Abstract
RepA, the replication initiator protein of Pseudomonas pPS10 plasmid, is made of two winged-helix (WH) domains. RepA dimers undergo a structural transformation upon binding to origin DNA sequences (iterons), resulting in monomerization and α-helix into β-strand conversion. This affects the N-terminal domain (WH1) and generates a metastable intermediate. Here it is shown that the interaction of short dsDNA oligonucleotides, including iteron or operator RepA targets, with the isolated WH1 domain promotes the assembly of different nanostructures. These range from irregular aggregates to amyloid spheroids and fibers. Their intrinsic order inversely correlates with the extent of the transformation induced by each DNA sequence on RepA. However, DNA is not a constituent of the assembled fibers, in agreement with the protein-only principle for amyloid structure. Thus, the RepA-WH1 domain on DNA binding mimics the behavior of the mammalian prion protein. The stretch of amino acids responsible for WH1 aggregation has been identified, leading to the design of mutants with enhanced or reduced amyloidogenicity and the synthesis of a peptide that assembles into a cross-β structure. RepA amyloid assemblies could have a role in the negative regulation of plasmid replication. This article underlines the potential of specific nucleic acid sequences in promoting protein amyloidogenesis at nearly physiological conditions.
Keywords: amyloidogenesis, conformational changes, DNA binding, fiber assembly, RepA protein
The formation of protein assemblies with amyloid properties is a subject of intense research because of their involvement in severe human diseases (1, 2). An unsolved issue is the nature of the signal(s) responsible for amyloidogenesis, which includes proteolytic processing of a functional precursor and mutations, and also hyperphosphorylation (3) or the binding to elements of the cell envelope (4) or nucleic acids (5–8). The major (if not the only) component of any mature amyloid assembly is protein (9, 10). The studies of amyloidogenesis have been extended to model systems based in proteins not involved in any disease (1).
Our current understanding of protein aggregation into amyloids suggests that some partially unfolded intermediates that populate the funnel-shaped folding energy landscape of virtually any protein aggregate into a stable, low-energy conformation distinct from the protein native state (1, 2). Supramolecular assembly is a common functional resource for proteins by using an ample repertoire of protein–protein interfaces. However, the family of structures responsible for the unique staining (11) and diffraction (12) properties of any protein amyloid is known as cross-β: a β-sheet of indefinite length is formed by the stacking, orthogonal to its axis, of β-strands from each protein monomer (13). Amyloidogenesis often includes the refolding as β-strands of protein regions that, although compatible with such secondary structure, are found as α-helices or coils in the native structure (14–18). In other cases, nearly native protein folds contribute with short sequence stretches that, gaining self-interaction, assemble into fibrils (13).
RepA (230 amino acid residues) is the replication protein of the Pseudomonas pPS10 plasmid (reviewed in ref. 19). RepA dimers are repressors of repA transcription through binding to an inversely repeated operator sequence, whereas RepA monomers are proficient in recognition of directly repeated sequences (iterons) found at the origin of replication. In common with many amyloidogenic proteins, RepA undergoes an α-helix into β-sheet conversion, which affects a three-helix bundle in its N-terminal winged-helix dimerization domain (WH1; RepA residues 1–132) (20). Such a conformational change requires the dissociation of RepA dimers and the activation of WH1 as a second DNA binding interface (subsidiary to the C-terminal WH2 domain) (21). Binding of a RepA dimer to a single iteron is sufficient to induce both protein dissociation and the coupled structural transformation, whereas binding to the operator has no major effect on the structure of dimers (22). The conversion of RepA-WH1 dimers into monomers implies partial unfolding of the protein subunits and an increase in their solvent-accessible surface (19), resulting in a transient metastable intermediate prone to aggregation (22, 23). Normally isolated WH1 dimers are thermostable and proteolytically resistant (21). This complex behavior of RepA has been extensively characterized by CD and fluorescence spectroscopies (21–23), hydrodynamics (22), x-ray crystallography (20), and modeling (19).
Here I describe the identification of a protein stretch in RepA-WH1 with the potential to aggregate through β-strand formation. Mutations in a single residue of the motif were introduced, leading to enhanced or reduced WH1 aggregation. Combining these WH1 mutants with binding to short dsDNA oligonucleotides including the natural targets for the domain when present in full-length RepA (22), it is possible to modulate the formation of assemblies with diverse sizes and shapes (irregular, spheroidal, and fibrillar) exhibiting typical amyloid properties (11). A synthetic peptide including the optimized amyloidogenic motif in WH1 assembles into fibers with the characteristic biophysical features of an amyloid cross-β structure. The extent to which each DNA sequence transforms the RepA structure (22, 23) inversely correlates with the order of the resulting WH1 amyloid assemblies. However, DNA is not part of the fibers, thus acting as a real inducer, rather than as a structural component, of the aggregates. The results of this study provide important insights into a way to modulate the assembly of distinct protein amyloid structures by interaction with short, defined DNA sequences.
Results
A Protein Sequence Stretch Potentially Involved in RepA Aggregation.
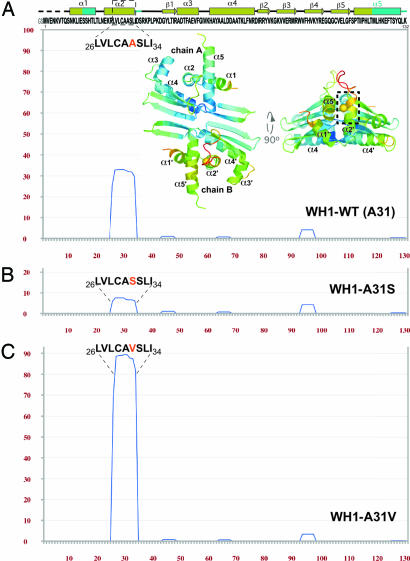
Although the dimeric RepA-WH1 domain is soluble to millimolar concentrations (20, 21), its titration in the micromolar range with 11-bp oligonucleotides comprising the two distinct targets for WH1, either the spacer between the two 8-bp half repeats constituting the repA operator (opsp) or the 5′ half of a 22-bp iteron repeat (5′iter) (19), resulted in massive protein precipitation (data not shown). Thus, a search for sequence stretches in WH1 prone to aggregation was carried out by means of TANGO (http://tango.crg.es), an algorithm designed to detect protein regions with a propensity to aggregate through β-sheets (17). The sequence between residues 26 and 34 (LVLCAASLI) shows a net tendency toward β-aggregation (Fig. 1A), similar to that found in typical amyloid proteins (15, 17). Those residues map to the C terminus of helix α2, one of the elements in WH1 undergoing structural transformation to a β-strand. This region and the following loop make up the most flexible portion (highest B-factors) in its crystal structure (Protein Data Bank ID code 1HKQ) (Fig. 1A) (19, 20).
Fig. 1.
A sequence stretch in RepA-WH1 has a potential for aggregation. (A) Plot of the TANGO β-aggregation score (17) for the sequence of WH1 domain (pH 8). Its secondary structure (elements affected by conformational changes in cyan) and two orthogonal views (PyMOL, www.pymol.org) of the crystal structure of the dimer (20) are shown. Rainbow coloring of the model reflects the B-factors distribution (bluish lower and reddish higher). The side chain of Ala-31 (highlighted in orange) is shown as a CPK sphere (Cβ atom). Dashed boxes mark residues 26–34. The TANGO prediction plots for the WH1-A31S (B) and A31V (C) mutants are also displayed.
To test this prediction, two different mutants were constructed by substituting Ala-31 in WH1: A31S (Fig. 1B), designed to decrease its aggregation tendency, and A31V (Fig. 1C), which would increase the aggregation potential of WH1 to the levels found, e.g., in the Alzheimer's β-amyloid peptide (15, 17). All WH1 proteins were then purified, their average secondary structure [supporting information (SI) Fig. 6A] and thermal stability (SI Fig. 6A Inset) were estimated by CD spectroscopy, and their association state was determined by gel filtration analysis (SI Fig. 6B). None of the mutants appeared to have its biophysical properties altered (21): they are dimers with a nearly identical average secondary structure, remaining essentially folded even at 90°C.
Combining Single WH1 Mutations with Binding to Discrete DNA Sequences Results in Distinct Nanoassemblies.
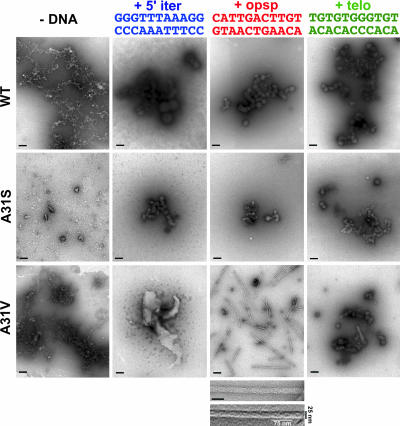
A search for conditions influencing aggregation in vitro was then carried out with the three WH1 domain variants (WT, A31S/V). Proteins were incubated with equimolar concentrations of the specific 11-bp dsDNA oligonucleotides mentioned above, or a nonspecific sequence of the same length (telo), related to one previously tested with full-length RepA (22). Aggregation was explored by means of transmission electron microscopy (TEM) on negatively stained samples (Fig. 2). After a month of incubation in the absence of any DNA, heterogeneous reticular aggregates were evident for both WT and A31V mutant domains, whereas simpler aggregated particles were present in the A31S mutant. When the incubation was performed with the 5′iter dsDNA, large spheroids (up to 200 nm in diameter, Ø) appeared in WH1-WT, with smaller spheroids (≈50 nm Ø) in WH1-A31S. The aggregates in WH1-A31V were much larger and had an irregular shape for A31V. With the opsp DNA sequence, the WT and A31S domains gave smaller spheroids than those with 5′iter. However, WH1-A31V in the presence of opsp DNA mainly assembled into straight, unbranched ≈25-nm-wide fibers. These are composed of two identical coiled fibrils with a variable pitch that, in the segments most tightly twisted, span ≈75 nm. With the nonspecific dsDNA (telo) all three proteins produced small spheroids. The pronounced lag in the appearance of the fibers suggests a nucleation-dependent process (1, 13): after the first week of incubation, small irregular particles give way to short fibrils, which, in 2–4 weeks, grow into the fibers displayed in Fig. 2. Longer incubations (2–6 months) increase the length of the fibers and result in their bundling into thick filaments (data not shown). In conclusion, these experiments show that certain DNA sequences have the ability to induce the formation of nanoassemblies by WH1.
Fig. 2.
Electron micrographs (negative stain) of purified RepA-WH1 proteins (rows) incubated with equimolar amounts (35 μM) of 11-bp dsDNA oligonucleotides (columns) spanning the sequences naturally bound by the domain as a dimer (opsp) or as monomer (5′iter) or a nonspecific sequence (telo). Color coding for the DNAs is the same as used in refs. 22 and 23 and in Fig. 4. Magnification bars in each field span 200 nm. Below the WH1-A31V plus opsp field is shown a close-up view of a fiber (scale bar: 50 nm) above the result of embossing and filtering (Gaussian) the same image to highlight its two component fibrils.
DNA Is Not a Component of WH1-A31V Fibrillar Assemblies.
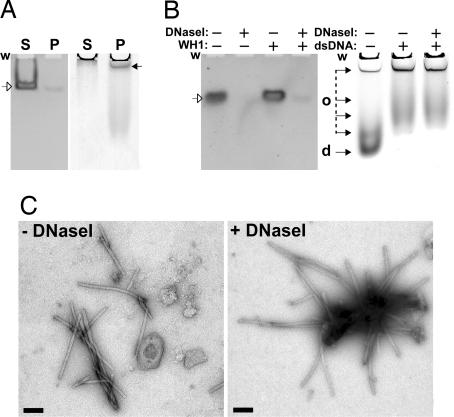
Because the binding of each of the individual RepA-WH domains to their target sequences has very low affinity and is expected to be transient (21, 23), the fibrillar assemblies were analyzed to establish whether they contained dsDNA. First, ultracentrifugation of mature fiber preparations containing equimolar concentrations of opsp dsDNA and WH1-A31V yielded DNA essentially in the soluble fraction (Fig. 3A Left) whereas most of the aggregated protein was in the pellet (Fig. 3A Right). This result indicates that DNA is excluded from the protein fibers. Second, electrophoretic analysis of duplicate samples in native gels showed no retarded (or retained in the well) dsDNA band in the presence of WH1-A31V (Fig. 3B, compare first and third lanes). This is consistent with a transient/unstable protein–DNA interaction. Third, extensive digestion of mature fiber preparations with DNaseI efficiently degraded most of opsp DNA, reflecting the inability of WH1-A31V to protect DNA against the nuclease (Fig. 3B Left, fourth lane). On the contrary, according to both native protein gel electrophoresis (Fig. 3B Right) and TEM (Fig. 3C), the fibers were unaltered by the enzymatic removal of DNA, strongly indicating that WH1 is their only macromolecular component. Incubation of WH1-A31V with each of the individual opsp DNA strands led to large spherical aggregates (data not shown). When fresh WH1-A31V, with no dsDNA added, was seeded (1/500 to 1/50, vol/vol) with sonicated (10) WH1-A31V plus opsp fibers, the resulting aggregates were very large and irregular, instead of fibrillar (data not shown). These results, together with those from the previous section, point to dsDNA as inducer of RepA-WH1 self-assembly.
Fig. 3.
The dsDNA oligonucleotide opsp is not part of the WH1-A31V fibers that it promotes. (A) Nondenaturing polyacrylamide gels showing the supernatant (S) and pellet (P) fractions after ultracentrifugation of mature samples including WH1-A31V and opsp dsDNA. (Left) Ethidium bromide staining of dsDNA. (Right) Coomassie blue detection of protein. (B) The same type of gels (and staining) with samples previously digested (+) or not (−) with DNaseI. The wells (w), undigested dsDNA (open arrows), and protein (filled arrows) dimers (d) and various oligomers (o) are marked. (C) Electron micrographs (negative staining) of WH1-A31V plus opsp fibers, either undigested (− DNaseI) or after DNA removal (+ DNaseI). (Scale bars: 200 nm.)
The WH1-WT and A31V Aggregates Exhibit Amyloid Properties.
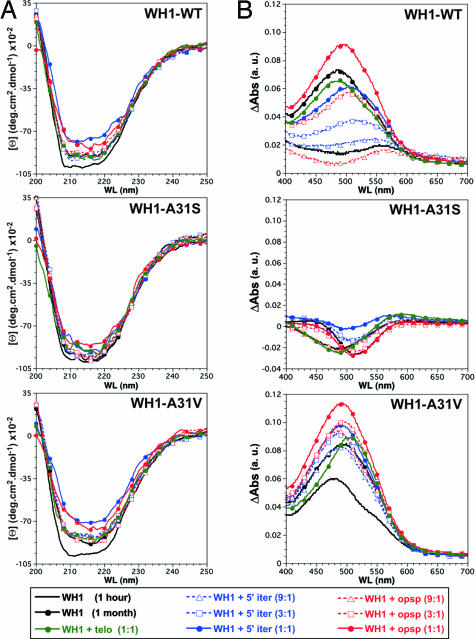
To be regarded as amyloid, any protein aggregate has to exhibit landmark properties such as a significant content in β-sheet secondary structure and the ability to bind Congo red (CR) stain (11). Thus, the average secondary structure of the distinct WH1 aggregates was compared based on their CD spectra (Fig. 4A), and their ability to bind CR was explored by differential absorption spectroscopy (Fig. 4B). These measurements were carried out on 1-month-old samples, set up with the three variants of WH1 and DNA sites. The results, once the spectral contributions of DNA and buffer were subtracted, support the amyloid character of WH1-WT and A31V aggregates: compared with fresh protein samples, they show a reduction in the CD ellipticity signal together with a broadening of its minima at ≈215 nm (a sign for a net increase in β-sheet structure) and a prominent CR absorption band at ≈500 nm (attributable to dye binding). However, the aggregates formed by WH1-A31S exhibit a less pronounced change in their CD spectra and virtually no CR binding, indicating that they are not amyloids. The response elicited by the opsp oligonucleotide is, in terms of CR binding, more intense than that promoted by the other two sequences, being most marked in the WH1-A31V fibers. In contrast, the changes that opsp DNA produces in protein secondary structure are less pronounced than those promoted by 5′iter. This is in agreement with previous results on the binding of full-length RepA to longer oligonucleotides including the same sequences (22). The nonspecific oligonucleotide telo elicits spectral responses on the order of those for the samples incubated in the absence of DNA, suggesting that, although it promotes protein aggregation (22), it does not enhance amyloidogenesis. In conclusion, the spheroids and fibers assembled, upon interaction with dsDNA oligonucleotides, by WH1-WT or the WH1-A31V mutant have amyloid nature.
Fig. 4.
The aggregates made by WH1-WT and its A31V variant exhibit amyloid features, but not the mutant with a reduced tendency to β-aggregation (A31S; see Fig. 1). (A) Far-UV CD spectra of the WH1 proteins titrated with dsDNA oligonucleotides. (B) CR binding, as measured by differential absorption spectroscopy. Both assays were carried out on replicas of the samples visualized in Fig. 2.
Assembly of the Amyloid Motif in WH1-A31V into Cross-β Fibers.
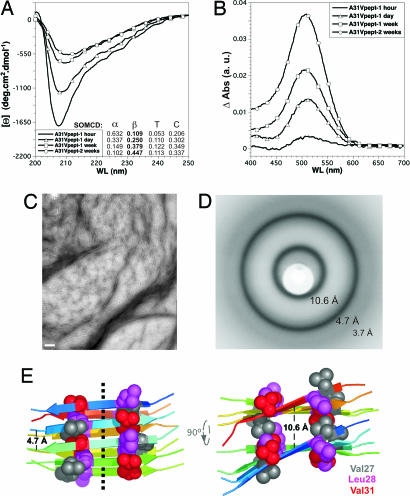
It was then explored whether the optimized amyloid motif in WH1-A31V (L26VLCAVSLI34) could have a role in building the fibers. A peptide was synthesized spanning those residues in RepA-A31V, plus flanking Arg-25 and Asp-35 to improve peptide solubility in aqueous buffers. An additional N-terminal Tyr was also added to allow quantification by UV absorption, but with no expected contribution to peptide aggregation, because of the breaking effect of Arg-25 on amyloidogenicity (24). Incubation of the peptide at identical concentration and in the same buffer assayed for the whole WH1-A31V led to a clear increase (up to ≈34%) in its contents in β-sheet secondary structure (CD spectroscopy) (Fig. 5A) and to a net enhancement of its amyloid character (CR binding) (Fig. 5B). Both features reached their highest values after 2 weeks of incubation, when TEM revealed the presence of bundles of short (≈100–200 nm) fibrils (Fig. 5C). Saturated suspensions of the peptide, after slow drying, led to aggregate deposits that exhibited the x-ray diffraction rings typical for unoriented cross-β amyloid assemblies (12): strong reflections at ≈4.7 and ≈10.6 Å (that would correspond to the distances between contiguous β-strands in a sheet and between adjacent β-sheets, respectively) plus a weaker ring at ≈3.7-Å resolution (probably a higher-order reflection along the fiber axis) (Fig. 5D). No higher-resolution rings were detected because of the limited order of the peptide assemblies. These results suggest a contribution from the L26VLCAVSLI34 sequence to the assembly of the RepA-WH1 domain into amyloid fibrils, which would probably be arranged around a β-sheet axial cord (Fig. 5E).
Fig. 5.
A peptide comprising the sequence with enhanced β-aggregation tendency in WH1-A31V assembles into amyloid fibers with a cross-β core. Shown is the incubation course of the peptide, in the absence of DNA, followed by far-UV CD (A) and CR (B) binding. A Inset estimates the evolution of average secondary structure composition. (C) Electron micrograph of the fiber bundles assembled by the peptide after 2 weeks of incubation. (Scale bar: 200 nm.) (D) X-ray fiber diffraction image of dehydrated peptide assemblies. Average resolution of each of the three visible rings is indicated. (E) Two orthogonal views of a model for a minimal assembly of the WH1-A31V peptide, built of two layers of antiparallel, right-handed twisted β-strands. The dotted line in Left indicates the axis of the resulting fibril. CPK spheres correspond to side chain atoms of residues (magenta and gray) making hydrophobic contacts with Val-31 (red). Elongation and bundling of peptide fibrils (C) would be achieved by successive addition of strands and sheets, respectively.
Discussion
In this report it is shown that transient binding of the WH1 domain of the bacterial plasmid replication protein RepA to small dsDNA fragments with defined sequence drives its assembly into distinct amyloid nanostructures. These can be further modulated by single point mutations in Ala-31, a WH1 residue in a region not involved in DNA binding (20). In WH1 dimers, the amyloidogenic region is mostly folded into an α-helix (C terminus of α2) (Fig. 1A) (19, 20). Sequences with the potential for β-aggregation in proteins are commonly found in α-helices (16, 17). This could be a means to imprint a protein stretch for an ulterior conformational rebuilding.
The findings reported here suggest that ligand-induced transient and local destabilization of a protein fold could expose amyloidogenic residues, prone to aggregation, in regions distal to the binding pocket. The occurrence of just a few intermolecular contacts, in a quasi-native folding intermediate, would be sufficient to drive the assembly of amyloids (25–28). In the light of the previous work on DNA-induced transformation of RepA (20, 22, 23), there is an inverse correlation between the magnitude of the structural change elicited by each dsDNA sequence (iteron ≥ nonspecific > operator) and the internal order of the resulting WH1-A31V assemblies (irregular < spheroids < fibrils). The exposure to the solvent of hydrophobic residues in WH1, associated with 5′iter binding and monomerization (19), must lead to uncontrolled aggregation. The resulting spheroids resemble bacterial inclusion bodies, which share some of the features of amyloids, albeit with much reduced internal order (29). More subtle structural changes, coupled to opsp DNA binding (19–23), would lead to the ordered assembly of dimeric WH1-A31V into fibers. Alternatively, the inducer effect of opsp could consist in shielding the electropositive patch at the DNA binding interface in WH1 (20), thus avoiding electrostatic repulsion and facilitating its tight package into the fibers through the amyloidogenic motif. In a sense, the dsDNA fragments assayed fulfill some of the requirements for being considered molecular chaperones because they assist the folding and assembly of WH1 into amyloid nanostructures (30).
Besides RepA-WH1, nucleic acid-induced amyloid aggregation has been previously reported for the conversion of the soluble mammalian prion protein (PrPc) into its aggregated pathogenic form (PrPsc) (5–8). However, a mixture of fibrillar and spheroidal assemblies was obtained, probably because of the very long DNA/RNA molecules tested and thus their heterogeneous sequences (6, 8). Although the results discussed in this article do not necessarily imply that nucleic acids are involved in prion conversion (the elusive “factor-X”; ref. 9), they suggest that these natural ligands could also be potential promoters of amyloid aggregation in other proteins.
A full characterization of the structure of WH1-A31V fibers is needed. In advance of that, a few structural principles emerge: (i) DNA is not physically part of the fibers (Fig. 3), as expected from the protein-only principle of amyloid structure (1) and from the small length of the oligonucleotides assayed, being unable to build a template long and continuous enough to be then coated with protein. (ii) Because the binding to the operator sequence does not dissociate RepA (22, 23), the dimensions of each of the two fibrils (Fig. 2) should accommodate a discrete number of WH1 dimers per turn. (iii) The amyloidogenic motif in WH1 could have a role, as suggested by mutations (Figs. 1 and 2) and the A31V peptide (Fig. 5), in assembling a cross-β fiber backbone. However, a contribution from the antiparallel β-sheet responsible for WH1 dimerization (20) to amyloidogenesis cannot be excluded, because sheets from neighboring dimers could stack orthogonal to the fiber axis, constituting a second fibril-forming segment (31). The tentative model proposed for the peptide fibers (Fig. 5E) falls into the class-7 (“face-to-back” stacked sheets, with the edges of the antiparallel strands up) of cross-β spines, according to the classification recently proposed from the structures of 13 amyloid peptide microcrystals (31). (iv) If the rest of WH1-A31V remains natively folded in the fibers, these would fit into the “gain of interaction through a cross-β spine” class of amyloids (13). WH1 amyloidogenic stretch is part of a very stable subdomain (α1–α2–α5) that brings together both protein ends (20). Thus, a “three-dimensional domain-swapped” type of assembly (13) seems unlikely, because it would imply extensive unfolding/refolding, the kind of structural transformation that occurs upon iteron-induced monomerization, not upon operator binding (19–23).
Bacterial genomes, while developing efficient chaperone networks, have successfully avoided the accumulation of genes coding for amyloidogenic proteins (24). There are very few naturally amyloidogenic proteins in bacteria, mostly secreted to an extracellular location, as the CsgA curly fibrils that have a role in biofilm formation (32). In terms of the role for RepA protein in pPS10 replication, it is noteworthy that A31V, the mutation enhancing the amyloidogenicity of WH1, was the most frequently isolated in a search for mutations expanding the plasmid host range beyond Pseudomonas, improving the interaction between RepA and host factors (33). An implication of the findings discussed here is that protein–protein contacts carried out through the amyloidogenic region in WH1 would involve the formation of a β-sheet. This could be the case for the replication inhibitory handcuffed arrays, in which two plasmids are bridged together through interactions between multiple origin-bound RepA molecules, thus hindering origin firing (reviewed in ref. 34).
Beyond the interest of this report in the quest for natural factors contributing to trigger the aggregation of proteins into amyloids, the ability to build at will protein supramolecular assemblies with defined morphologies is a crucial issue in nanotechnology. It also opens the way to the design of a simple, intracellular, and nonpathogenic bacterial model system for high-throughput approaches to the screening of small molecules that, by targeting its universal structural basis, would inhibit amyloid aggregation.
Materials and Methods
Mutagenesis, Expression, and Purification of RepA-WH1.
Site-directed mutagenesis of the amyloidogenic motif in WH1 was performed by PCR on the pPSEC1 vector (22) by using the following oligonucleotides (mutations underlined): A31S, 5′-CTCATCGAATCGTCACATACGTTGACACTCAATGAGAAACGCCTAGTGCTATGCGCTTCGTCTTTGATCGATTCACGTAA; A31V, 5′-CTCATCGAATCGTCACATACGTTGACACTCAATGAGAAACGCCTAGTGCTATGCGCTGTGTCTTTGATCGATTCACGTAA.
The reverse primer used was ΔC133-R (HindIII site underlined; stop codon in lowercase): 5′-GCTAAGCTtcaCTTGAGCTGATAGCTGGTGAA. A second PCR round was performed on the fragments amplified by using the same reverse primer, but an overlapping forward oligonucleotide reconstructing the 5′ end of repA gene plus a SacII site (underlined; Met-1 codon in lowercase): 5′-GCTCCGCGGGGCAGCatgGTCGAGAACAAAGTCACGCAGTCCAATAAACTCATCGAATCGTCAC.
The PCR products were cloned into the expression vector pRGrectac-NHis, a variant of pRGrecA-NHis (21) that includes a ClaI cassette with the Ptac promoter. Constructs were verified by DNA sequencing (ABI3730; Applied Biosystems). Protein expression was performed in the Escherichia coli strain BL21, carrying the pLysS plasmid to lyse cells after freezing–thawing. Protein purification was carried out under native conditions as described (21), but using the high-density IDA matrix manufactured by Agarose Bead Technologies for Ni2+ affinity (gradient to 0.4 M imidazole) and substituting ammonium sulfate and acetate by sodium sulfate and phosphate, respectively, in the SP-Sepharose chromatography that follows the thrombin cleavage of the His-6 tag (21). Protein concentration was worked out in 5.4 M GuHCl, assuming a molar extinction coefficient (280 nm) of 12,950 M−1·cm−1 (www.expasy.ch/tools/protparam.html) per RepA-WH1 chain. Protein stocks (70 μM, after Amicon concentration) were kept at −70°C in 0.2 M Na2SO4, 15 mM Na2HPO4 (pH 6), 5 mM 2-mercaptoethanol, and 10% (vol/vol) glycerol.
Setup for RepA-WH1 Amyloidogenesis in Vitro.
Screening for conditions resulting in WH1 aggregation was performed in Eppendorf tubes by mixing 50 μl of protein stock (after centrifugation at 15,700 × g for 5 min) with the same volume of solutions composed of different salts, buffers, and additives. Samples were left to stand at 4°C for 2–4 weeks. The mixing solution leading to the protein aggregates discussed here was 60 mM Tris·HCl (pH 8), 8 mM MgSO4, 14% PEG4000, and 6% MPD. When indicated, 11-bp DNA oligonucleotides (either 5′iter, opsp, or telo) (Fig. 2) were included in the mixing solution. They were synthesized (1 μmol; Applied Biosystems Model 3400), purified, and annealed (200 μM stocks) as described (21).
Looking for DNA in WH1-A31V Fibers.
A total of 50 μl of a 1-month-old WH1-A31V plus opsp (35 μM) preparation was spun down (100,000 × g for 1 h at 4°C), and then 10-μl aliquots of the supernatant and the resuspended pellet were processed as follows: The first sample was loaded onto a 15% polyacrylamide gel (19:1 acrylamide:bisacrylamide, 0.5× TBE), and electrophoresis was carried out at 100 V for 30 min before ethidium bromide staining and UV detection (Gel Doc 2000; Bio-Rad). The second one was run in a native 6% polyacrylamide (29:1) protein gel, in which SDS and the stacking gel had been omitted, with the polarity of the electrodes inverted (WH1 theoretical pI = 9.42), at 150 V for 3 h. The gel was stained with Coomassie blue. In addition, two 10-μl aliquots of an untreated preparation of mature fibers were digested with 1.0 unit of DNaseI (Worthington) for 1 h at 25°C and then analyzed by electrophoresis as just described. A third aliquot was processed for TEM (see below).
CD Spectroscopy.
Spectra were acquired with a Jasco-720 spectropolarimeter using 0.1-cm path-length quartz cuvettes (Hellma). The cell holder was kept at 5°C with a Peltier module. For comparing the average secondary structure of the WH1 proteins, 200-μl samples were made by diluting each protein stock to 15 μM in 0.1 M Na2SO4 and 15 mM Na2HPO4 (pH 6). CD spectra were acquired between 260 and 195 nm (20 nm/min scan speed). Five spectra (10 for the A31V peptide) were averaged for each sample, and the buffer was subtracted as a blank. Raw data were transformed to molar ellipticity [θ] and plotted with KaleidaGraph software version 3.6.2 (Synergy). For thermal denaturation, temperature was increased from 5°C to 90°C (at 20°C/h) monitoring variations in the ellipticity at 222 nm. Samples analyzed in Fig. 4A were diluted 1/5 in water before spectra acquisition. When appropriate, spectra for free dsDNAs were subtracted as described (22). Secondary structure contents for the A31V peptide were estimated by means of SOMCD (35) at http://geneura.ugr.es/cgi-bin/somcd/som.cgi.
Differential CR Absorption Spectroscopy.
A 7 mg·ml−1 CR (Sigma) solution in water was clarified at 15,700 × g for 5 min before 1/100 dilution (to 0.1 mM) in 0.1 M Na2SO4 and 20 mM Tris·HCl (pH 8). Aliquots (985 μl) were dispensed into plastic cuvettes (1-cm path length). A total of 15 μl from each WH1 (with or without DNA) or peptide sample was added, gently mixed, and left to equilibrate at room temperature for 30 min. Differential UV absorption spectra were acquired between 400 and 700 nm (1-nm steps) in an Amersham Ultrospec-3300pro spectrophotometer by using as a reference blank a protein-free sample of the buffer used in the amyloidogenesis assays (with or without DNA) diluted in the CR solution. Two consecutively acquired spectra were averaged by using KaleidaGraph.
TEM.
Collodion-carbon-coated and glow-discharged copper grids (300 mesh) were placed on 4-μl drops of each WH1 amyloidogenic solution displayed on a Parafilm sheet. After 1.5 min, grids were washed twice by rising through 5-μl drops of milli-Q water before being left for 2 min in 2% (wt/vol) uranyl acetate. Excess solution was then removed with filter paper and left to dry further. Grids were then examined in a JEOL JEM-1230 transmission electron microscope operating at 100 kV.
Synthesis, Purification, and Assembly of an Amyloidogenic RepA-WH1 Peptide.
The 12-residue peptide NH2-YR25LVLCAVSLID35-COOH, including the amyloidogenic motif in WH1-A31V (underlined), was made in a PerkinElmer Synergy synthesizer (Fmoc chemistry, 25 μmol), and its purity was estimated to be ≥98% by means of MALDI-TOF mass spectrometry. The peptide was dissolved (2 mM) in 6 M GuHCl, 50 mM Tris·HCl (pH 8), and 50 mM 2-mercaptoethanol and diluted to 35 μM in 0.1 M Na2SO4 and 20 mM Tris·HCl (pH 8). Samples were then incubated at 4°C.
X-Ray Diffraction on Peptide Fibers.
The A31V peptide was resuspended to saturation in 10 mM Tris·HCl (pH 8) and then transferred to a quartz capillary tube (1 mm Ø) where it was left to dry for a week. A series of x-ray diffraction still images, rotated by 0°, 45°, and 90°, were exposed (90 min each) at room temperature. The experimental setup consisted of an Enraf-Nonius FR-591 rotating anode (Cu-Kα), operating at 40 kV, and a MarResearch-345 detector, with sample-to-plate distances ranging from 150 to 300 mm. The average resolution corresponding to each diffraction ring was determined by means of the software distributed with the image plate.
Modeling Amyloid Peptide Assemblies.
A four-stranded β-sheet model for the A31V peptide was manually built by means of Swiss-PdbViewer (36), keeping an average interstrand distance of ≈4.7 Å (Fig. 5D). An antiparallel arrangement of the peptide strands was selected to accommodate the electrostatic interactions between charged N-terminal (+) and C-terminal (−) residues. Contiguous strands were right-hand twisted by ≈10° to get the lowest energy conformation for the resulting β-sheet. An extra sheet was then layered, spaced from the first one by ≈10.6 Å (Fig. 5D). The model was then energy-minimized by using Tinker v.4.2 (http://dasher.wustl.edu/tinker) with the AMBER99 force field to a rmsd of 0.1 kcal·mol−1·Å−1 (final global energy: −3,839 kcal·mol−1).
Supplementary Material
Acknowledgments
I am indebted to the members of the electron microscopy facility at Centro de Investigaciones Biológicas for their assistance and training, to M. J. Mate for help with x-ray diffraction, and to A. Prieto for mass spectrometry on the A31V peptide. The DNA sequencing and the oligonucleotide/peptide synthesis services at Centro de Investigaciones Biológicas are acknowledged for their valuable technical support. Thanks are also due to the members of my group for encouragement and to J. M. Andreu, D. Rhodes, and L. Chapman for critical reading of the manuscript. This work has been financed by Spanish Ministry of Education and Science Grants BMC2003-00088 and BFU2006-00494.
Abbreviations
- CR
Congo red
- TEM
transmission electron microscopy
- WH
winged-helix.
Footnotes
This article is a PNAS Direct Submission.
The author declares no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702006104/DC1.
References
- 1.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Jahn TR, Radford SE. FEBS J. 2005;272:5962–5970. doi: 10.1111/j.1742-4658.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 3.Avila J. FEBS Lett. 2006;580:2922–2927. doi: 10.1016/j.febslet.2006.02.067. [DOI] [PubMed] [Google Scholar]
- 4.Deleault NR, Geoghegan JC, Nishina K, Kascsak R, Williamson BA, Supattapone S. J Biol Chem. 2005;280:26873–26879. doi: 10.1074/jbc.M503973200. [DOI] [PubMed] [Google Scholar]
- 5.Cordeiro Y, Machado F, Juliano L, Juliano MA, Brentani RR, Foguel D, Silva JL. J Biol Chem. 2001;276:49400–49409. doi: 10.1074/jbc.M106707200. [DOI] [PubMed] [Google Scholar]
- 6.Nandi PK, Leclerc E, Nicole J-C, Takahashi M. J Mol Biol. 2002;322:153–161. doi: 10.1016/s0022-2836(02)00750-7. [DOI] [PubMed] [Google Scholar]
- 7.Deleault NR, Lucassen RW, Supattapone S. Nature. 2003;425:717–720. doi: 10.1038/nature01979. [DOI] [PubMed] [Google Scholar]
- 8.Nandi PK, Nicole J-C. J Mol Biol. 2004;344:827–837. doi: 10.1016/j.jmb.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 9.Prusiner SB. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castilla J, Saá P, Hetz C, Soto C. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson MR. Methods. 2004;34:151–160. doi: 10.1016/j.ymeth.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 12.Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CCF. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 13.Nelson R, Eisenberg D. Adv Protein Chem. 2006;73:235–282. doi: 10.1016/S0065-3233(06)73008-X. [DOI] [PubMed] [Google Scholar]
- 14.Kallberg Y, Gustafsson M, Persson B, Thyberg J, Johansson J. J Biol Chem. 2001;176:12945–12950. doi: 10.1074/jbc.M010402200. [DOI] [PubMed] [Google Scholar]
- 15.Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 16.López de la Paz M, Serrano L. Proc Natl Acad Sci USA. 2004;101:87–92. doi: 10.1073/pnas.2634884100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 18.Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. J Mol Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Giraldo R, Fernández-Tresguerres ME. Plasmid. 2004;52:69–83. doi: 10.1016/j.plasmid.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Giraldo R, Fernández-Tornero C, Evans PR, Díaz-Orejas R, Romero A. Nat Struct Biol. 2003;10:565–571. doi: 10.1038/nsb937. [DOI] [PubMed] [Google Scholar]
- 21.Giraldo R, Andreu JM, Díaz-Orejas R. EMBO J. 1998;17:4511–4526. doi: 10.1093/emboj/17.15.4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Díaz-López T, Lages-Gonzalo M, Serrano-López A, Alfonso C, Rivas G, Díaz-Orejas R, Giraldo R. J Biol Chem. 2003;278:18606–18616. doi: 10.1074/jbc.M212024200. [DOI] [PubMed] [Google Scholar]
- 23.Díaz-López T, Dávila-Fajardo C, Blaesing F, Lillo MP, Giraldo R. J Mol Biol. 2006;364:909–920. doi: 10.1016/j.jmb.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Rousseau F, Serrano L, Schymkowitz JWH. J Mol Biol. 2006;355:1037–1047. doi: 10.1016/j.jmb.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 25.Kammerer RA, Kostrewa D, Zurdo J, Detken A, García-Echevarría C, Green JD, Müller SA, Meier BH, Winkler FK, Dobson CM, Steinmetz MO. Proc Natl Acad Sci USA. 2004;101:4435–4440. doi: 10.1073/pnas.0306786101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambashivan S, Liu Y, Sawaya MR, Gingery M, Eisenberg D. Nature. 2005;437:266–269. doi: 10.1038/nature03916. [DOI] [PubMed] [Google Scholar]
- 27.Jahn TR, Parker MJ, Homans SW, Radford SE. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 28.Plakoutsi G, Bemporad F, Calamai M, Taddei N, Dobson CM, Chiti F. Structure (London) 2006;14:993–1001. doi: 10.1016/j.str.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Carrió M, González-Montalbán N, Vera A, Villaverde A, Ventura S. J Mol Biol. 2005;347:1025–1037. doi: 10.1016/j.jmb.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 30.True HL. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 31.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 32.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maestro B, Sanz JM, Díaz-Orejas R, Fernández-Tresguerres E. J Bacteriol. 2003;185:1367–1375. doi: 10.1128/JB.185.4.1367-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulsson J, Chattoraj DK. Mol Microbiol. 2006;61:9–15. doi: 10.1111/j.1365-2958.2006.05229.x. [DOI] [PubMed] [Google Scholar]
- 35.Unneberg P, Merelo JJ, Chacón P, Morán F. Proteins. 2001;42:460–470. doi: 10.1002/1097-0134(20010301)42:4<460::aid-prot50>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 36.Guex N, Peitsch MC. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.