, the electron density is weak; maintaining continuity of the backbone chain during model building necessitated reducing the contour level to values as low as 0.6 σ. The individual temperature factors range from 21 to 99 Å2, with an average value of 56 Å2 (SI Fig. 6). The average B-factor of the α-domain is 46 Å2, whereas that of β-domain is 75 Å2.
Fig. 1.
Ribbon diagram of the IF-Cbl complex with the Cbl molecule shown in ball and stick. This diagram was produced by using CCP4MG (23).
The α-domain consists of an intertwined α6/α6 helical barrel; the β-domain contains mostly β strands. The helical barrel is formed by an inner core of six parallel even-numbered α-helices (α2, α4, α6, α8, α10, and α12) surrounded by an outer shell of six parallel odd-numbered α-helices (α1, α3, α5, α7, α9, and α11) that run in the opposite direction, as observed in squalene-hopene cyclase (21) and TC (18) (SI Fig. 7a). The barrel is capped at the bottom by a peptide segment (260–273) that contains a short 3/10 helix. The α-domain is cross-linked by three disulfide bridges (Cys-8-Cys-228, Cys-85-Cys-270, and Cys-125-Cys-164), all distant from the Cbl-binding site. The first bridge links the domain's N-terminal portion to the N terminus of outer helix α11, the second links the domain's C-terminal end to the C terminus of inner helix α4, and the third links the C termini of inner helices α6 and α8. The β-domain contains strands β1, β2, β7, and β6 that form an antiparallel β-sheet. Stacked on this sheet are helix α13 and strands β3, β4, and β5 that run antiparallel to each other and roughly perpendicular to the first β-sheet (SI Fig. 7b). A glycosylation site has been identified at residue Asn-395 of the β-domain, and two molecules of N-acetylglucosamine per full-length molecule have been modeled into the electron density. Additional density extends beyond the second sugar, suggesting that more sugar molecules may be present.
Cbl Binding.
The Cbl molecule is bound at the interface between the α- and β-domains of IF with the corrin ring of Cbl located close to and oriented approximately parallel to the central axis of the α-barrel. A close-up view of Cbl bound to IF, represented by a molecular surface made up of neighboring residues, is shown in SI Fig. 8. The cobalt ion is coordinated by the four nitrogen atoms of the corrin ring and by the N3B atom of the dimethylbenzimidazole ring on the α-side of the corrin ring as the fifth ligand (SI Fig. 5) at a distance of 2.3 Å. The sixth coordination site of the cobalt appears to be vacant, and the β-side of the corrin ring is completely exposed to bulk solvent, making no interactions with any protein side chains or water molecules. A β-hairpin formed by residues 343–352, connecting strands β3 and β4 of the β-domain, covers the dimethylbenzimidazole group of Cbl and maintains it in a hydrophobic environment. Upon binding to IF, all but 19% of solvent-accessible surface area of Cbl is buried within the protein.
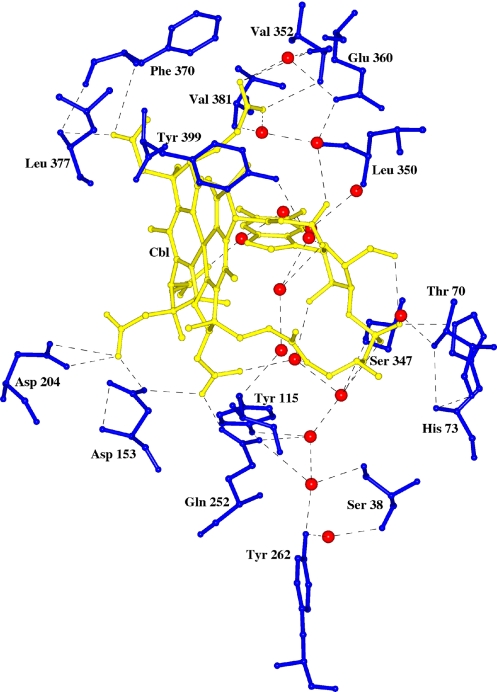
There are five residues: His-73, Tyr-115, Asp-153, Asp-204, and Gln-252 of the α-domain, and four residues, Ser-347, Val-352, Phe-370, and Leu-377 of the β-domain, that form direct hydrogen bonds with Cbl (Fig. 2 and SI Fig. 9). There are also several water-mediated hydrogen bonds formed between Cbl and three residues of the α-domain, Ser-38, Thr-70, and Tyr-262, and four residues of the β-domain, Leu-350, Glu-360, Val-381, and Tyr-399. In the case of Ser-38 and Tyr-262, four water molecules form a channel connecting these side chains to oxygen atoms of the ribose and pyrophosphate portions of Cbl (Fig. 2 and SI Fig. 9). In addition to these linkages through the Cbl molecule, the α- and β-domains are linked directly by one salt bridge and four hydrogen bonds (Lys-365 NZ-Glu-110 OD1; Lys-365 NZ-Ser-105 OG, Asn-36 ND-Ser-347 OG; Asn-246 ND2-W348 O; and Asn-246 ND2-Asp-383 OD2) (data not shown).
Fig. 2.
Environment around the Cbl molecule at the binding site of IF. Cbl is in gold, and the water molecules are shown as red spheres. This diagram was produced by using CCP4MG (23).
Quaternary Structure.
The four molecules in the asymmetric unit are packed together, so that each of the full-length molecules makes extensive contact with one of the truncated molecules to form an incomplete dimer (SI Fig. 10). The α-domains within each pair of full-length and truncated molecules are related to each other by a local two-fold axis. They are stacked on one another with the axes of their α-barrels inclined by ≈120° and pointed in opposite directions. The interface between pairs of α-domains within an incomplete dimer comprises a buried surface area of ≈1,250 Å2 per molecule, and there are ≈24 hydrogen bonds connecting them. The α-subunits of the complete and incomplete molecules are very similar in structure, having rmsd for equivalent Cα atoms between them of ≈0.48 Å. The largest deviations occur between residues 100–108, a loop region between helices α5 and α6, and vary from 0.7 to 1.2 Å.
The two full-length molecules in the asymmetric unit also make contact with each other through their α-domains. The buried surface area at this interface is ≈500 Å2 per molecule, and there is one salt bridge between the molecules. Additional contacts between molecules related by crystallographic symmetry are generally limited to 300–500 Å2 and involve from one to eight hydrogen-bonding interactions.
Comparison of Trans-Cbl and IF.
The structure of another Cbl transport protein, TC, was solved recently from human and bovine sources (18). They share 73% sequence identity and have a similar molecular architecture. The sequence identity between IF and human TC is 27%, whereas between IF and bovine TC, it is 29%. Because the rmsd between bovine and human TC is only 1.2 Å, and the monoclinic form of bovine TC diffracted to higher resolution (2 Å) than did human TC (3.2 Å), a detailed comparison will be limited to human–IF and bovine–TC [Protein Data Bank (PDB) ID code 2BB6].
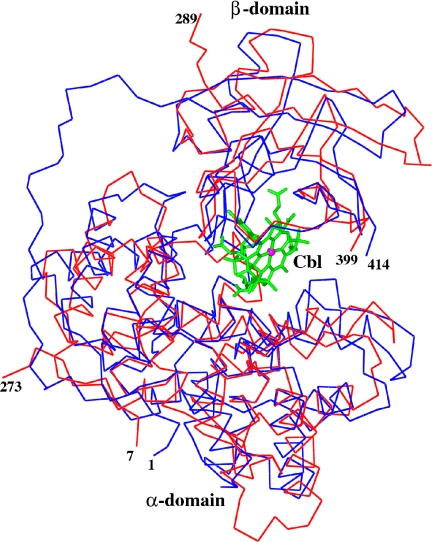
TC is composed of an α- and a β-domain of similar structure to IF. The rmsd between α-domains of IF and TC is 2.16 Å, with 219 matched Cα− atoms (SI Fig. 11) based on the secondary structure matching (22) carried out by using CCP4MG (23). Both TC and IF adopt an α6-α6 barrel motif. The 3/10 helix at the C-terminal end of the α-domain is present in both proteins. TC contains a large loop between residues 67 and 81, which is absent in IF, and there are differences in the length and orientation of the various helices between TC and IF in the α-domains. The β-domains of IF and TC align more closely than do the α-domains, with rmsd of 1.13 Å for 96 matched Cα atoms. The only major difference in the β-domain is a loop formed by residues 302–309 in IF, which extends away from the structure relative to TC. The rmsd between the complete molecules of IF and TC is 2.02 Å for 312 matched Cα-atoms (Fig. 3 and SI Fig. 12). However, the structural similarity does not extend beyond monomers.
Fig. 3.
Superposition of TC (blue) with IF (red). The Cbl is shown in ball and stick. This diagram was produced by using CCP4MG (23).
The Cbl molecule is bound to TC between the α- and β-domains in a manner very similar to that in IF, with its cobalt ion coordinated by the four nitrogen atoms of the corrin ring and by the N3B atom of the dimethylbenzimidazole ring as the fifth ligand on the α-side of the corrin ring. However, the sixth coordination site of the cobalt, rather than being unoccupied as in IF, is occupied by the Nε2 atom of a histidine side chain, at position 175 in TC, which is located on a loop between helices α7 and α8.
Of 16 residues in IF involved in the binding of Cbl, nine interact directly with it. Of these, four residues (Tyr-115, Asp-153, Gln-252, and Leu-377) are conserved between IF and TC, maintaining the same relative positions and interactions with Cbl in the two proteins. Four of the remaining five IF residues that interact directly with Cbl, His-73, Asp-204, Val-352, and Phe-370 are replaced in TC by Gln-68, Asn-227, Leu-368, and Val-384, which make similar side- or main-chain interactions with Cbl. The last of these five residues, Ser-347, which forms a hydrogen bond between its Oγ atom and a Cbl phosphate oxygen, is replaced in TC by Leu-363. However, the adjacent residue, Ser-362, forms a water-mediated hydrogen bond to the same Cbl phosphate oxygen. The net result of these substitutions is to reduce the number of charged groups by two at the active site of TC compared with IF.
Of the seven residues in IF that interact with Cbl through water molecules, only five maintain these interactions at equivalent positions in TC. Three of these are Thr-70, Leu-350, and Val-381 in IF and are replaced by residues with similar properties, Ser-83, Pro-366, and Ile-396, that make similar side- or main-chain water-mediated interactions with TC. In addition, Ser-38 and Tyr-262 of IF, which are connected to Cbl through a channel of four water molecules, are conserved in both proteins. The residues Tyr-399 and Glu-360 of IF are replaced by nonpolar residues Trp-414 and Ala-376 in TC.
In IF, the environment of the dimethylbenzimidazole group of Cbl is crowded, as in TC, whereas on the opposite side of the corrin ring, unlike in TC, it is quite open, there being no histidine residue in IF available to coordinate with the Co2+ ion of Cbl. The five-coordinate nature of the cobalt ligation in IF suggests that the cobalt ion is in the 2 oxidation state. The reduction of Co(III) to Co(II) might have been induced by extensive x-ray exposure during data collection, as observed in human TC (18) and glutamate mutase (24). Helix 7 of IF (residues 128–145) is displaced along its axis by ≈2.5 Å compared with TC, and the following loop, from 146 to 150, is oriented differently from the corresponding loop in TC. In TC, this loop, from 168 to 176, is four residues longer and contains the histidine ligand to the cobalt ion. The shortening of this loop by four residues in IF compared with TC and the longitudinal displacement of helix α7 cause the loop to relocate far from the corrin ring.
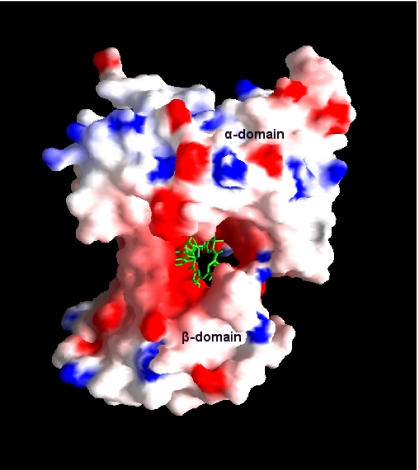
The electrostatic potential surface of the IF-Cbl complex is shown in Fig. 4. The binding site for Cbl of IF is dominated by negatively charged residues in contrast to TC and forms a channel between the α- and β-domain. In TC, the Cbl-binding site is dominated by neutral-charged residues, and the channel is not as predominant as in the IF. However, the overall electrostatic potential surface of IF-Cbl is neutral in nature compared with human and bovine TC. The potential surface diagram of IF-Cbl (Fig. 4) clearly indicates that the binding site for Cbl is broad and open on both sides of the molecule compared with TC. Upon binding to IF, the solvent accessibility area of Cbl is reduced to ≈19% compared with ≈7% in TC (23). This difference does not seem to influence the ability of Cbl to bind to these proteins, as shown by recent studies involving binding of fluorescent analogues and surface plasmon resonance, which show that the affinity of Cbl is similar for all three Cbl-binding proteins (25, 26). However, the absence of a His coordination bond and wide binding site in IF might allow Cbl to move in and out of IF freely compared with TC. In fact, Cbl can transfer at a low rate from IF to TC at neutral pH, but not from TC to IF (27).
Fig. 4.
Electrostatic surface potential of the IF. Positive, negative, and neutral potentials are in blue, red, and white, respectively. Cbl is shown as sticks in green. This diagram was produced by using GRASP (54).
Comparison of IF with a Homology IF Model.
Homology models of IF (PDB ID code 2CKT.pdb) and of HC (PDB ID code 2CKV.pdb) were recently constructed based on the x-ray structure of TC (18, 28). These models have provided valuable insight on the relative binding affinity of Cbl to the two homologous structures in the absence of experimental results. Inherent drawbacks of such homology models, however, are bias for the experimental model and the inability in general to include water molecules in the models. The rmsd between the x-ray structure and the modeled IF is 2.29 Å with 310 Cα atoms. Comparison of the homology IF with the experimental model shows that the predicted β-domain matches much more closely than the α-domain, consistent with the greater similarity of TC and IF for this domain. In the α-domain, the even-numbered inner helices of the homology IF model match the x-ray structure closely and are in perfect register (SI Fig. 7a). The odd-numbered outer helices, however, match the x-ray structure less well, and four of them (1, 3, 5, and 7) are partially or wholly out of register by between one and three residues. Two important loops, residues 146–150 between helices α7 and α8 and residues 100–115 between helices α5 and α6, are substantially different in conformation in the x-ray structure. The first of these loops is predicted to cover the sixth coordination site of Cbl occupied by His-175 in TC and the second of these to partially block access to amide side chain on pyrrole ring C. Both of these sites are quite accessible in the IF structure. In addition, the homology IF model shows distinct differences from the x-ray structure for residues 25–62, a putative interaction site for human CUB (29) (see below). This segment extends from the middle of helix α1 to the end of helix α3. Approximately two-thirds of these residues in the homology IF are out of register with respect to the x-ray structure. In the homology IF, ≈7% of the solvent-accessible area of Cbl remains accessible, a value similar to that observed in TC; in the experimental model of IF, the value is ≈19%. In general, the homology IF model follows the TC more closely than the experimental IF model, because the rmsd of Cα atom between them is 1.20 Å.
There are few significant differences in the positions of the binding-site residues between the two models. Of nine binding-site residues that interact directly with Cbl, four conserved residues (Tyr-115, Asp-153, Gln-252, and Leu-377) and three nonconserved residues (Ser-347, Val-352, and Phe-370) deviate by 0.5–1.0 Å. The two remaining nonconserved residues, His-73 and Asp-204, deviate by 1.8 and 1.3 Å, respectively. All but one of these interactions (Ser-347) were correctly predicted in the homology model, which probably reflects the conserved nature of the Cbl-binding site in the three proteins. Of the seven residues that interact with Cbl through water molecules, the two residues that are conserved between IF and TC (Ser-347 and Tyr-262) deviate between 0.4 and 0.9 Å; the other five residues deviate between 1.2 and 3.4 Å.
Discussion
An oligomerization study of recombinant IF from Arabidopsis thaliana (30) indicated that dimers are formed only with uncleaved full-length IF, and only if Cbl is bound. The presence of an intact linker region is required for dimerization to occur, but the dimers form only at a relatively high concentrations of IF (KD = ≈1 mM), well above that required under physiological conditions. The crystal structure of IF contains pairs of molecules that contact each other through their α-domains, forming a two-fold related “crystallographic dimer” (SI Fig. 10). If this mode of interaction represented the form of the complete dimer that occurs in solution, it would leave the Cbl moiety and the β-domains at opposite ends of the dimer, and it is unclear how the absence of one of the β-domains, its Cbl cofactor, and the covalent interdomain linker would influence the dimerization process. Furthermore, it would be inconsistent with the proposed dimerization model (30).
A more likely possibility is that proteolysis of IF had occurred during the process of crystallization. Based on the oligomerization study, cleavage of IF during crystallization would destabilize any dimers present. Examination of the crystal packing in an artificial unit cell arrangement in which a β-domain plus Cbl was positioned on the α-domain of each of the two truncated molecules indicated that each of the artificially positioned β-domains overlapped extensively with symmetry related α-domains of both halves of the same dimer, i.e., βB overlapped with both α∗A and α∗B and βD overlapped with α∗C and α∗D, where ∗ indicates a symmetry-related molecule, and the subscripts indicate the molecule involved. No such overlap occurred for the β-domains of the full-length molecules. Thus, the crystal packing allows only half of the four IF molecules to be present in full-length form; for the remaining half of the molecules, only truncated α-domains can be accommodated in the lattice. It is possible that only partial proteolysis of IF had occurred during crystallization, and that the uncleaved IF with bound Cbl cocrystallized with cleaved α-domains unable to bind Cbl. However, it is more likely that cleavage would proceed to completion rather than stopping halfway during the long time it took to grow the crystals. Furthermore, the electron density of the C terminus of all four α-domains ends abruptly at the same residue, Asp-273, and the orientation of the backbone chain is the same in each. The average B-factor for the β-domains is very high compared with those of the α-domains (SI Fig. 6), which indicate a greater mobility for the β-domain. This clearly indicates that the β-domains can be cleaved at the slightest provocation during the process of crystallization.
It has been shown in solution studies that independent α- and β- domains of IF can still form an effective complex when Cbl binds to it (30). The apparent cleavage of IF in the crystals is consistent with its susceptibility to proteolysis by cathepsin L, an intracellular ileal protease (31), and by plant proteases during expression in Arabidopsis thalises (30), despite its resistance to luminal proteases in vivo (5, 14). In the present study, IF was expressed in yeast, which is well known to contain proteases, although the freshly prepared recombinant material remained intact for relatively short periods. However, the relatively long incubation time at high protein concentration (≈10 mg/ml) during crystallization could have led to complete cleavage into α- and β-domains, had more than trace amounts of protease been present.
As has been observed in a number of Cbl-binding proteins such as methionine synthase (32), methyl-malonyl CoA mutase (4), glutamate mutase (24), and trans-Cbl II (18), the Cbl binds to IF at the interface of two domains. In TC, histidine forms a coordination bond with Cbl on the β-side, but in methionine synthase, the histidine forms a coordination bond with Cbl on the α-side. However, there is no such coordination bond formation by histidine in IF, and the β-side of Cbl is empty, devoid of any ordered water molecules. Kinetics studies have indicated a low affinity of Cbl for the α- compared with the β-domain, although the α-domain is essential for the retention of bound Cbl (33). The same study suggested that Cbl binds first to the β-domain, and that subsequently the α-domain approaches it in a second step, thereby forming the ligand-binding site. Hydrogen bond analysis of the IF active site indicates that an equal number of hydrogen bonds are formed between the Cbl and the α- and β-domains. However, there is a β-strand from the β-domain that covers the α-side of Cbl and forms main-chain hydrogen bonds to it. This formation of main-chain hydrogen bonds might be responsible for the greater affinity of the β-domain toward the Cbl. All of the hydrogen bonds between the β-domain and Cbl are formed by residues located between residues 347 and 399, which clearly confirm an earlier study showing that cleavage of the C-terminal 12% of the molecule abolished Cbl binding (29).
CUB is a large multidomain protein (460 kDa), which contains 27 tandem CUB domains that harbor the IF-Cbl-binding site (34, 35). It has been proposed that a site on IF for binding the CUB receptor might lie within the segment between residues 25 and 62 (29). CUB recognizes both the full-length Cbl-saturated IF complex and the Cbl-saturated cleaved IF complex but not the isolated α- or β-domains, even if they are saturated with Cbl (33). This led to the prediction that the CUB-binding site on IF might be formed by regions present in both the α- and β-domains. The crystal structures of several CUB domains are known (36–39), and these show that their overall electrostatic potential surfaces are neutral, as is IF (this work). Comparison of the IF-Cbl complex with the CUB domain structures suggests that the nature of these interactions is primarily nonpolar or neutral hydrophilic and not electrostatic.
In the IF structure, residues 25–62 contain two α-helical segments, namely α2 and α3 (SI Fig. 13). The α2 helix formed by residues 37–47 blocks access to the domain interface channel, which encompasses the Cbl-binding site. It may be that helix α2 can block the internal channel after binding the Cbl molecule, but this cannot be confirmed with the present structure.
Although human and rat IF share 80% sequence identity, human IF does not bind to the rat IF receptor (40). Only 6 of the 37 residues implicated in the receptor-binding region (residues 25–62) differ between human and rat IF (29). They are S31E, A33D, Y34L, G47S, A48T, and K52E (SI Fig. 14). A homology model was constructed of an IF chimera containing the rat sequence for residues 25–62 and the human sequence for the rest of the molecule (see Materials and Methods). It is apparent that the 3D structure of IF is essentially unchanged as the result of the amino acids substitutions. However, these six substitutions drastically alter the nature of the interaction between the two IF-Cbl complexes with their respective receptors. This change in the distribution of nonpolar, neutral hydrophilic, and electrostatic residues may be one of the major reasons why the interactions of human IF with the rat receptor are greatly impaired.
Because the homology model for HC was derived from the experimental TC model (28), some of the salient features of the binding site may be missing, as observed in the homology IF model, although an overall view of the binding site is demonstrated (18, 28). Of nine residues that interact directly with Cbl in IF, the four residues (Tyr-115, Asp-153, Gln-252, and Leu-377) conserved between IF and TC are also conserved in HC. In addition, Ser-347 of IF is conserved in HC. Of the remaining four residues, Val-352 and Phe-370 of IF are replaced by residues with similar properties, Ile-363 and Leu-381. The last two residues, His-73 and Asp-204, are replaced by Glu-71 and Asn-217. Of the seven residues that interact with Cbl through water molecules in IF, only one residue, Tyr-399 is retained in HC. Four of the remaining six residues, Ser-38, Thr-70, Leu-350, and Val-381, are replaced by residues of similar properties, Asn-33, Ser-69, Pro-361, and Ala-392. The remaining two residues, Tyr-262 and Glu-360, are replaced by nonpolar residues, Phe and Ala.
In general, the binding affinity of Cbl analogues may be governed predominantly by steric and electrostatic interactions, as observed elsewhere (28). On comparing the Cbl-binding site of IF with TC and HC, it is known that only IF contains His-73 and Asp-204, which interact directly with Cbl and Glu 360 through the water molecules. It may be possible that protonation/deprotonation of Asp-His, which interact directly with Cbl through hydrogen bonding, can influence the binding affinities of Cbl. Because both His-73 and Asp-204 are part of the α-domain, they would play a major role in stabilization of the complex. At or below pH 6, these residues could become protonated, which would lead to changes in the hydrogen-bonding pattern and, in turn, movement of helices. This could lead to a decrease in affinity of the Cbl. The numbers of charged active-site residues at the binding site of IF, TC, and HC are six, four, and three, respectively. The dissociation constant of cobinamide (Cbl without the benzimidazole moiety) for TC is 1 nM (41), a value that is intermediate between that found for IF and HC. Thus, the electrostatic charge at the active site could account for some of the differences in binding of cobamide among the Cbl-binding proteins.
It has been speculated that intracellular transfer of Cbl from IF to TC could occur in a neutral cellular compartment, because Cbl-binding affinity decreases rapidly with decrease in pH (27). The crystals of IF, human TC, and bovine TC were grown at pH 6, 7.5, and 8.5, respectively. Unlike in bovine TC, no His-Co coordination bond was found in human TC (18). Although lack of this bond was attributed to shortening of the loop by three residues in human TC compared with bovine TC and reduction of Co(III) to Co(II) as a result of x-ray-induced radiation damage (18), the difference in pH at the time of crystallization cannot be ruled out. The pKa of histidine can vary from 6.4 to 7.4 in different protein structures depending on the environment (42). It is possible that the His is in the neutral state in bovine TC, because the coordination occurs through its unprotonated nitrogen, whereas it is protonated in human TC. Similarly, a pH effect on the displacement of helix α7 and the following loop 146–150 of IF in comparison to bovine TC cannot be ruled out, modifying formation of a sixth ligand for Co2+. However, a comparison of the experimental IF and TC models with the homology HC model does not show any systematic pH-dependent difference in the molecular geometry. Thus, differences in ligand binding at the active site between the three Cbl-binding proteins appear more likely related to steric and electrostatic charge effects than to pH alone.
Conclusions
Although IF is highly glycosylated and was difficult to crystallize, its structure provides abundant information and insight concerning its functional properties, including identification of a major sugar-binding site. The x-ray IF model is locally different from the known TC and homology IF models, especially in the N-terminal domain. Most of the odd-numbered outer helices of the N-terminal domain in the x-ray model are partially or wholly out of alignment by one to three residues with respect to the homology model. Because residues 25–62, implicated for receptor binding, differ substantially between the x-ray and homology IF models, these differences will clearly influence interactions with the receptor site. Furthermore, the rat/human chimera model for IF indicates that changes in the nature of the residues in the receptor-implicated region may be one of the major reasons for the noninterchangeability of rat and human IF receptors.
Comparisons among IF, TC, and HC have revealed the exclusive presence of two polar side chains in the binding site for Cbl in IF. This difference could affect the stability of the IF-B12 complex at low pH. The x-ray IF model provides the details of all possible interactions with Cbl, especially those involving water molecules, thereby enabling accurate visualization of possible modification sites at the corrin ring of Cbl. The x-ray model of IF clearly identifies the β-side of the corrin ring, which is completely exposed to solvent, and the side chain of pyrrole ring C can accommodate bioconjugates or Cbl analogues much larger in size, because these are more accessible than predicted by the homology model. The identification of water molecules in IF also provides greater detail for receptor-binding site interactions at the N-terminal end of the protein. IF has been a target for drug discovery projects that would allow uptake of small molecules linked either with Cbl (43) or with a small peptide that would bind to the receptor. This latter approach to improved absorption of biologically active substances has recently been used with other peptide backbones.
Materials and Methods
Human IF has been cloned, sequenced, and expressed in Pichia pastoris as described (44, 45). Vitamin B12 (Cyanocobalamin; Sigma–Aldrich, St. Louis, MO) (SI Fig. 5) was added to apo IF in a 3:1 molar ratio, and the final concentration of IF was 10 mg/ml in 10 mM phosphate buffer, pH ≈6.8. Pink crystals were obtained in sitting drops from 10% PEG 20000/100 mM Mes, pH 6.0/20 mM CaCl2 with 9 mM BaCl2 as an additive at 18°C in 4–6 weeks time.
Data were recorded to 2.5-Å resolution from a flash-cooled crystal 0.4 × 0.15×≈ 0.08 mm in size (with 18% glycerol containing mother liquor) near the absorption edge for cobalt. A second crystal of similar size was soaked with 0.2 mM K2PtCl4 for 10 min, and a complete data set to 3.2-Å resolution was collected near the absorption edge of platinum. Both data sets were collected on a ADSC Q315 detector at the Advanced Photon Source, Argonne National Laboratory on Beamline NE-CAT 8BM, and were processed by using HKL2000 (46).
Twenty-six platinum sites were then located by using difference-Patterson and difference-Fourier methods with ShelxD (47) and the CCP4 program suite (48). Initial phases using both data sets were calculated by SIRAS using BP3 (48) and improved with solvent flattening in RESOLVE (49); in the process, the resolution was extended to 2.6 Å. The resulting electron density map revealed clear main-chain density and substantial side-chain detail. After skeletonization using COOT (50) and using the model of TC (18) as a reference, the path of the main chain could be traced for one of the two complete molecules of IF in the asymmetric unit. Although broken, the density for the other complete molecule and the two incomplete molecules was good enough to calculate rotation matrices for noncrystallographic symmetry (NCS) averaging (50). The NCS averaged map clearly showed density for the Cbl cofactor for both complete molecules, which was confirmed from the positions of cobalt atoms located in an anomalous scattering difference map from the native data (49). The (2Fo−Fc) electron density map for Cbl is shown in SI Fig. 15. Refinement at 2.6-Å resolution was then carried out by using CNS (51) with NCS restraints. During the refinement, model building was carried out by using COOT (50) and Turbo-Frodo (52). Phase recombination and density modification using the CCP4 program suite (48) and RESOLVE (49) were carried out to remove errors in the model, because the refinement was stalled when R and Rfree were ≈37.9 and 39.5, respectively. An anomalous difference-Fourier map was calculated to verify the positions of Cys and Met residues in the model. However, the loop regions connecting domains α and β in the two complete molecules appeared highly disordered and were impossible to trace. Several cycles of simulated annealing, positional, and temperature factor refinement (51) reduced the R and Rfree to 26.3% and 28.6%, respectively. At this point, water molecules were added, and the model was refined further until R and Rfree values converged to 21.3% and 24.9%, respectively (51).
The final model contains four molecules of IF, with two complete molecules containing both α- and β-domains and two incomplete molecules with only an α-domain. The two Cbl molecules identified corresponded to the complete molecules. Four sugar molecules were also located, two attached to Asn-395 of each complete molecule along with 459 water molecules. The stereochemistry of the model was analyzed by using PROCHECK (53). The x-ray data, structure solution, and refinement statistics are listed in SI Table 1.
The chimera rat–human IF model was generated (SI Fig. 14) in silico by mutating six residues (S31E, A33D, Y34L, G47S, A48T, and K52E) implicated in receptor binding and subjecting them to geometrically restrained refinement by using COOT (50).
Supplementary Material
Acknowledgments
This work is supported by the NE-CAT facility at the Advanced Photon Source (Award RR-15301) from the National Center for Research Resources, National Institutes of Health; and Grants P01 DK-33487 (to D.H.A.) and GM20530 (to F.S.M.) from the National Institutes of Health. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Science, under contract DE-AC02-06CH11357.
Abbreviations
- IF
intrinsic factor
- Cbl
cobalamin
- TC
trans-Cbl II
- HC
haptocorrin
- CUB
cubilin
- PDB
Protein Data Bank.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2PMV).
This article contains supporting information online at www.pnas.org/cgi/content/full/0703228104/DC1.
References
- 1.Brown KL. Chem Rev. 2005;105:2075–2149. doi: 10.1021/cr030720z. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee R. Chemistry and Biochemistry of B12. New York: Wiley; 1999. [Google Scholar]
- 3.Ludwig M, Matthews R. Annu Rev Biochem. 1997;66:269–313. doi: 10.1146/annurev.biochem.66.1.269. [DOI] [PubMed] [Google Scholar]
- 4.Mancia F, Keep NH, Nakagawa A, Leadlay PF, McSweeney S, Rasmussen B, Bosecke P, Diat O, Evans PR. Structure (London) 1996;4:339–350. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- 5.Nexo E. In: Vitamin B12-Proteins. Krautler B, Angoni D, Golding BT, editors. Weinheim, Germany: Wiley; 1998. pp. 461–475. [Google Scholar]
- 6.Allen RH. Prog Hematol. 1975;9:57–84. [PubMed] [Google Scholar]
- 7.Li N, Seetharam S, Seetharam B. Biochem Biophys Res Commun. 1995;208:756–764. doi: 10.1006/bbrc.1995.1402. [DOI] [PubMed] [Google Scholar]
- 8.Hewitt JE, Gordon MM, Taggart RT, Mohandas TK, Alpers DH. Genomics. 1991;10:432–440. doi: 10.1016/0888-7543(91)90329-d. [DOI] [PubMed] [Google Scholar]
- 9.Johnston J, Yang-Feng T, Berliner N. Genomics. 1992;12:459–464. doi: 10.1016/0888-7543(92)90435-u. [DOI] [PubMed] [Google Scholar]
- 10.Allen RH, Seetharam B, Podell E, Alpers DH. J Clin Invest. 1978;61:47–54. doi: 10.1172/JCI108924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marcoullis G, Parmentier Y, Nicolas JP, Jimenez M, Gerard P. J Clin Invest. 1980;66:430–440. doi: 10.1172/JCI109873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen RH, Mehlman CS. J Biol Chem. 1973;248:3660–3669. [PubMed] [Google Scholar]
- 13.Dieckgraefe BK, Seetharam B, Banaszak L, Leykam JF, Alpers DH. Proc Natl Acad Sci USA. 1988;85:46–50. doi: 10.1073/pnas.85.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seetharam B. Annu Rev Nutr. 1999;19:173–195. doi: 10.1146/annurev.nutr.19.1.173. [DOI] [PubMed] [Google Scholar]
- 15.Seetharam B, Bose S, Li N. J Nutr. 1999;129:1761–1764. doi: 10.1093/jn/129.10.1761. [DOI] [PubMed] [Google Scholar]
- 16.Quadros EV, Regec AL, Khan KM, Quadros E, Rothenberg SP. Am J Physiol. 1999;277:G161–G166. doi: 10.1152/ajpgi.1999.277.1.G161. [DOI] [PubMed] [Google Scholar]
- 17.Quadros EV, Nakayama Y, Sequeira JM. Biochem Biophys Res Commun. 2005;327:1006–1010. doi: 10.1016/j.bbrc.2004.12.103. [DOI] [PubMed] [Google Scholar]
- 18.Wuerges J, Garau G, Geremia S, Fedosov SN, Petersen TE, Randaccio L. Proc Natl Acad Sci USA. 2006;103:4386–4391. doi: 10.1073/pnas.0509099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolhouse JF, Allen RH. J Clin Invest. 1977;60:1381–1392. doi: 10.1172/JCI108899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seetharam B, Alpers D. In: Handbook of Physiology: Intestinal Absorption and Secretion Section. Field M, Frizell R, editors. Bethesda: Am Physiol Soc; 1990. pp. 437–461. [Google Scholar]
- 21.Wendt KU, Poralla K, Schulz GE. Science. 1997;277:1811–1815. doi: 10.1126/science.277.5333.1811. [DOI] [PubMed] [Google Scholar]
- 22.Krissinel E, Henrick K. Acta Crystallogr D. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 23.Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Acta Crystallogr D. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- 24.Champloy F, Gruber K, Jogl G, Kratky C. J Synchrotron Radiat. 2000;7:267–273. doi: 10.1107/S0909049500006336. [DOI] [PubMed] [Google Scholar]
- 25.Fedosov SN, Grissom CB, Fedosova NU, Moestrup SK, Nexo E, Petersen TE. FEBS J. 2006;273:4742–4753. doi: 10.1111/j.1742-4658.2006.05478.x. [DOI] [PubMed] [Google Scholar]
- 26.Cannon MJ, Myszka DG, Bagnato JD, Alpers DH, West FG, Grissom CB. Anal Biochem. 2002;305:1–9. doi: 10.1006/abio.2002.5647. [DOI] [PubMed] [Google Scholar]
- 27.Brada N, Gordon MM, Wen J, Alpers DH. J Nutr Biochem. 2001;12:200–206. doi: 10.1016/s0955-2863(00)00129-7. [DOI] [PubMed] [Google Scholar]
- 28.Wuerges J, Geremia S, Randaccio L. Biochem J. 2007;403:431–440. doi: 10.1042/BJ20061394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang LH, Chokshi H, Hu CB, Gordon MM, Alpers DH. J Biol Chem. 1992;267:22982–22986. [PubMed] [Google Scholar]
- 30.Fedosov SN, Fedosova NU, Berglund L, Moestrup SK, Nexo E, Petersen TE. Biochemistry. 2004;43:15095–15102. doi: 10.1021/bi048924c. [DOI] [PubMed] [Google Scholar]
- 31.Gordon MM, Howard T, Becich MJ, Alpers DH. Am J Physiol. 1995;268:G33–G40. doi: 10.1152/ajpgi.1995.268.1.G33. [DOI] [PubMed] [Google Scholar]
- 32.Drennan CL, Huang S, Drummond JT, Matthews RG, Ludwig ML. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 33.Fedosov SN, Fedosova NU, Berglund L, Moestrup SK, Nexo E, Petersen TE. Biochemistry. 2005;44:3604–3614. doi: 10.1021/bi047936v. [DOI] [PubMed] [Google Scholar]
- 34.Moestrup SK, Kozyraki R, Kristiansen M, Kaysen JH, Rasmussen HH, Brault D, Pontillon F, Goda FO, Christensen EI, Hammond TG, Verroust PJ. J Biol Chem. 1998;273:5235–5242. doi: 10.1074/jbc.273.9.5235. [DOI] [PubMed] [Google Scholar]
- 35.Kristiansen M, Kozyraki R, Jacobsen C, Nexo E, Verroust PJ, Moestrup SK. J Biol Chem. 1999;274:20540–20544. doi: 10.1074/jbc.274.29.20540. [DOI] [PubMed] [Google Scholar]
- 36.Romao MJ, Kolln I, Dias JM, Carvalho AL, Romero A, Varela PF, Sanz L, Topfer-Petersen E, Calvete JJ. J Mol Biol. 1997;274:650–660. doi: 10.1006/jmbi.1997.1423. [DOI] [PubMed] [Google Scholar]
- 37.Romero A, Romao MJ, Varela PF, Kolln I, Dias JM, Carvalho AL, Sanz L, Topfer-Petersen E, Calvete JJ. Nat Struct Biol. 1997;4:783–788. doi: 10.1038/nsb1097-783. [DOI] [PubMed] [Google Scholar]
- 38.Varela PF, Romero A, Sanz L, Romao MJ, Topfer-Petersen E, Calvete JJ. J Mol Biol. 1997;274:635–649. doi: 10.1006/jmbi.1997.1424. [DOI] [PubMed] [Google Scholar]
- 39.Feinberg H, Uitdehaag JC, Davies JM, Wallis R, Drickamer K, Weis WI. EMBO J. 2003;22:2348–2359. doi: 10.1093/emboj/cdg236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seetharam B, Bakke JE, Alpers DH. Biochem Biophys Res Commun. 1983;115:238–244. doi: 10.1016/0006-291x(83)90995-6. [DOI] [PubMed] [Google Scholar]
- 41.Fedosov SN, Petersen TE, Nexo E. Biochemistry. 1995;34:16082–16087. doi: 10.1021/bi00049a023. [DOI] [PubMed] [Google Scholar]
- 42.Cantor C, Schimmel P. Biophys Chem. San Francisco: Freeman; 1980. Part I. [Google Scholar]
- 43.Chalasani KB, Russell-Jones GJ, Yandrapu SK, Diwan PV, Jain SK. J Control Rel. 2007;117:421–429. doi: 10.1016/j.jconrel.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 44.Wen J, Kinnear MB, Richardson MA, Willetts NS, Russell-Jones GJ, Gordon MM, Alpers DH. Biochim Biophys Acta. 2000;1490:43–53. doi: 10.1016/s0167-4781(99)00218-3. [DOI] [PubMed] [Google Scholar]
- 45.Gordon MM, Russell-Jones G, Alpers DH. Methods Enzymol. 1997;281:255–261. doi: 10.1016/s0076-6879(97)81031-2. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 48.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.Terwilliger TC, Berendzen J. Acta Crystallogr D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 52.Roussel A, Cambillau C. Silicon Graphics Geometry Partners Directory. Mountain View, CA: Silicon Graphics; 1989. pp. 77–78. [Google Scholar]
- 53.Laskowski R, Thornton J, Moss D, MacArthur M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 54.Honig B, Nicholls A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.