Abstract
Retinoid X receptors (RXRα, -β, and -γ) occupy a central position in the nuclear receptor superfamily, because they form heterodimers with many other family members and hence are involved in the control of a variety of (patho)physiologic processes. Selective RXR ligands, referred to as rexinoids, are already used or are being developed for cancer therapy and have promise for the treatment of metabolic diseases. However, important side effects remain associated with existing rexinoids. Here we describe the rational design and functional characterization of a spectrum of RXR modulators ranging from partial to pure antagonists and demonstrate their utility as tools to probe the implication of RXRs in cell biological phenomena. One of these ligands renders RXR activity particularly sensitive to coactivator levels and has the potential to act as a cell-specific RXR modulator. A combination of crystallographic and fluorescence anisotropy studies reveals the molecular details accounting for the agonist-to-antagonist transition and provides direct experimental evidence for a correlation between the pharmacological activity of a ligand and its impact on the structural dynamics of the activation helix H12. Using RXR and its cognate ligands as a model system, our correlative analysis of 3D structures and dynamic data provides an original view on ligand actions and enables the establishment of mechanistic concepts, which will aid in the development of selective nuclear receptor modulators.
Keywords: crystal structure, ligand design, nuclear receptor, agonist, antagonist
Nuclear Receptor (NR)-controlled gene expression relies on a mechanism in which NRs recruit coregulators that are part of multiprotein complexes. These complexes correspond to chromatin-modifying and transcription-initiating machineries that act at target gene promoters in a precisely timed and sequential fashion (1). The binding of a ligand to the ligand-binding domain (LBD) of NRs constitutes the initial step of this regulatory process. In this context, the C-terminal helix H12 of LBDs plays a key role, because its position, which depends on the bound ligand, determines the type of coregulator recruited by the receptor (2). Structural studies have shown that in agonist-bound NR LBDs, H12 adopts the so-called “active” or “holo” conformation and provides a binding surface for short NR interaction motifs of coactivators (3). In contrast, antagonists prevent H12 from adopting the holo position (4).
Therapeutically, retinoid X receptor (RXR)-selective ligands, referred to as rexinoids, are used in cancer therapy, and previously uncharacterized rexinoid-based therapeutic paradigms are currently being explored. In addition, rexinoids have promise for use in the therapy of metabolic diseases (5, 6), but important side effects associated with existing compounds limit their use. Improved understanding of the biological role and the structural biology of RXR (7, 8) will allow the synthesis of selective modulators that might overcome the limitations of current drugs. Here, we describe the rational design and functional characterization of a spectrum of RXR modulators and discuss the opportunity to use these ligands as pharmacological tools. Moreover, using x-ray crystallography and fluorescence anisotropy, we elucidate the molecular basis of their mechanisms and suggest a structural and dynamic model of partial agonist action.
Results and Discussion
Rational Design of RXR-Selective Modulators.
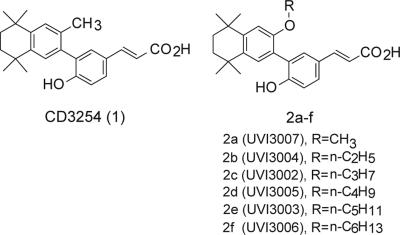
Contrary to the fairly large collection of existing RXR agonists (9–11), only a few antagonists have been reported (12–16). To develop RXR modulators, we selected as a lead compound CD3254 (compound 1), a potent and selective RXR agonist (17) that contains two positions suitable for chemical modification located in ortho position to the biaryl bond (Fig. 1). To guide rational ligand design, we built a model of RXRα LBD bound to compound 1 and identified the methyl group as the most appropriate position for substitution [supporting information (SI) Fig. 6]. Replacement by a side chain of six atoms was predicted to prevent H12 from adopting the active position and hence generate an RXR-selective antagonist. The corresponding synthetic route (SI Fig. 7) provided a series of alkyl ether analogs with chain lengths ranging from C1 to C6 (Fig. 1).
Fig. 1.
Structures of the agonist CD3254 (compound 1) and of the series of alkyl ether analogs 2a–f.
Conversion of CD3254 into Partial and Full RXR Antagonists.
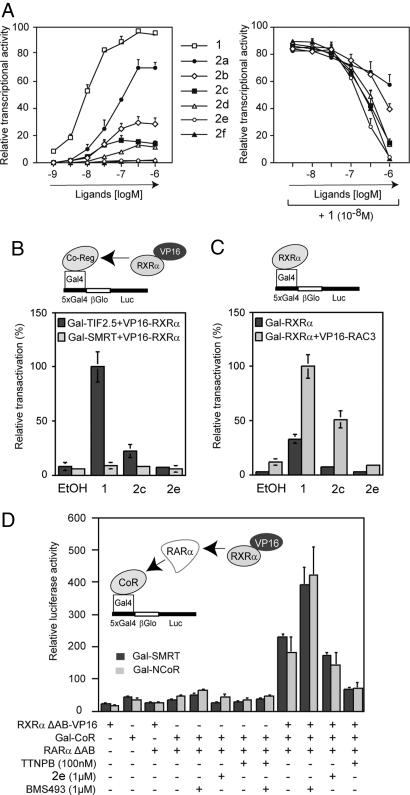
Transient transactivation experiments (Fig. 2A) revealed a progressive transition from agonist via mixed agonist/antagonist to full antagonist upon extension of the aliphatic side chain. Indeed, 2a and 2b are progressively weaker agonists that exhibit some antagonist activity relative to CD3254 (compound 1), whereas 2c induces only ≈15% of the transactivation seen with the parent compound but acquires strong antagonist activity. Further extension of the side chain reduces the transactivation capacity of the rexinoid such that 2e is transcriptionally inactive. However, 2e and 2f retain RXR interaction capacity and consequently act as RXR antagonists. Coregulator recruitment assays confirm this transition from agonist to antagonist (Fig. 2B). Although agonist 1 induces TIF2 coactivator association, 2c does so only weakly, and 2e is inactive. The antagonist 2e does not acquire inverse agonist activity, because there is no gain in SMRT corepressor binding. The 2e-induced RXR antagonist conformation is insensitive to changes in coactivator expression levels, whereas the agonist activity of CD3254 (compound 1) and the weak agonist activity of the partial agonist 2c can be largely enhanced when the coactivator RAC3 is present at high levels (Fig. 2C). These results suggest that 2c may act as a cell-specific RXR agonist or antagonist, depending on the cellular coactivator expression levels. “Sandwich” two-hybrid experiments show that, in the context of the retinoic acid receptor (RAR)-RXR heterodimer, 2e does not affect the corepressor interaction capacity of the RARα subunit as, for example, does the RAR inverse agonist BMS493 or the RAR agonist TTNPB (Fig. 2D).
Fig. 2.
RXR agonist/antagonist potential of new compounds. (A) HeLa cells stably transfected with the reporter recombinant 5xGal4-βGlo-Luc and Gal4-RXRβ were incubated with increasing concentrations of compounds to assess their RXR agonist potential (Left) or with 10 nM CD3254 (compound 1) and increasing concentrations of the compounds to assess their RXR antagonist potential (Right). (B) Mammalian two-hybrid assays were performed in HeLa cells to assess the influence of both 2c and 2e at 1 μM on interaction between RXR and both coregulators TIF2 (coactivator) and SMRT (corepressor) in a cellular context. (C) Mammalian two-hybrid assays were performed in COS cells to reveal the partial agonist activity of 2c. Compounds were used at 1 μM. (D) Mammalian “sandwich” two-hybrid assays in HeLa cells to assess the influence of ligands on corepressor interaction in the context of the RXR-RAR heterodimer. Note that BMS493 acts as an RAR inverse agonist by stabilizing corepressor interaction, whereas the RAR agonist TTNPB destabilizes the corepressor complex.
Impact of Ligand Binding on RXR Structural Dynamics.
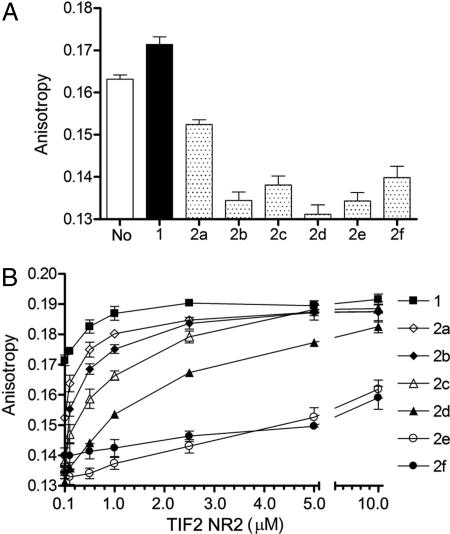
Because recent reports suggest that the functional consequences of ligand binding to NRs are mediated largely through modulation of the structural dynamics of their C-terminal region (2), we studied the impact of ligands 2a–f on RXR helix H12 mobility. Attaching a fluorescein moiety to the C terminus of RXRα through intein chemistry (18, 19) allowed analysis of the dynamic properties of RXR in RARα-RXRα heterodimers. Steady-state fluorescence anisotropy measurements showed that addition of agonist 1 slightly increases anisotropy, indicating stabilization of H12, presumably in the active conformation (Fig. 3A). In contrast, binding of ligands 2a–f decreases anisotropy, revealing a higher mobility of H12 in the presence of these compounds. Moreover, addition of coactivator transcriptional intermediary factor 2 NR box 2 (TIF2 NR2) peptide (20) causes a slight dose-dependent increase of the anisotropy of the heterodimer bound to CD3254 (compound 1), indicating that peptide binding further stabilizes the active conformation (Fig. 3B). With the mixed agonists/antagonists 2a–d, addition of peptide strongly increases anisotropy, suggesting that TIF2 NR2 reduces helix H12 mobility by shifting the equilibrium toward the holo form. Remarkably, at the highest peptide concentrations, the dynamics of H12 is comparable to that observed with the agonist-bound protein. In the presence of antagonists 2e–f, even high doses of TIF2 peptide fail to stabilize H12. Time-resolved fluorescence anisotropy studies confirm the steady-state observations and reveal that both the fraction of RXR molecules with a stabilized holo-H12 and the time scale of H12 dynamics depend on the nature of the bound ligand (SI Table 1). Together, these data provide direct experimental evidence for the differential effects of various classes of NR ligands on H12 dynamics. Moreover, they reveal a mechanism according to which mixed agonists/antagonists can “sense” intracellular coregulator levels and act as cell-selective modulators with agonist or antagonist properties depending on the cellular context (21).
Fig. 3.
RXRα structural dynamics monitored by steady-state fluorescence anisotropy. (A) Using an RARα-RXRα LBD heterodimer in which a fluorescent dye is specifically attached to the C terminus of RXRα, we measured anisotropy values in absence of added ligand (No) and in the presence of saturating concentrations of the RXR agonist CD3254 (compound 1), mixed agonists/antagonists 2a–d or antagonists 2e–f. Of note, it is very likely that the heterodimer used as a reference (No) is not truly unliganded, because it has been previously reported that bacterially expressed RXR is able to bind endogenous fatty acids. (B) Similar experiments were carried out in the presence of increasing concentrations of the NR interaction motif 2 peptide of the coactivator TIF2 (TIF2 NR2).
Structural Basis for Agonist to Antagonist Transition.
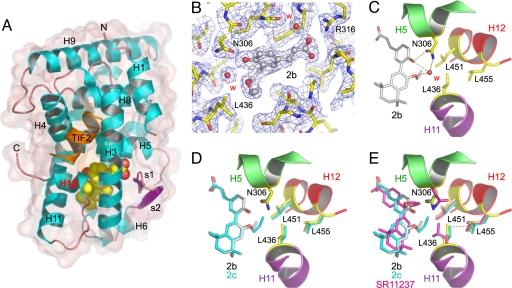
To gain structure-based insight into the mechanism of action of these modulators, we solved the structures of RXRα LBD in complex with 2a, 2b, 2c, and TIF2 NR2 (SI Table 2). The structures display the canonical agonist conformation with H12 capping the ligand-binding pocket (LBP) and TIF2 NR2 bound to the so-called AF-2 surface (Fig. 4A). A particular feature of compounds 2a–f is the presence of two oxygen atoms attached to the tetrahydronaphthyl and cinnamic acid aromatic rings (Fig. 1). In the structures with 2a and 2b, these oxygen atoms are involved in a network of hydrogen bonds that stabilizes a water molecule in a predominantly hydrophobic environment (Fig. 4C). In both structures, the water molecule occupies a well defined position, as indicated by the clear electron density (Fig. 4B) and the B-factor values of 11.77 Å2 and 24.15 Å2 (mean B values for all atoms 15.44 Å2 and 25.18 Å2 in the 2a and 2b complexes). In contrast, no electron density is observed for the corresponding water molecule in the 2c complex. The reason for this absence is that, to maintain favorable interactions with LBP residues, the longer side chain of 2c rotates around the oxygen atom by an angle of 42° and adopts a conformation in which the oxygen electron lone pairs are unfavorably positioned to be engaged in a hydrogen bond (Fig. 4D). Comparison of these LBP structures with that of the previously reported (22) RXRα LBD bound to the agonist SR11237 reveals some residue reorientations, the most significant one affecting L436 in H11 (Fig. 4E). To accommodate the water molecule present in the 2a and 2b complexes and/or the side chains of 2b and 2c, L436 rotates toward H12. However, in this conformation, the interaction distance between the Cδ atoms of L436 and L455 in H12 (3.38 Å, 3.46 Å, and 3.68 Å in the 2c, 2b, and 2a complexes) is significantly shorter than the sum of the van der Waals radii for two interacting methyl groups (3.84 Å) (23), thereby generating repulsive forces accounting for the destabilization of the holo conformation in solution. Interestingly, the differences observed for these interaction distances in the various complexes suggest that the steric constraints exerted by 2c on H12 are stronger than those imposed by 2b and 2a. Indeed, functional analyses show that 2c displays weaker agonist activity than 2b, which is itself less efficient than 2a (Fig. 2A). Together, these data reveal that the mixed activity of 2a relies on a water-mediated mechanism involving the repositioning L436 (H11), which, through a steric clash with L455 (H12), lowers the association strength between holo-H12 and the LBD surface. With 2b, 2c, and most likely 2d, a direct interaction with L436 accounts for the destabilization of H12, the effect being proportional to the chain length. Thus, L436 and more generally all residues that are in contact with holo-H12 and whose conformation can be affected by the bound ligand (i.e., L436 and W305 in RXRα) should be considered as target residues for the design of new NR modulators. With 2e–f, for which no crystals could be obtained, modeling studies indicate that their inhibitory effect results from an interference of their long side chains with L451 of H12 (SI Fig. 8).
Fig. 4.
Structures of RXRα LBD in complex with partial agonists. (A) Overall structure of RXRα LBD in complex with 2a, 2b, or 2c. The ligand is represented by red (oxygen atoms) and yellow (carbon atoms) van der Waals spheres. Helices and β-strands are numbered from N to C terminus. Together, helices H3, H4, and H12 define the activation function 2 (AF-2) surface to which the TIF2 NR2 peptide is bound. (B) 2Fo−Fc density (1σ) for the LBP of RXRα bound to 2b. W indicates a water molecule. (C) Closeup view showing the hydrogen bond network that stabilizes a water molecule in close proximity of L436. An identical hydrogen bond network is observed in the complex with 2a (not shown). (D) Superposition of the RXR LBP with 2b and 2c. (E) Comparison with the structure of RXRα LBD bound to SR11237 (Protein Data Bank ID code 1MVC). To accommodate the particular features of 2a–c, L436 must adopt a conformation that differs from that found in the presence of the agonist. The dashed line between L436 and L455 indicates a short distance.
UVI3003 as a Tool to Reveal RXR Function.
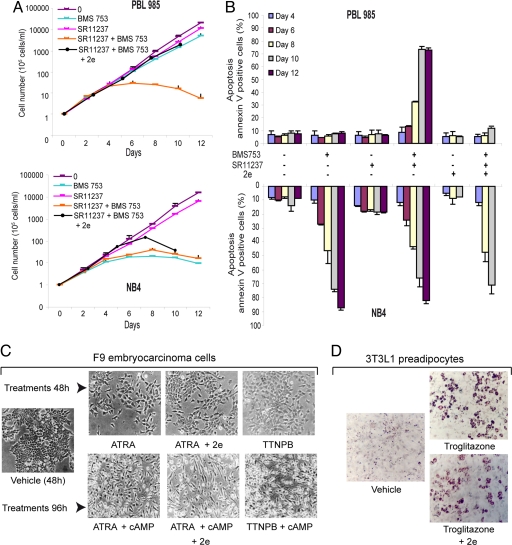
The availability of a high-affinity RXR-selective full antagonist provided the possibility to assess the contribution of RXR transactivation in the context of RXR heterodimers by pharmacological means. Comparing the responsiveness of promyelocytic NB4 cells expressing the PML-RARα oncofusion protein (24, 25) with that of the HL60 subclone PLB985 (26) revealed major differences in the growth and differentiative and apoptogenic response toward RAR and RXR-selective ligands. Indeed, NB4 cells cease proliferation (Fig. 5A), differentiate (not shown) and undergo apoptosis (Fig. 5B) in the presence of an RARα agonist (BMS753) alone, whereas PLB985 cells require in addition the presence of an RXR agonist (SR11237) (Fig. 5 A and B). The RXR antagonist 2e (UVI3003) fully confirmed these results, because its addition derepressed PLB985 growth inhibition by the combined action of RAR and RXR agonists but did not significantly affect growth inhibition or apoptosis in NB4 cells exposed to RAR agonists (not shown) or RAR and RXR agonists (Fig. 5 A and B). No similar critical effect of the RXR antagonist was seen when F9 embryo carcinoma cells were differentiated to primitive or parietal endodermal cells using all-trans retinoic acid (ATRA) or ATRA plus cAMP, respectively, or when 3T3L1 preadipocyte cells were differentiated by troglitazone (Fig. 5 C and D). Together, these data reveal UVI3003 (2e) as a tool to test the contribution of RXR to transactivation by a given RXR heterodimer. This is particularly important in cases where endogenous rexinoids (27) may contribute to a cell biological or physiological phenomenon, or when RXR is actively engaged in signaling (28, 29). The comparison of RXR responsiveness of PLB985 and NB4 cells suggests the challenging possibility that the PML-RARα fusion protein may have obliterated the RXR requirement for apoptosis, even though the leukemogenic species corresponds to higher-order heterooligomers composed of acute promyelocytic leukemia (APL) oncofusion proteins and RXR (30, 31). Indeed, that RARα ligands alone are sufficient to induce NB4 apoptosis may explain the efficiency of the retinoic acid therapy in APL patients. Moreover, the results obtained with PLB985 cells suggest that certain non-APL leukemias may benefit from the combined treatment with RAR and RXR agonists.
Fig. 5.
UVI3003 (2e) as tool to reveal the role of RXR in cell physiology. (A) Proliferation of PLB 985 or NB4 cells after exposure to the RARα-selective agonist BMS753 (1 μM) or the RXR agonist SR11237 (1 μM) alone or in combination with BSM753 or in combination with BMS753 and 2e (10 μM). (B) FACS quantification of apoptotic Annexin V-positive/propidium iodide-negative PLB 985 or NB4 cells after treatments. BMS753, SR11237, and 2e was used at 1, 1, and 10 μM, respectively. (C) Induction of differentiation of F9 cells into primitive endoderm (Upper) after exposure for 48 h to the RAR agonists TTNPB (10 nM) or ATRA (1 μM) alone or in combination with 2e (10 μM) or into parietal endoderm (Lower) after exposure for 96 h to 8CPT-cAMP (50 μM) and TTNPB (10 nM) or ATRA (1 μM), and 2e (10 μM). (D) Induction of differentiation of 3T3L1 cells after exposure to the PPAR agonist troglitazone alone or in combination with 2e (10 μM).
Concluding Remarks.
Despite the fact that RXR plays a major role as a promiscuous heterodimerization partner (7, 8), and in contrast to the related RAR for which multiple small-molecule effectors are available, a restricted repertoire of ligands exists for RXR. We have generated selective RXR modulators with distinct pharmacological profiles. Such ligands that are able to positively or negatively and in a cell-specific manner modulate RXR transcriptional activity and thus affect several signaling pathways not only have interesting therapeutic perspectives (5, 6) but also promise to be extremely useful in understanding broader RXR signaling networks and its functional implications with multiple partners. This is of particular interest in the context of aberrant signaling, as is the case for oncofusion protein–RXR complexes that cause APL (30). Importantly, several RXR-based therapeutic paradigms for leukemia have been discovered (refs. 28 and 29, and H.G., unpublished results), thus emphasizing the importance of tools to assess the impact of RXR in heterooligomers.
Using fluorescence spectroscopy, we have been able to quantify the impact of ligand binding on H12 motion. The combination of these dynamic data with the static snapshots obtained using x-ray crystallography has revealed the mechanism underlying the effect of ligands whose activity could not be explained simply by structural considerations. Indeed, although the structures of the complexes with 2a, 2b, and 2c display helix H12 locked in the same holo position, fluorescence anisotropy data show they differ from each other by the “lifetime” of their active conformation in solution. Interestingly, crystal structures reveal that these ligands impair H12 mobility indirectly, by modifying the conformation of a residue (L436), which is involved in a stabilizing interaction with holo-H12. Because 2a, 2b, and 2c induce slightly different L436 conformations, their impact on holo-H12 stability is different, thereby providing a rational basis for the distinct functional outcome of these mixed agonists/antagonists. We have presented a comprehensive model for the molecular mechanism of partial agonists that combines structural and dynamic concepts. These mechanistic insights gained using RXR as a model of NR/ligand interaction likely correspond to conserved mechanisms that, we believe, will aid in the design of selective NR modulators with varying degrees of agonist or antagonist activities and the promise to separate therapeutically desired from unwanted toxic effects.
Materials and Methods
Chemistry.
Experimental procedures for the synthesis of all new compounds are available in SI Text.
Transfections and Determination of RXR Activity.
Transient transfections and two-hybrid assays were performed as described (32, 33). HeLa cells were established according to Chen et al. (34) and were used as described in SI Text.
Preparation of the RARα-RXRα LBD Heterodimer for Fluorescence Anisotropy Experiments.
Human RXRα LBD (residues 223–462) was expressed in Escherichia coli BL21(DE3) as a fusion with an inducible self-splicing intein (Sce VMA) and a chitin-binding domain using the vector pTYB1 (New England Biolabs, Ipswich, MA). Labeling was performed as described (18, 19). The histidine-tagged LBD of human RARα (residues 176–421 in a pET15b vector) was expressed and purified as described for RXRα LBD in SI Text. Fractions containing RARα LBD were pooled, dialyzed against buffer B (10 mM Tris·HCl, pH 8.0/150 mM NaCl/5 mM DTT/1 mM EDTA/10% glycerol), and incubated overnight with the fluorescein-labeled RXRα LBD. The RARα-RXRα heterodimer was further purified by using a superdex 75 26/60 gel filtration column (Amersham Biosciences, Piscataway, NJ). Fractions containing the purified heterodimer were pooled and concentrated to 117 μM. The labeled-heterodimer concentration was estimated to 1.12 μM by using the characteristic maximum absorption wavelength (λ = 490 nm) of fluorescein.
Fluorescence Anisotropy Measurements.
Steady-state fluorescence anisotropy assays were performed with a BEACON 2000 polarization instrument (Panvera, Madison, WI) regulated at 4°C. The labeled-heterodimer concentration was set to 0.725 nM by dilution with buffer B. The excitation wavelength was 490 nm, with emission measured at 530 nm. The TIF2 NR2 coactivator peptide (686-KHKILHRLLQDSS-698) was added to protein samples containing 20 μM of ligand to a final concentration of 10 μM, and then the sample was diluted successively with buffer B supplemented with 0.725 nM of heterodimer and 20 μM of ligand. At least three independent measurements were made for each sample. Time-resolved fluorescence anisotropy data were obtained as described in SI Text.
Protein Expression, Purification, and Crystallization.
Human RXRα LBD was expressed and purified as described in SI Text. Before crystallization, the protein was mixed with a 3-fold molar excess of ligand 2a, 2b, or 2c and a 5-fold molar excess of TIF2 NR2 peptide (686-KHKILHRLLQDSS-698). The complexes were incubated overnight at 4°C and concentrated to ≈15 mg·ml−1. Crystals were obtained by vapor diffusion at 20°C. The well buffers contained 16% PEG 3350; 0.1 M Tris·HCl, pH 8.5; 1 M ammonium acetate (2a); 14% PEG 10000; 0.1 M Tris·HCl, pH 8.5; 1 M ammonium chloride (2b); or 24% PEG 4000; 0.1 M Tris·HCl, pH 7.5 (2c). Crystals were of space group P43212.
Data Collection and Structure Solution.
The protein crystals were mounted from mother liquor onto a cryoloop (Hampton Research, Aliso Viejo, CA), soaked in the reservoir solution containing an additional 20% glycerol, and quickly frozen in liquid nitrogen. Diffraction data were collected by using an ADSC (Poway, CA) Quantum 4 detector at the ID14-EH2 beamline of European Synchrotron Radiation Facility (France) for the complex with 2a (1.8-Å resolution) and at the ID14-EH4 beamline of ESRF for complexes with 2b (2.2-Å resolution) and 2c (2.2-Å resolution). Diffraction data were processed by using MOSFLM (35) and scaled with SCALA from the CCP4 program suite (36). The structures were solved by using the previously reported structure 1MVC (22) of which the ligand and the coactivator peptide were omitted. Initial Fo−Fc difference maps had strong signals for the ligands and the TIF2 NR2 peptide, which could be fitted accurately into electron densities. The structures were modeled with O (37) and refined with REFMAC5 (36) by using rigid-body, least-squares, and individual B-factor refinements. The final models exhibit very good geometry with 94.9% (2a), 95.4% (2b), and 94.9% (2c) of the residues in the most-favored regions of the Ramachandran plot and no residue in the disallowed regions. Experimental electron density Fo−Fc maps of all ligands, calculated by using the refined model with the ligands omitted, are shown in SI Fig. 9.
Cell Culture and Analysis of Apoptosis.
F9 cells, grown in tissue culture plates coated with 0.1% gelatin, and preadipogenic 3T3L1 cells were maintained in DMEM supplemented with 10% FCS and 1 mg·liter−1 gentamicin and 2 mM glutamine. For F9 cells, collagen IV expression was monitored by semiquantitative RT-PCR (SI Fig. 10 and SI Text). PLB 985 and NB4 leukemia cell lines were cultured in RPMI medium 1640 supplemented with 10% FCS/2 mM glutamine/25 mM Hepes buffer/40 μg·ml−1 gentamycin in a humidified incubator at 37°C and 5% CO2. Apoptosis of leukemia cells was quantified by propidium iodide-annexin V double staining on a FACScan (Becton Dickinson, Franklin Lakes, NJ) flow cytometer.
Supplementary Material
Acknowledgments
We thank Jean-François Guichou, Yvan Boublik (Centre de Biochimie Structurale, Montpellier, France), and Emmanuel Margeat (Centre de Biochimie Structurale, Montpellier, France) for help with fluorescence experiments. We acknowledge the technical assistance of the beamline managers at the European Synchrotron Radiation Facility (Grenoble, France). We thank Michelle Lieb and Catherine Huck (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France) for their contribution to the NB4 and PLB985 experiments and Claudine Gaudon and Audrey Bindler (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France) for the RXR reporter cells and assistance in the analysis. We thank Galderma (Sophia-Antipolis, France) for providing the CD3254. This work was supported by funds from the European Community [QLK-2002-02029 “Anticancer Retinoids” and LSHC-CT-2005-518417 “EPITRON” (to A.R.d.L. and H.G.), the Spanish Ministerio de Educación y Ciencia (SAF04-07131, FEDER, to A.R.d.L.); and Juan de la Cierva Contract (to F.R.-B.)], the Institut National du Cancer (H.G.), and the Ligue contre le Cancer (“Equipe labellisée la Ligue”) (H.G.).
Abbreviations
- RXR
retinoid X receptor
- LBD
ligand-binding domain
- NR
nuclear receptor
- TIF2 NR2
transcriptional intermediary factor 2 NR box 2
- ATRA
all-trans retinoic acid
- APL
acute promyelocytic leukemia
- RAR
retinoic acid receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2P1T, 2P1U, and 2P1V).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705356104/DC1.
References
- 1.Perissi V, Rosenfeld MG. Nat Rev Mol Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- 2.Nagy L, Schwabe JW. Trends Biochem Sci. 2004;29:317–324. doi: 10.1016/j.tibs.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Renaud JP, Moras D. Cell Mol Life Sci. 2000;57:1748–1769. doi: 10.1007/PL00000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourguet W, Germain P, Gronemeyer H. Trends Pharmacol Sci. 2000;21:381–388. doi: 10.1016/s0165-6147(00)01548-0. [DOI] [PubMed] [Google Scholar]
- 5.Liby KT, Yore MM, Sporn MB. Nat Rev Cancer. 2007;7:357–369. doi: 10.1038/nrc2129. [DOI] [PubMed] [Google Scholar]
- 6.Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 7.Germain P, Chambon P, Eichele G, Evans RM, Lazar MA, Leid M, De Lera AR, Lotan R, Mangelsdorf DJ, Gronemeyer H. Pharmacol Rev. 2006;58:760–772. doi: 10.1124/pr.58.4.7. [DOI] [PubMed] [Google Scholar]
- 8.Mark M, Ghyselinck NB, Chambon P. Annu Rev Pharmacol Toxicol. 2006;46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- 9.Dawson MI. Curr Med Chem Anticancer Agents. 2004;4:199–230. doi: 10.2174/1568011043352975. [DOI] [PubMed] [Google Scholar]
- 10.Kagechika H, Shudo K. J Med Chem. 2005;48:5875–5883. doi: 10.1021/jm0581821. [DOI] [PubMed] [Google Scholar]
- 11.de Lera AR, Bourguet W, Altucci L, Gronemeyer H. Nat Rev Drug Discov. 2007;6:811–820. doi: 10.1038/nrd2398. [DOI] [PubMed] [Google Scholar]
- 12.Lala DS, Mukherjee R, Schulman IG, Koch SS, Dardashti LJ, Nadzan AM, Croston GE, Evans RM, Heyman RA. Nature. 1996;383:450–453. doi: 10.1038/383450a0. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi B, Ohta K, Kawachi E, Fukasawa H, Hashimoto Y, Kagechika H. J Med Chem. 2002;45:3327–3330. doi: 10.1021/jm0255320. [DOI] [PubMed] [Google Scholar]
- 14.Cavasotto CN, Liu G, James SY, Hobbs PD, Peterson VJ, Bhattacharya AA, Kolluri SK, Zhang XK, Leid M, Abagyan R, et al. J Med Chem. 2004;47:4360–4372. doi: 10.1021/jm030651g. [DOI] [PubMed] [Google Scholar]
- 15.Stebbins JL, Jung D, Leone M, Zhang XK, Pellecchia M. J Biol Chem. 2006;281:16643–16648. doi: 10.1074/jbc.M600318200. [DOI] [PubMed] [Google Scholar]
- 16.Ohta K, Iijima T, Kawachi E, Kagechika H, Endo Y. Bioorg Med Chem Lett. 2004;14:5913–5918. doi: 10.1016/j.bmcl.2004.09.035. [DOI] [PubMed] [Google Scholar]
- 17.Bernardon JM. Int Patent Appl. 1997 WO9733881. [Google Scholar]
- 18.Chong S, Mersha FB, Comb DG, Scott ME, Landry D, Vence LM, Perler FB, Benner J, Kucera RB, Hirvonen CA, et al. Gene. 1997;192:271–281. doi: 10.1016/s0378-1119(97)00105-4. [DOI] [PubMed] [Google Scholar]
- 19.Kallenberger BC, Love JD, Chatterjee VK, Schwabe JW. Nat Struct Biol. 2003;10:136–140. doi: 10.1038/nsb892. [DOI] [PubMed] [Google Scholar]
- 20.Voegel JJ, Heine MJ, Tini M, Vivat V, Chambon P, Gronemeyer H. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith CL, O'Malley BW. Endocr Rev. 2004;25:45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 22.Egea PF, Mitschler A, Moras D. Mol Endocrinol. 2002;16:987–997. doi: 10.1210/mend.16.5.0823. [DOI] [PubMed] [Google Scholar]
- 23.Li AJ, Nussinov R. Proteins. 1998;32:111–127. [PubMed] [Google Scholar]
- 24.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 25.Kakizuka A, Miller WH, Jr, Umesono K, Warrell RP, Jr, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 26.Yin W, Rossin A, Clifford JL, Gronemeyer H. Oncogene. 2006;25:3735–3744. doi: 10.1038/sj.onc.1209410. [DOI] [PubMed] [Google Scholar]
- 27.de Urquiza AM, Liu S, Sjoberg M, Zetterstrom RH, Griffiths W, Sjovall J, Perlmann T. Science. 2000;290:2140–2144. doi: 10.1126/science.290.5499.2140. [DOI] [PubMed] [Google Scholar]
- 28.Benoit GR, Flexor M, Besancon F, Altucci L, Rossin A, Hillion J, Balajthy Z, Legres L, Segal-Bendirdjian E, Gronemeyer H, Lanotte M. Mol Endocrinol. 2001;15:1154–1169. doi: 10.1210/mend.15.7.0654. [DOI] [PubMed] [Google Scholar]
- 29.Altucci L, Rossin A, Hirsch O, Nebbioso A, Vitoux D, Wilhelm E, Guidez F, De Simone M, Schiavone EM, Grimwade D, et al. Cancer Res. 2005;65:8754–8765. doi: 10.1158/0008-5472.CAN-04-3569. [DOI] [PubMed] [Google Scholar]
- 30.Zeisig BB, Kwok C, Zelent A, Shankaranarayanan P, Gronemeyer H, Dong S, So CW. Cancer Cell. 2007;12:36–51. doi: 10.1016/j.ccr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Zhu J, Nasr R, Peres L, Riaucoux-Lormiere F, Honore N, Berthier C, Kamashev D, Zhou J, Vitoux D, Lavau C, et al. Cancer Cell. 2007;12:23–35. doi: 10.1016/j.ccr.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 32.Vivat V, Zechel C, Wurtz JM, Bourguet W, Kagechika H, Umemiya H, Shudo K, Moras D, Gronemeyer H, Chambon P. EMBO J. 1997;16:5697–5709. doi: 10.1093/emboj/16.18.5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Germain P, Iyer J, Zechel C, Gronemeyer H. Nature. 2002;415:187–192. doi: 10.1038/415187a. [DOI] [PubMed] [Google Scholar]
- 34.Chen JY, Penco S, Ostrowski J, Balaguer P, Pons M, Starrett JE, Reczek P, Chambon P, Gronemeyer H. EMBO J. 1995;14:1187–1197. doi: 10.1002/j.1460-2075.1995.tb07102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leslie AGW. Newsl Protein Crystallogr. 1992;26:27–33. [Google Scholar]
- 36.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.