Abstract
The genes coding for the lactose permease and beta-galactosidase, two proteins involved in the metabolism of lactose by Lactobacillus bulgaricus, have been cloned, expressed, and found functional in Escherichia coli. The nucleotide sequences of these genes and their flanking regions have been determined, showing the presence of two contiguous open reading frames (ORFs). One of these ORFs codes for the lactose permease gene, and the other codes for the beta-galactosidase gene. The lactose permease gene is located in front of the beta-galactosidase gene, with 3 bp in the intergenic region. The two genes are probably transcribed as one operon. Primer extension studies have mapped a promoter upstream from the lactose permease gene but not the beta-galactosidase gene. This promoter is similar to those found in E. coli with general characteristics of GC-rich organisms. In addition, the sequences around the promoter contain a significantly higher number of AT base pairs (80%) than does the overall L. bulgaricus genome, which is rich in GC (GC content of 54%). The amino acid sequences obtained from translation of the ORFs are found to be highly homologous (similarity of 75%) to those from Streptococcus thermophilus. The first 460 amino acids of the lactose permease shows homology to the melibiose transport protein of E. coli. Little homology was found between the lactose permease of L. bulgaricus and E. coli, but the residues which are involved in the binding and the transport of lactose are conserved. The carboxy terminus is similar to that of the enzyme III of several phosphoenolpyruvate-dependent phosphotransferase systems.
Full text
PDF

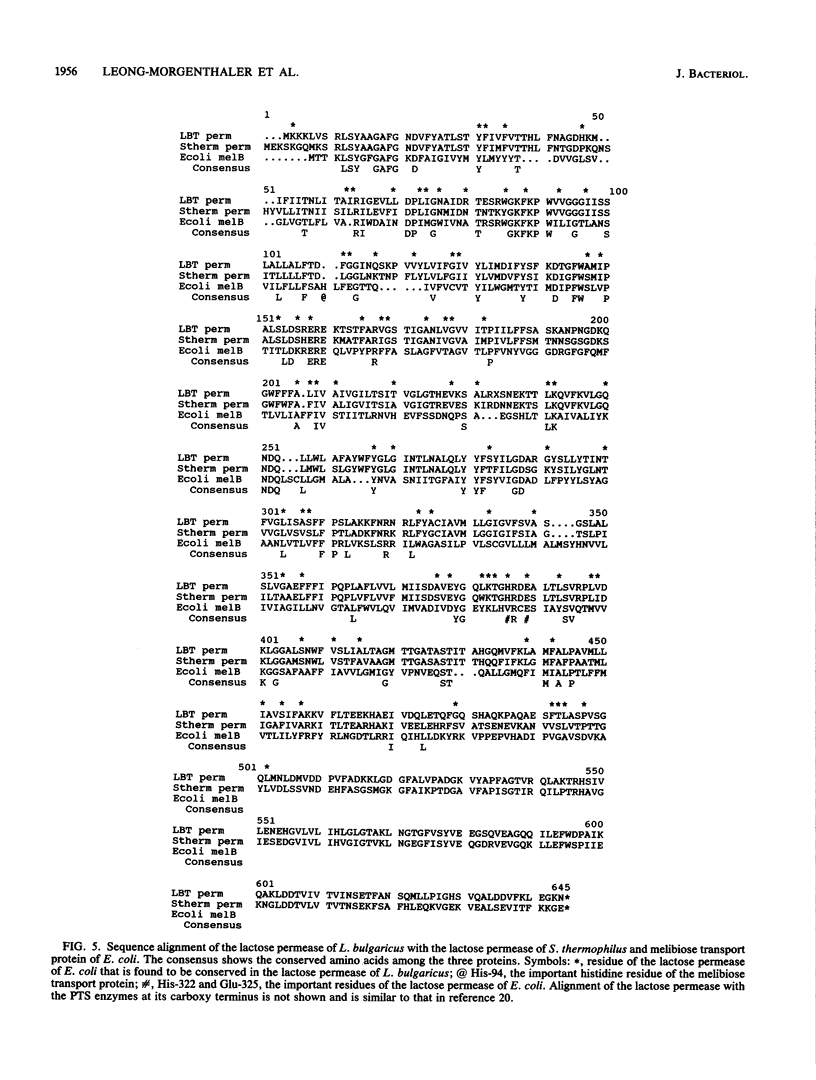

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brooker R. J. Characterization of the double mutant, Val-177/Asn-322, of the lactose permease. J Biol Chem. 1990 Mar 5;265(7):4155–4160. [PubMed] [Google Scholar]
- Carrasco N., Püttner I. B., Antes L. M., Lee J. A., Larigan J. D., Lolkema J. S., Roepe P. D., Kaback H. R. Characterization of site-directed mutants in the lac permease of Escherichia coli. 2. Glutamate-325 replacements. Biochemistry. 1989 Mar 21;28(6):2533–2539. doi: 10.1021/bi00432a028. [DOI] [PubMed] [Google Scholar]
- Cupples C. G., Miller J. H., Huber R. E. Determination of the roles of Glu-461 in beta-galactosidase (Escherichia coli) using site-specific mutagenesis. J Biol Chem. 1990 Apr 5;265(10):5512–5518. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M. Digestion of DNA: size fractionation. Methods Enzymol. 1987;152:183–189. doi: 10.1016/0076-6879(87)52019-5. [DOI] [PubMed] [Google Scholar]
- Hickey M. W., Hillier A. J., Jago G. R. Transport and metabolism of lactose, glucose, and galactose in homofermentative lactobacilli. Appl Environ Microbiol. 1986 Apr;51(4):825–831. doi: 10.1128/aem.51.4.825-831.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottinger H., Ohgi T., Zwahlen M. C., Dhamija S., Söll D. Allele-specific complementation of an Escherichia coli leuB mutation by a Lactobacillus bulgaricus tRNA gene. Gene. 1987;60(1):75–83. doi: 10.1016/0378-1119(87)90215-0. [DOI] [PubMed] [Google Scholar]
- Jacquet M. A., Ehrlich R., Reiss C. In vivo gene expression directed by synthetic promoter constructions restricted to the -10 and -35 consensus hexamers of E. coli. Nucleic Acids Res. 1989 Apr 25;17(8):2933–2945. doi: 10.1093/nar/17.8.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback H. R. Site-directed mutagenesis and ion-gradient driven active transport: on the path of the proton. Annu Rev Physiol. 1988;50:243–256. doi: 10.1146/annurev.ph.50.030188.001331. [DOI] [PubMed] [Google Scholar]
- Menick D. R., Carrasco N., Antes L., Patel L., Kaback H. R. lac permease of Escherichia coli: arginine-302 as a component of the postulated proton relay. Biochemistry. 1987 Oct 20;26(21):6638–6644. doi: 10.1021/bi00395a012. [DOI] [PubMed] [Google Scholar]
- Nakayama M., Fujita N., Ohama T., Osawa S., Ishihama A. Micrococcus luteus, a bacterium with a high genomic G + C content, contains Escherichia coli-type promoters. Mol Gen Genet. 1989 Sep;218(3):384–389. doi: 10.1007/BF00332399. [DOI] [PubMed] [Google Scholar]
- Nelson S. O., Schuitema A. R., Benne R., van der Ploeg L. H., Plijter J. S., Aan F., Postma P. W. Molecular cloning, sequencing, and expression of the crr gene: the structural gene for IIIGlc of the bacterial PEP:glucose phosphotransferase system. EMBO J. 1984 Jul;3(7):1587–1593. doi: 10.1002/j.1460-2075.1984.tb02015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill M. C. Escherichia coli promoters. I. Consensus as it relates to spacing class, specificity, repeat substructure, and three-dimensional organization. J Biol Chem. 1989 Apr 5;264(10):5522–5530. [PubMed] [Google Scholar]
- Osumi T., Saier M. H., Jr Regulation of lactose permease activity by the phosphoenolpyruvate:sugar phosphotransferase system: evidence for direct binding of the glucose-specific enzyme III to the lactose permease. Proc Natl Acad Sci U S A. 1982 Mar;79(5):1457–1461. doi: 10.1073/pnas.79.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri K. G., Waygood E. B. Sequence of cloned enzyme IIN-acetylglucosamine of the phosphoenolpyruvate:N-acetylglucosamine phosphotransferase system of Escherichia coli. Biochemistry. 1988 Aug 9;27(16):6054–6061. doi: 10.1021/bi00416a034. [DOI] [PubMed] [Google Scholar]
- Poolman B., Royer T. J., Mainzer S. E., Schmidt B. F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989 Jan;171(1):244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postma P. W., Lengeler J. W. Phosphoenolpyruvate:carbohydrate phosphotransferase system of bacteria. Microbiol Rev. 1985 Sep;49(3):232–269. doi: 10.1128/mr.49.3.232-269.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourcher T., Sarkar H. K., Bassilana M., Kaback H. R., Leblanc G. Histidine-94 is the only important histidine residue in the melibiose permease of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jan;87(1):468–472. doi: 10.1073/pnas.87.1.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premi L., Sandine W. E., Elliker P. R. Lactose-hydrolyzing enzymes of Lactobacillus species. Appl Microbiol. 1972 Jul;24(1):51–57. doi: 10.1128/am.24.1.51-57.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Püttner I. B., Sarkar H. K., Padan E., Lolkema J. S., Kaback H. R. Characterization of site-directed mutants in the lac permease of Escherichia coli. 1. Replacement of histidine residues. Biochemistry. 1989 Mar 21;28(6):2525–2533. doi: 10.1021/bi00432a027. [DOI] [PubMed] [Google Scholar]
- Rogers M. J., Ohgi T., Plumbridge J., Söll D. Nucleotide sequences of the Escherichia coli nagE and nagB genes: the structural genes for the N-acetylglucosamine transport protein of the bacterial phosphoenolpyruvate: sugar phosphotransferase system and for glucosamine-6-phosphate deaminase. Gene. 1988;62(2):197–207. doi: 10.1016/0378-1119(88)90558-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B. F., Adams R. M., Requadt C., Power S., Mainzer S. E. Expression and nucleotide sequence of the Lactobacillus bulgaricus beta-galactosidase gene cloned in Escherichia coli. J Bacteriol. 1989 Feb;171(2):625–635. doi: 10.1128/jb.171.2.625-635.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J. H., Clewell D. B. Complete nucleotide sequence of macrolide-lincosamide-streptogramin B-resistance transposon Tn917 in Streptococcus faecalis. J Bacteriol. 1985 Nov;164(2):782–796. doi: 10.1128/jb.164.2.782-796.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. Lactic acid bacteria: model systems for in vivo studies of sugar transport and metabolism in gram-positive organisms. Biochimie. 1988 Mar;70(3):325–336. doi: 10.1016/0300-9084(88)90205-2. [DOI] [PubMed] [Google Scholar]
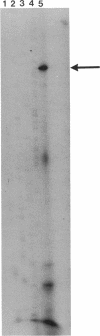
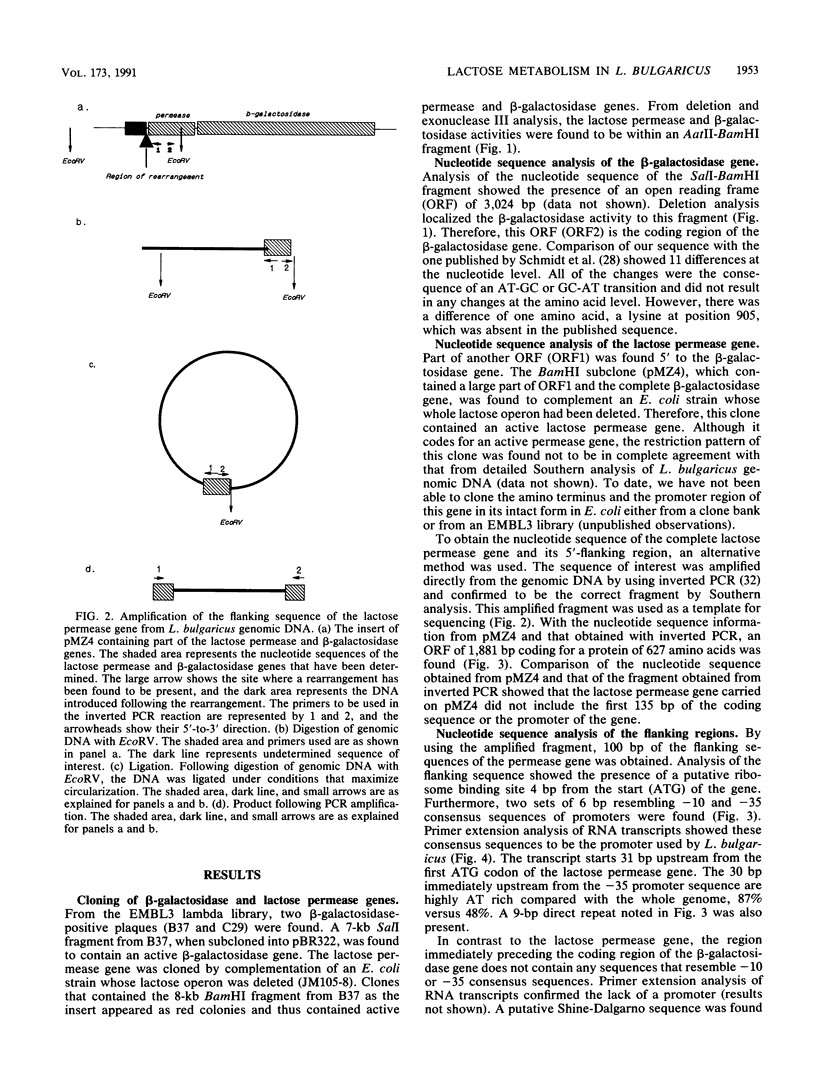
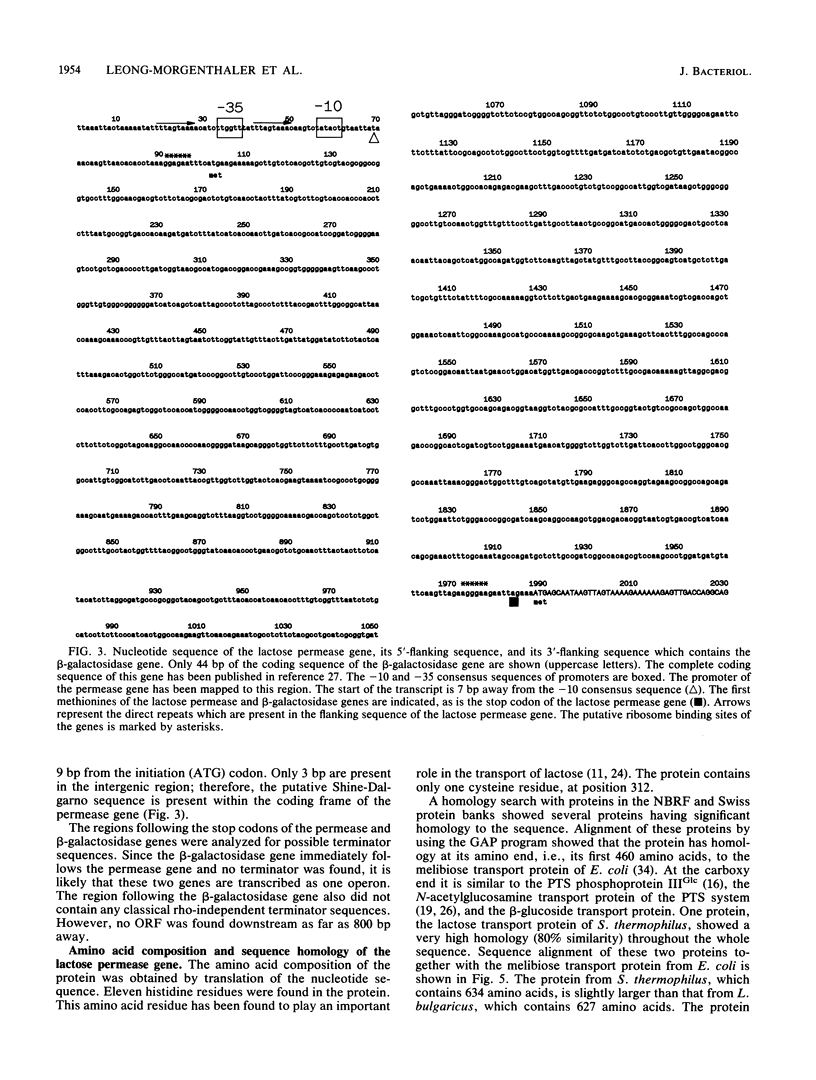
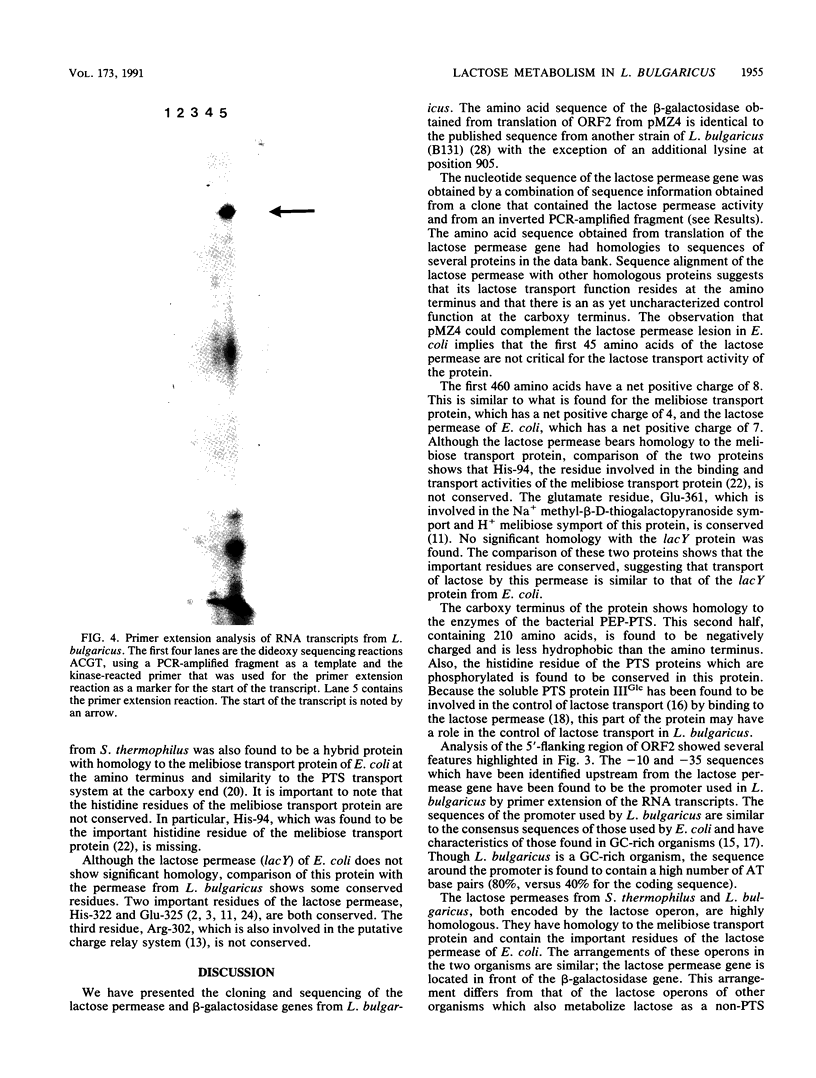
- Triglia T., Peterson M. G., Kemp D. J. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988 Aug 25;16(16):8186–8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yazyu H., Shiota-Niiya S., Shimamoto T., Kanazawa H., Futai M., Tsuchiya T. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J Biol Chem. 1984 Apr 10;259(7):4320–4326. [PubMed] [Google Scholar]