Abstract
Acid β-glucosidase (GCase) is a soluble lysosomal enzyme responsible for the hydrolysis of glucose from glucosylceramide and requires activation by the small nonenzymatic protein saposin C (sapC) to gain access to the membrane-embedded glycosphingolipid substrate. We have used in situ atomic force microscopy (AFM) with simultaneous confocal and epifluorescence microscopies to investigate the interactions of GCase and sapC with lipid bilayers. GCase binds to sites on membranes transformed by sapC, and enzyme activity occurs at loci containing both GCase and sapC. Using FRET, we establish the presence of GCase/sapC and GCase/product contacts in the bilayer. These data support a mechanism in which sapC locally alters regions of bilayer for subsequent attack by the enzyme in stably bound protein complexes.
Keywords: atomic force microscopy, confocal microscopy, FRET, interfacial catalysis, lipid storage disease
Gaucher disease is a common lysosomal storage disease characterized by the presence of engorged macrophages in the liver, spleen, and bone marrow (1, 2). The disorder is caused by abnormal accumulations of glucosylceramide (GlcCer) in these tissues because of the inability to catabolize this lipid within lysosomes. Typically, the breakdown of membrane glycosphingolipids requires the combined action of a hydrolytic enzyme and a nonenzymatic “activator” protein, which, in the case of GlcCer, are acid β-glucosidase (GCase, EC 3.2.1.45) and saposin C (sapC). In the absence of activator and acidic phospholipids, the enzyme does not have direct access to the glycosphingolipid, which is tightly packed within the lipid bilayer. Imiglucerase [Cerezyme (Genzyme, Cambridge, MA), recombinant GCase expressed in mammalian cell cultures] is used in enzyme replacement therapy of Gaucher patients (3).
The activator proteins saposin A, B, C, and D are homologous, soluble, nonenzymatic proteins that interact with lysosomal membranes and facilitate the breakdown of glycosphingolipids by specific hydrolases (2, 4, 5). Recent findings have also implicated saposins in glycolipid antigen presentation by CD1 molecules, where saposins are thought to provide the means by which glycolipids are extracted from membranes and loaded onto CD1 molecules (6–8). Current models for enzyme activation address the location of saposin-mediated lipid–hydrolase interactions and define the “solubilizer” and “liftase” modes of action. In the former model, target lipid molecules are extracted from bilayers by saposins and presented to cognate enzymes as soluble protein–lipid complexes. In contrast, the “liftase” model involves the binding of enzyme to the bilayer surface where saposin molecules facilitate the access to the glycosphingolipid substrates. Different saposins and enzymes are hypothesized to fall into a particular category of enzyme activation. For example, saposin B is thought to be a detergent-like lipid solubilizer (9, 10), whereas sapC may act as a liftase at the bilayer surface (2, 11, 12). Strong binding of sapC to lipid bilayers was shown by using coprecipitation assays (13) and NMR spectroscopic titrations (14). Simultaneous atomic force microscopy (AFM)/fluorescence microscopy allowed the visualization of saposin binding to planar bilayers and the resulting membrane transformation (15–18). Although sapC has no enzymatic activity, sapC-transformed areas contain narrow channels surrounding islets of the original bilayer in a manner strikingly similar to the effects of secreted phospholipase A2 (19) on lipid bilayers. Like GCase, phospholipases are water-soluble enzymes that act on membrane-embedded lipids, some using a membrane-docking C2 domain distinct from the catalytic domain (20, 21). Pancreatic lipase is another example of a soluble enzyme that hydrolizes lipids in aggregated forms. The enzyme requires the action of a small activator protein, colipase, which anchors the enzyme to lipid particles and stabilizes its active conformation (22). Conceptually, some aspects of the lipase/colipase pair may be similar in the GCase/sapC association acting according to the “liftase” model.
Despite extensive in vivo and in vitro studies, it remains unclear whether the activation of GCase by sapC can be exclusively assigned to either model. An understanding of saposin-mediated glycosphingolipid hydrolysis requires the characterization of GCase interactions with saposin- and substrate-containing lipid bilayers. In this study, we present the simultaneous visualization of sapC and GCase action on model membranes by AFM and confocal fluorescence imaging. Using a membrane-bound fluorogenic substrate in the combined microscope, we directly observe enzyme activation at the membrane interface. Finally, we establish GCase/sapC and GCase/product contacts by FRET within the bilayer. Our results are fully consistent with a membrane-bound reaction involving a form of interfacial catalysis.
Results
GCase Membrane Localization.
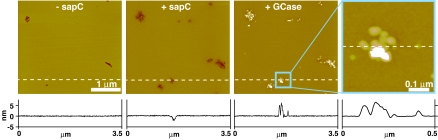
The addition of sapC to a supported bilayer resulted in the lowering of the membrane from a small number of nucleation sites that are preferentially initiated from the edges of membrane defects, as described (15, 16) (Fig. 1). At concentrations <1 μM, these areas were lowered by ≈1.5 nm, probably because of partial lipid removal, and contained stably bound sapC (15). After the addition of GCase to a final molar ratio of 1:3 (GCase:sapC), material accumulated exclusively on areas that were previously remodeled by sapC to levels that were 2–8 nm higher than the unaffected bilayer (Fig. 1). We also observed the occasional presence of larger aggregates in these areas, which are seen as white features in Fig. 1. Although sapC alone has the potential to form raised plaques and aggregates atop remodeled areas, these appear only at sapC concentrations >5 μM (15). We therefore presume that the material observed upon the addition of the enzyme is composed of GCase or a mixture of GCase and sapC. Because the diameter of GCase is ≈5 nm (23), the change in height is consistent with the presence of one to two molecules of the enzyme. The features that arose from the addition of GCase did not appear in areas unaffected by sapC action.
Fig. 1.
AFM imaging of sapC and GCase interactions with a supported planar bilayer. The series of height images were acquired after the sequential addition of sapC (0.2 μM) and A488-GCase (0.07 μM), as labeled. The images depict the equilibrium state of the bilayer at the end of an evolution period after protein addition. The rightmost frame is a higher-resolution image of the GCase-affected area marked by the cyan box in the lower-resolution frame. The lateral XY scale of the images is given by the thick white bars. Pseudocolors are used to represent the height data, with darker areas corresponding to lower features. The height profiles corresponding to the dashed lines are all plotted with the same vertical scale.
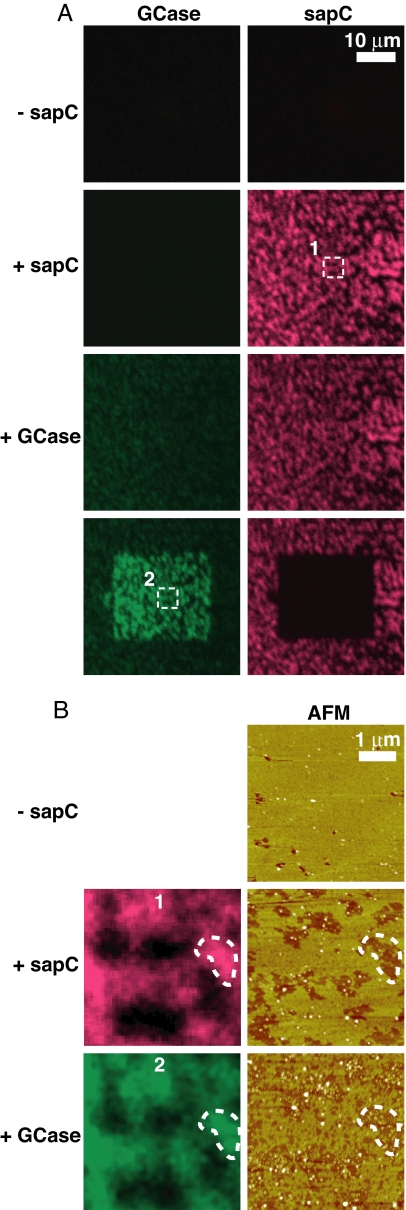
To confirm the identity of the proteins bound to the supported lipid bilayer, we used combined in situ AFM/multiprobe confocal fluorescence imaging (Fig. 2). sapC was labeled with the Alexa Fluor 546 (A546) fluorophore at residue 22. This position is in a short loop between helices α1 and α2 of the protein and is the site normally glycosylated in the natural protein (24). GCase was chemically labeled using an amine-reactive Alexa Fluor 488 (A488) succinimidyl conjugate. Images of A546-labeled sapC (A546-sapC) and A488-labeled GCase (A488-GCase) fluorescence were collected in separate channels (red and green, respectively). Similar to the experiment in Fig. 1, the supported planar bilayer had a model lysosomal lipid composition with some initial defects. As shown (15), fluorescent sapC localized to the lipid surface in saposin-induced lowered areas [Fig. 2 and supporting information (SI) Fig. 7]. After the addition of A488GCase, granular material similar to that observed in high-resolution AFM scans (Fig. 1) was detected in areas of saposin-induced bilayer lowering (Fig. 2B) along with weak A488-GCase fluorescence in the GCase channel (Fig. 2A).
Fig. 2.
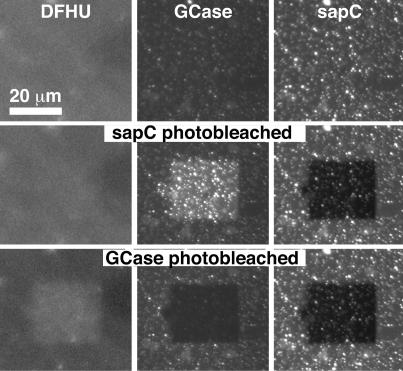
Binding of labeled GCase to lipid bilayers remodeled by sapC. (A) The first three rows of confocal microscopy images correspond to the sequential addition to a supported planar bilayer (−sapC) of 0.5 μM A546-sapC (+sapC) and 0.05 μM A488-GCase (+Gcase). Left and Right images are, respectively, A488-GCase (green) and A546-sapC (red) channel images of the same bilayer area. (Bottom) After A546-sapC and A488-GCase addition, A546-sapC (FRET acceptor) was photobleached within a 25 × 25-μm square using intense 546-nm HeNe laser illumination (Bottom Right), resulting in an increase in FRET donor A488-GCase fluorescence in the area corresponding to photobleached A546-sapC (Bottom Left). (B) The confocal images labeled 1 and 2 are magnifications of the respective areas enclosed in dashed boxes in A. Tapping mode AFM height images of the corresponding area are given in Right. The AFM height image before protein addition is given in Top. Pseudocolors are used to represent the AFM height data, with darker areas corresponding to lower features. Image scales are given by white bars in each image.
The low intensity of the A488-GCase signal made it difficult to correlate the sapC and GCase fluorescence distributions. Because the “liftase” mechanism implies a direct sapC/GCase interaction, we expected that the fluorophores on the two proteins would be within their Förster radius (25, 26), resulting in FRET between the A488-GCase donor and the A546-sapC acceptor. In this scenario, FRET would reduce the A488-GCase fluorescence. We tested this hypothesis by photobleaching the A546-sapC with the confocal microscope laser and observed a strong increase of A488-GCase fluorescence (Fig. 2A). Thus, sapC and GCase are in close contact on the membrane surface. The enhanced A488-GCase intensity allowed clear correlation between the A546-sapC and A488-GCase fluorescence distributions (Fig. 2B), confirming the assignment of the AFM features.
GCase Enzyme Activity.
We used a membrane-bound fluorogenic substrate analog to image and assay GCase activity in bilayers. The compound 6,8-difluoro-4-heptadecylumbelliferyl β-d-glucopyranoside (DFUG) is similar to soluble umbelliferyl-based glucosides used as GCase substrates in previous studies (12, 27, 28) but contains a C17 acyl chain that anchors the molecule to the membrane. DFUG is an excellent analogue of the natural GlcCer substrate for measuring membrane-bound GCase activity, because hydrolysis of the glucoside headgroup yields the fluorescent compound 6,8-difluoro-7-hydroxy-4-heptadecylcoumarin (DFHU), which also remains in the lipid bilayer.
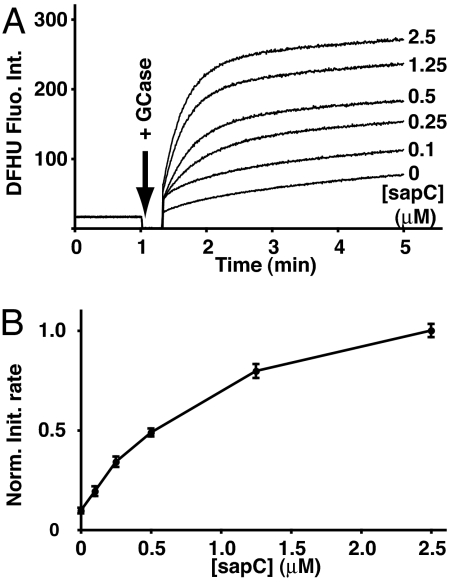
In the absence of activator protein, GCase had a low but detectable level of enzyme activity when incubated with liposomes containing DFUG (Fig. 3). Preincubation of the liposomes with varying amounts of sapC before the addition of fixed amounts of enzyme resulted in a dose-dependent increase in the GCase activity. The time curves resemble typical activity rates when increasing amounts of substrate are mixed with enzyme, but in this case, the total amount of DFUG substrate is constant, and only the activator concentrations are changing. Thus, sapC is effectively increasing the amount of substrate that is available for hydrolysis in this assay. There is a 10-fold increase in the initial rate of hydrolysis at 2.5 μM sapC, which is similar to the ≈7-fold increase in GCase activity when incubated with the same concentration of sapC and liposomes containing radiolabeled GlcCer (12). No substrate hydrolysis was observed in the absence of GCase, regardless of the saposin concentration.
Fig. 3.
GCase activity assay using the liposome-anchored DFUG substrate. (A) Kinetics of DFHU accumulation after addition of 0.05 μM GCase to liposomes preincubated with 0, 0.1, 0.25, 0.5, 1.25, and 2.5 μM sapC, as indicated. The total lipid concentration in each reaction was 100 μM, including 10 mol % DFUG. Reaction product DFHU fluorescence intensities are given in arbitrary units. The arrow indicates the time at which the enzyme was added to each sample, before mixing and the resumption of fluorescence measurements. (B) Dependence of the GCase initial reaction rates on sapC concentration. The rates were determined from the kinetics in A and normalized relative to the highest rate. Error bars correspond to ±3 standard deviations.
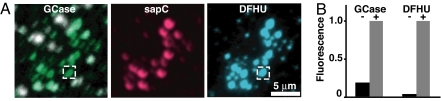
These observations were reproduced with supported bilayers in the confocal microscope. A488-GCase was added to DFUG-containing membranes in the absence of sapC. Isolated spots of intense A488-GCase fluorescence (white in Fig. 4 and SI Fig. 8) correspond to protein bound directly to exposed mica surfaces in areas of large membrane defects and serve as an internal negative control. Weak but measurable A488-GCase fluorescence was observed in areas of intact membrane, indicating low concentrations of enzyme at the membrane surface. Upon the addition of A546-sapC, there was a significant increase in the amount of GCase bound at the membrane, giving rise to new localized areas of enzyme (Fig. 4A, “GCase,” green patches). In the DFHU channel, fluorescence was seen only in areas that contained both sapC and GCase (Fig. 4). Thus, sapC resulted in GCase translocation to concentrated patches in the bilayer, and enzyme activation by sapC was confirmed by the colocalization of the A488-GCase, A546-sapC, and DFHU signals.
Fig. 4.
sapC-induced GCase binding and activation. (A) Pseudocolor fluorescence images of a lipid bilayer after consecutive additions of 0.1 μM A488-GCase and 5 μM sapC containing 10% A546-sapC. (Left) Overlay of A488-GCase confocal images before and after sapC addition to the bilayer (SI Fig. 8), showing newly accumulated A488-GCase after sapC addition (green areas) and the original areas of GCase accumulation before adding sapC (white patches). The A546-sapC confocal image (Center) and reaction product DFHU epifluorescence image (Right) of the same area show fluorescence localizations matching the newly accumulated A488-GCase fluorescence (Left). (B) Localized A488-GCase and enzyme product (DFHU) fluorescence intensities before (−) and after (+) sapC addition to the bilayer. Fluorescence intensities were quantified from the dashed box areas and normalized relative to the + intensity in each channel. The areas correspond to the position of a sapC-induced GCase spot. The images from which the pre-sapC intensities were quantified are not shown.
GCase-sapC–DFHU Interactions.
The results described above demonstrate the translocation of soluble GCase to the bilayer in areas remodeled by sapC, with the concurrent accumulation of the reaction product DFHU at sapC/GCase sites. Next, we used FRET between A546-sapC and A488-GCase and between A488-GCase and DFHU, as a sensitive tool to determine the presence of associations between sapC, GCase and membrane-bound DFHU. Fig. 5 shows that the fluorescence from membrane-bound A488-GCase increases when the FRET acceptor A546-sapC is photobleached by the confocal microscope laser (Fig. 5 Middle, “Gcase” image). The FRET efficiency, E, for the pair A546-sapC/A488-GCase is 0.65, which indicates very close proximity. Similarly, DFHU fluorescence experiences a net increase when A488-GCase is photobleached in turn (Fig. 5 Bottom, “DFHU” image). This confirms the close proximity of GCase, sapC and the product of the hydrolase reaction, DFHU, in the bilayer. Because there is no spectral overlap between the absorption spectrum of A546 and the emission spectrum of the coumarin derivative, there is no direct FRET between the labeled sapC and DFHU. However, we have described FRET between sapC and labeled lipid molecules in sapC transformed bilayers (15).
Fig. 5.
Sequential A546-sapC/A488-GCase and A488-GCase/DFHU FRET. (Top) Epifluorescence DFHU (Left) and confocal A488-GCase (Center) and A546-sapC (Right) images of a lipid bilayer after simultaneous exposure to 0.1 μM A488-GCase and 1 μM sapC containing 50% A546-sapC. (Top) Images before any photobleaching. (Middle) A546-sapC was photobleached within a 25 μm x 25 μm square using intense 543 nm HeNe laser illumination (Right). (Bottom) A488-GCase was then photobleached within a 25 × 25-μm square using intense Ar-ion 488 nm laser illumination (Center). Conditions were chosen to achieve extensive bilayer coverage with sapC and GCase. At this resolution, the sapC and GCase coverage appears relatively uniform. (The bright spots correspond to aggregates of sapC and GCase as seen in Fig. 1 and were confirmed by AFM scans of the same area.) DFUG hydrolysis occurs predominantly in the areas of uniform GCase/sapC coverage.
Discussion
GCase is a soluble enzyme that requires a facilitator to access its membrane-embedded substrate in the hydrolysis of glucose headgroups. Purified GCase was previously shown to interact only weakly with liposomes and to exhibit a low level of liposomal GlcCer hydrolysis (29). Unlike the pancreatic lipase/colipase system (22), there is no indication of an interaction between sapC and purified delipidated GCase in solution in the absence of membrane lipids (30–33). The association of the proteins and hydrolase activation is recovered when lipids, especially negatively charged lipids, are added (30–33). [Earlier GCase preparations were shown to bind Sepharose-coupled sapC (34, 35), and sapC induced the lowering of the GCase Km value for 4-methylumbelliferyl-β-d-glucopyranoside (MUG) hydrolysis (35). However, the same preparations were shown to contain considerable amounts of lipids (35).] Acidic pH and the presence of the negatively charged lipids 1,2-diacyl-sn-3-phosphoinositol (PI) or bis(monoacylglycero)phosphate (BMP), which are abundant in intralysosomal compartment (36), are required for the activation of GCase activity by sapC (12, 27). In our DFUG assay, the activation of GCase by sapC was maximal in the pH range 4.8–5.0 and required PI or BMP (SI Fig. 9), as expected.
The sapC preparations used in this study were not glycosylated, and the consequences of covalent modifications at position Asn-22 (the single glycosylation site in sapC) deserve comment. Two recent reports have shown that glycosylated recombinant saposins A (37) and B (38) expressed in yeast differ from the unglycosylated forms in their ability to solubilize lipids from immobilized liposomes. However, earlier reports have shown that nonglycosylated saposins retain their lipid-binding and enzyme activation effects (39), and recombinant sapC expressed in Escherichia coli was shown to have similar properties to sapC purified from natural tissues (40). Notably, E. coli-expressed saposin B (nonglycosylated) was as active as glycosylated human saposin B purified from urine in an in vitro activity assay (10) and could functionally complement saposin B-deficient human fibroblast cells (41). We have shown in a previous report (15) that both unglycosylated sapC and Alexa-labeled N22C sapC have similar lipid extraction activities on supported bilayers. Moreover, our previous (15) and current results (Figs. 1 and 2 and SI Fig. 7) show that both unlabeled sapC and the Alexa-labeled N22C mutant have similar bilayer transformation effects when observed by AFM imaging. As an additional test, we found that the GCase activation properties of Alexa-labeled sapC were similar to that of unlabeled sapC in the liposomal assay (SI Fig. 10). We conclude that the covalent addition of a 1,034-Da fluorophore at position Asn-22 has negligible effects on sapC activity in these assays. Further studies will be required to directly demonstrate any effects of sapC glycosylation in these assays.
Although sapC has the potential to solubilize membrane lipids (12, 15), solution studies with liposomes have led to conflicting conclusions in assigning the GCase/sapC pair to either the “solubilizer” or the “liftase” mode of action (12, 33). Our direct visualization of the events occurring at the membrane interface suggests that GCase hydrolyzes its substrate at the bilayer level with the help of sapC within a complex at the membrane surface. Although we cannot formally exclude the possibility that all or part of the product may be originating in solution and transferred back to the bilayer, that sapC brings GCase to concentrated areas at the bilayer with a subsequent increase in GCase activity in the same areas is strong support for an interfacial reaction. The increased enzyme activity may be due to several mechanisms, including increased levels of membrane-bound enzyme, a higher specific activity of the enzyme within an activator complex, and/or better access to the substrate. GlcCer, a membrane-embedded glycosphingolipid with a glucosyl headgroup, is the natural substrate for GCase. Assuming that the enzyme alone cannot penetrate the bilayer, the lipid substrate must be “lifted” for proper docking of the headgroup to the active site (42). We have shown that sapC induces a nucleated spread of membrane remodeling that is characterized by a reduced membrane thickness (15). sapC is present in the remodeled areas, and a particular model suggests that sapC removes the top leaflet, whereas its amphiphilic nature allows it to shield the hydrophobic tails of the lower acyl chains (15). The organization of lipids at the interface between intact bilayers and transformed areas likely exposes lipid molecules, including the GCase substrate (Fig. 6). The lipid fraction removed by sapC is presumably in a solubilized form and may potentially serve other functions, including loading lipid antigens onto CD1 molecules (6–8).
Fig. 6.
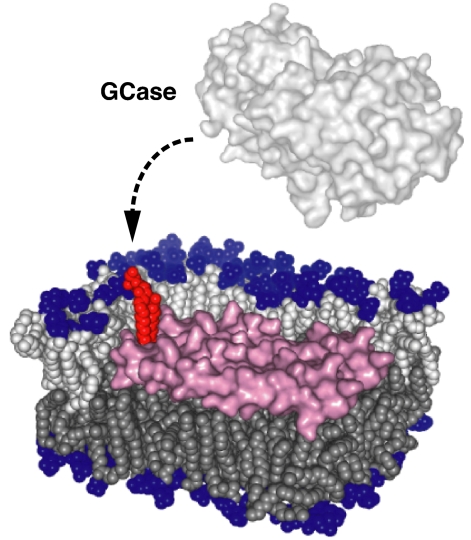
Schematic model for GCase binding to the membrane and saposin-mediated activation. GCase alone cannot extract membrane-embedded Glc-Cer from the bilayer. Saposin can help expose embedded lipids, including GlcCer (red), to soluble GCase molecules by creating perturbed edges between lowered and intact areas of the bilayer. Saposin molecules are colored in purple; lipid polar groups are blue. Acyl chains belonging to upper and lower leaflets are colored in light and dark gray, respectively.
The creation of perturbed bilayer edges could explain saposin-mediated GCase activation, but it is unclear whether the partially exposed substrate is sufficient for enzyme translocation. Electrostatic interactions may additionally be involved in securing GCase binding to the membrane. Crystal structures of GCase have consistently shown the presence of sulfate ion clusters bound in the vicinity of the active site (23, 28, 42, 43). The ions are thought to mimic negatively charged phospholipids that are also known to enhance enzyme activity (42). Close proximity of sapC and GCase molecules is confirmed by our results showing colocalization of labeled sapC and GCase fluorescences in the bilayer, as well as sapC/GCase FRET. However, because purified and delipidated GCase does not seem to interact with sapC in solution (30–33), GCase binding may depend on the specific recognition of membrane-bound conformations of sapC.
The sapC-induced increase in GCase activity may also be related to a higher intrinsic activity of the enzyme within an activator complex, possibly involving a conformational change in the enzyme (44), as seen in the case of pancreatic lipase. This is supported by the fact that sapC can increase the hydrolysis of soluble substrate analogs by up to 17-fold in the presence of large unilamellar vesicles (12, 33). However, this activation cannot be explained by an increase in the accessibility of catalytic pocket, because the crystal structure of isolated GCase showed that the active site is easily accessible to solution compounds (23). Alternatively, interactions with sapC and the bilayer leading to an altered configuration of the active site may account for the enhanced activity. Overall, these data provide a view of hydrolytically active sapC/GCase complexes at membrane surfaces and clarify the role of sapC in the treatment of Gaucher's disease by enzyme replacement therapy.
Materials and Methods
For descriptions of materials, cloning, expression, protein purification, fluorescence labeling, liposome preparation, AFM, and confocal epifluorescence imaging, refer to SI Text. Other procedures are described below.
Lipid Compositions.
A mixture of 1,2-diacyl-sn-3-phosphocholine (PC)/cholesterol/PI/1,2-diacyl-sn-3-phosphoethanolamine (PE)/GlcCer (50 mol %/20 mol %/10 mol %/10 mol %/10 mol %) was used to mimic lysosomal lipid composition in the experiment shown in Fig. 2. In the experiments shown in Fig. 1, a mixture of 1,2-dipalmitoyl-sn-3-phosphocholine/phosphatidylcholine/cholesterol/PI/phosphatidylethanolamin/GlcCer (25 mol %/25 mol %/20 mol %/10 mol %/10 mol %/10 mol %) was used. A mixture consisting of PC/cholesterol/PI/PE/DFUG (50 mol %/20 mol %/10 mol %/10 mol %/10 mol %) was used in the experiments shown in Figs. 3–5.
Liposome-Based GCase Activity Assay.
In this assay, GCase hydrolizes membrane-embedded DFUG to yield DFHU, which has a fluorescence emission peak at λmax = 450 nm. DFUG-containing liposomes were prepared as described above in 50 mM sodium acetate, pH 4.8/150 mM NaCl for use in the assay. Reaction mixtures were 150 μl in volume and contained 100 μM total lipids. The liposomes in each reaction were preincubated for 15 min with 0–2.5 μM sapC, as indicated. The reactions were performed at 20°C. The DFHU fluorophore was exited at λ = 330 nm, and maximum emission was sampled at 0.5-s intervals over a 10-min period using a Photon Technology International (Birmingham, NJ) QM-1 fluorescence spectrophotometer. Baseline fluorescence values were recorded for 1 min before the addition of 0.05 μM GCase to each reaction sample. Initial rate values were calculated by linear regression of the first 15 points, after GCase addition and mixing. Standard deviations were calculated from residual χ2 values.
FRET.
FRET was detected as an increase in donor fluorescence intensity after acceptor photobleaching (26, 45). A488-GCase/A546-sapC and DFHU/A488-GCase were used as FRET donor/acceptor pairs in the experiment shown in Fig. 5. The lipid bilayer containing 10 mol % DFUG (membrane-anchored, fluorogenic GCase substrate). To confirm FRET, a control experiment was performed with unlabeled sapC (SI Fig. 11). Consecutive A488-GCase/A546-sapC and DFHU/A488-GCase FRET experiments were performed by photobleaching within a 25 × 25-μm area A546-sapC followed by A488-GCase using intense HeNe 543- and Ar-ion 488-nm lasers, respectively. Images in the DFHU, A488-Gcase, and A546-sapC channels of a 70 × 70-μm area centered around the photobleached square were acquired before and after each photobleaching event. FRET efficiency, E, was calculated by using the equation E = 1 − Db/Da, where Db and Da are the donor fluorescence intensities before and after acceptor photobleaching, respectively (26). Db and Da were quantified from raw images in a 20 × 20-μm square within the photobleached area.
Supplementary Material
Acknowledgments
We thank Tim Edmunds for support and Genzyme for a gift of Cerezyme. This work was supported by a grant from the Canadian Institutes of Health Research (to G.G.P. and C.M.Y.).
Abbreviations
- sapC
saposin C
- A488
Alexa Fluor 488
- A546-sapC
A546-labeled sapC
- AFM
atomic force microscopy
- GCase
acid β-glucosidase
- GlcCer
glucosylceramide
- DFUG
6,8-difluoro-4-heptadecylumbelliferyl β-d-glucopyranoside
- DFHU
6,8-difluoro-4-heptadecylumbelliferyl β-d-glucopyranoside
- MUG
4-methylumbelliferyl-β-d-glucopyranoside
- PI
1,2-diacyl-sn-3-phosphoinositol.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. K.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704998104/DC1.
References
- 1.Futerman AH, van Meer G. Nat Rev Mol Cell Biol. 2004;5:554–565. doi: 10.1038/nrm1423. [DOI] [PubMed] [Google Scholar]
- 2.Kolter T, Sandhoff K. Annu Rev Cell Dev Biol. 2005;21:81–103. doi: 10.1146/annurev.cellbio.21.122303.120013. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski GA, Barton NW, Pastores G, Dambrosia JM, Banerjee TK, McKee MA, Parker C, Schiffmann R, Hill SC, Brady RO. Ann Intern Med. 1995;122:33–39. doi: 10.7326/0003-4819-122-1-199501010-00005. [DOI] [PubMed] [Google Scholar]
- 4.Sandhoff K, Kolter T. Trends Cell Biol. 1996;6:98–103. doi: 10.1016/0962-8924(96)80999-8. [DOI] [PubMed] [Google Scholar]
- 5.Vaccaro AM, Salvioli R, Tatti M, Ciaffoni F. Neurochem Res. 1999;24:307–314. doi: 10.1023/a:1022530508763. [DOI] [PubMed] [Google Scholar]
- 6.Winau F, Schwierzeck V, Hurwitz R, Remmel N, Sieling PA, Modlin RL, Porcelli SA, Brinkmann V, Sugita M, Sandhoff K, et al. Nat Immunol. 2004;5:169–174. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- 7.Zhou D, Cantu C, III, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, et al. Science. 2004;303:523–527. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang SJ, Cresswell P. Nat Immunol. 2004;5:175–181. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- 9.Yuan W, Qi X, Tsang P, Kang SJ, Illarionov PA, Besra GS, Gumperz J, Cresswell P. Proc Natl Acad Sci USA. 2007;104:5551–5556. doi: 10.1073/pnas.0700617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ahn VE, Faull KF, Whitelegge JP, Higginson J, Fluharty AL, Privé GG. Protein Expr Purif. 2003;27:186–193. doi: 10.1016/s1046-5928(02)00597-1. [DOI] [PubMed] [Google Scholar]
- 11.Sandhoff K, Kolter T. Philos Trans R Soc London Ser B. 2003;358:847–861. doi: 10.1098/rstb.2003.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilkening G, Linke T, Sandhoff K. J Biol Chem. 1998;273:30271–30278. doi: 10.1074/jbc.273.46.30271. [DOI] [PubMed] [Google Scholar]
- 13.Vaccaro AM, Ciaffoni F, Tatti M, Salvioli R, Barca A, Tognozzi D, Scerch C. J Biol Chem. 1995;270:30576–30580. doi: 10.1074/jbc.270.51.30576. [DOI] [PubMed] [Google Scholar]
- 14.de Alba E, Weiler S, Tjandra N. Biochemistry. 2003;42:14729–14740. doi: 10.1021/bi0301338. [DOI] [PubMed] [Google Scholar]
- 15.Alattia JR, Shaw JE, Yip CM, Privé GG. J Mol Biol. 2006;362:943–953. doi: 10.1016/j.jmb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 16.You HX, Yu L, Qi X. FEBS Lett. 2001;503:97–102. doi: 10.1016/s0014-5793(01)02700-4. [DOI] [PubMed] [Google Scholar]
- 17.You HX, Qi X, Grabowski GA, Yu L. Biophys J. 2003;84:2043–2057. doi: 10.1016/S0006-3495(03)75012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.You HX, Qi X, Yu L. Chem Phys Lipids. 2004;132:15–22. doi: 10.1016/j.chemphyslip.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 19.Grandbois M, Clausen-Schaumann H, Gaub H. Biophys J. 1998;74:2398–2404. doi: 10.1016/S0006-3495(98)77948-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Six DA, Dennis EA. Biochim Biophys Acta. 2000;1488:1–19. doi: 10.1016/s1388-1981(00)00105-0. [DOI] [PubMed] [Google Scholar]
- 21.Dessen A, Tang J, Schmidt H, Stahl M, Clark JD, Seehra J, Somers WS. Cell. 1999;97:349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 22.van Tilbeurgh H, Bezzine S, Cambillau C, Verger R, Carriere F. Biochim Biophys Acta. 1999;1441:173–184. doi: 10.1016/s1388-1981(99)00149-3. [DOI] [PubMed] [Google Scholar]
- 23.Dvir H, Harel M, McCarthy AA, Toker L, Silman I, Futerman AH, Sussman JL. EMBO Rep. 2003;4:704–709. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahn VE, Leyko P, Alattia JR, Chen L, Privé GG. Protein Sci. 2006;15:1849–1857. doi: 10.1110/ps.062256606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Förster T. Annal Phys. 1948;2:55–75. [Google Scholar]
- 26.Berney C, Danuser G. Biophys J. 2003;84:3992–4010. doi: 10.1016/S0006-3495(03)75126-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salvioli R, Tatti M, Scarpa S, Moavero SM, Ciaffoni F, Felicetti F, Kaneski CR, Brady RO, Vaccaro AM. Biochem J. 2005;390:95–103. doi: 10.1042/BJ20050325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liou B, Kazimierczuk A, Zhang M, Scott CR, Hegde RS, Grabowski GA. J Biol Chem. 2006;281:4242–4253. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 29.Vaccaro AM, Tatti M, Salvioli R, Ciaffoni F, Gallozzi E. Biochim Biophys Acta. 1990;1033:73–79. doi: 10.1016/0304-4165(90)90196-4. [DOI] [PubMed] [Google Scholar]
- 30.Sa Miranda MC, Aerts JM, Pinto RA, Magalhaes JA, Barranger JA, Tager JM, Schram AW. Biochim Biophys Acta. 1988;965:163–168. doi: 10.1016/0304-4165(88)90052-9. [DOI] [PubMed] [Google Scholar]
- 31.Aerts JM, Sa Miranda MC, Brouwer-Kelder EM, Van Weely S, Barranger JA, Tager JM. Biochim Biophys Acta. 1990;1041:55–63. doi: 10.1016/0167-4838(90)90122-v. [DOI] [PubMed] [Google Scholar]
- 32.Qi X, Grabowski GA. Biochemistry. 1998;37:11544–11554. doi: 10.1021/bi980785+. [DOI] [PubMed] [Google Scholar]
- 33.Vaccaro AM, Tatti M, Ciaffoni F, Salvioli R, Maras B, Barca A. FEBS Lett. 1993;336:159–162. doi: 10.1016/0014-5793(93)81631-9. [DOI] [PubMed] [Google Scholar]
- 34.Ho MW. FEBS Lett. 1975;53:243–247. doi: 10.1016/0014-5793(75)80029-9. [DOI] [PubMed] [Google Scholar]
- 35.Berent SL, Radin NS. Biochim Biophys Acta. 1981;664:572–582. doi: 10.1016/0005-2760(81)90134-x. [DOI] [PubMed] [Google Scholar]
- 36.Kobayashi T, Beuchat MH, Chevallier J, Makino A, Mayran N, Escola JM, Lebrand C, Cosson P, Gruenberg J. J Biol Chem. 2002;277:32157–32164. doi: 10.1074/jbc.M202838200. [DOI] [PubMed] [Google Scholar]
- 37.Locatelli-Hoops S, Remmel N, Klingenstein R, Breiden B, Rossocha M, Schoeniger M, Koenigs C, Saenger W, Sandhoff K. J Biol Chem. 2006;281:32451–32460. doi: 10.1074/jbc.M607281200. [DOI] [PubMed] [Google Scholar]
- 38.Remmel N, Locatelli-Hoops S, Breiden B, Schwarzmann G, Sandhoff K. FEBS J. 2007;274:3405–3420. doi: 10.1111/j.1742-4658.2007.05873.x. [DOI] [PubMed] [Google Scholar]
- 39.Hiraiwa M, Soeda S, Martin BM, Fluharty AL, Hirabayashi Y, O'Brien JS, Kishimoto Y. Arch Biochem Biophys. 1993;303:326–331. doi: 10.1006/abbi.1993.1291. [DOI] [PubMed] [Google Scholar]
- 40.Qi X, Leonova T, Grabowski GA. J Biol Chem. 1994;269:16746–16753. [PubMed] [Google Scholar]
- 41.Whitelegge JP, Ahn V, Norris AJ, Sung H, Waring A, Stevens RL, Fluharty CB, Privé G, Faull KF, Fluharty AL. Cell Mol Biol. 2003;49:799–807. [PubMed] [Google Scholar]
- 42.Brumshtein B, Wormald MR, Silman I, Futerman AH, Sussman JL. Acta Crystallogr D. 2006;62:1458–1465. doi: 10.1107/S0907444906038303. [DOI] [PubMed] [Google Scholar]
- 43.Premkumar L, Sawkar AR, Boldin-Adamsky S, Toker L, Silman I, Kelly JW, Futerman AH, Sussman JL. J Biol Chem. 2005;280:23815–23819. doi: 10.1074/jbc.M502799200. [DOI] [PubMed] [Google Scholar]
- 44.Qi X, Kondoh K, Krusling D, Kelso GJ, Leonova T, Grabowski GA. Biochemistry. 1999;38:6284–6291. doi: 10.1021/bi990079o. [DOI] [PubMed] [Google Scholar]
- 45.McLean PJ, Kawamata H, Ribich S, Hyman BT. J Biol Chem. 2000;275:8812–8816. doi: 10.1074/jbc.275.12.8812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.