Abstract
Plasmacytoid dendritic cells (pDC) are key players in viral immunity and produce IFN-α after HIV-1 exposure, which in turn regulates TNF-related apoptosis-inducing ligand (TRAIL) expression by CD4+ T cells. We show here that infectious and noninfectious HIV-1 virions induce activation of pDC into TRAIL-expressing IFN-producing killer pDC (IKpDC). IKpDC expressed high levels of activation markers (HLA-DR, CD80, CD83, and CD86) and the migration marker CCR7. Surprisingly, CXCR4 and CCR5 were down-regulated on IKpDC. We also show that HIV-1-induced IKpDC depended on Toll-like receptor 7 (TLR7) activation. HIV-1 or TLR7 agonistexposed IKpDC induced apoptosis of the CD4+ T cell line SupT1 via the TRAIL pathway. Furthermore, IFN-α produced after HIV-induced TLR7 stimulation was responsible for TRAIL expression and the down-regulation of both CXCR4 and CCR5 by IKpDC. In contrast, activation and migration markers were not regulated by IFN-α. Finally, IFN-α increased the survival of IKpDC. We characterized a subset of pDC with a killer activity that is activated by endosomal-associated viral RNA and not by infection.
Keywords: apoptosis, endocytosis, AT-2 HIV-1, CCR5, CXCR4
Plasmacytoid dendritic cells (pDC) are immature dendritic cells (DC) that participate in both innate and adaptative immunity (1, 2). pDC are specialized cells located in blood and lymphoid organs (2, 3) that produce up to 1,000-fold more IFN-α than other cell types in response to virus exposure (4). Viral activation of pDC can be regulated by either of two Toll-like receptors (TLR), TLR7 or TLR9 (5), which are considered to be the receptors that human pDC use for recognition of RNA/retroviruses (6, 7) and DNA (8), respectively.
The role of pDC and IFN-α in HIV-1 infection is not fully understood. Type I IFNs (IFN-α/β) provide essential innate immunity against viruses (3) and inhibit HIV-1 replication in vitro (9). The number of circulating pDC is decreased in HIV-1 infection (10), and the lack of IFN-α production was suggested to be responsible for HIV-1 disease progression (11, 12). In contrast, high plasma titers of IFN-α are found during acute HIV-1 infection and reappear during late-stage disease (13). Induction of type I IFNs could be a double-edged sword and might exert pathogenic in addition to protective effects in innate immunity. We previously reported that one consequence of IFN-α production by HIV-1-stimulated pDC is the expression of TNF-related apoptosis-inducing ligand (TRAIL) by monocytes (14). Furthermore, isolated pDC cultured with infectious or noninfectious HIV-1 particles produced large amounts of IFN-α (15, 16) that induced TRAIL expression by primary CD4+ T cells (17). TRAIL was shown to be involved in the selective induction of apoptosis in uninfected CD4+ T cells in both a human in vitro model (18) and an animal model using HIV-infected hu-PBL-NOD-SCID mice (19). We recently reported that the TRAIL/DR5 pathway contributed to selective apoptosis of CD4+ T cells in vitro and that levels of TRAIL and the percentages of CD4+ T cells expressing DR5 were elevated in blood of untreated HIV-infected patients (20). Recently, a report showed that influenza virus A regulated TRAIL expression by a human pDC cell line (GEN2.2), which became cytotoxic and induced apoptosis of a melanoma cell line (21). Because we have shown that HIV-1-exposed CD4+ T cells are sensitive to TRAIL-induced apoptosis (20), we questioned whether HIV-1 would induce TRAIL expression by pDC, resulting in apoptosis of CD4+ T cells.
We show in this study that HIV-1 induced expression of TRAIL and the activation and migration markers CD83 and CCR7 and turned pDC into IFN-producing killer pDC (IKpDC). By analyzing the expression of HIV-1 receptors on IKpDC, we found that the two major coreceptors of HIV-1, CXCR4 and CCR5, were down-regulated by aldrithiol-2 (AT-2) HIV-1 exposure. The transformation from pDC to IKpDC occurred through TLR7 after endocytosis of HIV-1 virions. In addition, IKpDC induced apoptosis of a CD4+ T cell line via the TRAIL pathway. Furthermore, we found that IFN-α was responsible for TRAIL expression and decreased CXCR4 and CCR5 on IKpDC, although the activation and migration markers were not regulated by IFN-α. Finally, we showed that stimulation of TLR7 induced survival of pDC and the production of TNF-α by IKpDC, a hallmark of immune cell activation. This study therefore identifies a subset of human DC with cytotoxic activity in HIV-1 infection, which may contribute to immunopathogenesis.
Results
Characterization of pDC After Exposure to AT-2 HIV-1.
Enriched pDC were cultured with noninfectious AT-2 HIV-1MN (CXCR4 coreceptor-specific) or AT-2 HIV-1ADA (CCR5 coreceptor-specific) particles or negative control microvesicles (mock) overnight. We tested TRAIL expression induced by these AT-2 HIV-1 particles on CD123+ BDCA2+ cells, markers that we used to define pDC (Fig. 1A). After AT-2 HIV-1 exposure, 50 ± 5% of pDC were TRAIL+ compared with 0.6 ± 0.5% (P = 0.006) when treated with microvesicles (pDCmock) (Fig. 1A), and the mean fluorescence intensity (MFI) was also increased in AT-2 HIV-treated pDC (MFI = 65 ± 19) compared with pDCmock (MFI = 15 ± 9; P = 0.003). The MFI of pDCmock was identical to pDC stained with isotype control (MFI = 15 ± 3) [supporting information (SI) Fig. 5A]. We also tested the effect of infectious HIV-1MN and HIV-1ADA, and we obtained similar results on TRAIL expression by pDC (SI Fig. 5A). We therefore define these TRAIL-expressing pDC as TRAIL+pDCHIV.
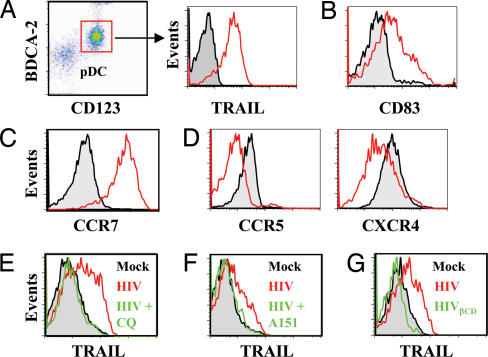
Fig. 1.
Characterization of pDC after treatment with AT-2 HIV-1. (A) Dot plot of BDCA4+ cells. pDC are defined as CD123+BDCA2+ cells. Shown is TRAIL expression on pDC cultured with microvesicles (pDCmock, black) compared with AT-2 HIV-1 (pDCHIV, red). (B) Increased CD83 expression on TRAIL+pDCHIV (red) compared with pDCMock (black). (C) Increased CCR7 expression on TRAIL+pDCHIV (red) compared with pDCMock (black). (D) Reduced expression of CCR5 on TRAIL+pDCHIV (red) compared with pDCMock (black). Shown is reduced expression of CXCR4 on TRAIL+pDCHIV (red) compared with pDCMock (black). (E) TRAIL expression on pDCmock (black) or pDCHIV (red) and pDCHIV plus CQ (green). (F) TRAIL expression on pDCmock (black) or pDCHIV (red) and pDCHIV plus A151 (green). (G) TRAIL expression on pDCmock (black), pDCHIV (red), and HIVβCD (green).
We investigated the expression of activation markers on pDC exposed to AT-2 HIV-1. CD83 was up-regulated on TRAIL+pDCHIV compared with pDCmock (P = 0.03) (Fig. 1B), as were CD80 and CD86 (P = 0.004 and P = 0.0001) (SI Fig. 5B). Similarly, the CCR7 migration marker was also increased after AT-2 HIV-1 exposure, and 97 ± 4% of TRAIL+pDCHIV were CCR7+ (Fig. 1C and SI Fig. 5C). Because the HIV-1 coreceptors CCR5 and CXCR4 are decreased on pDC of HIV+ patients (22) we tested for their expression in this system. We tested both AT-2 HIV-1MN (CXCR4 tropic) and AT-2 HIV-1ADA on coreceptor expression and obtained identical results. AT-2 HIV-1 induced down-regulation of both CCR5 and CXCR4 on TRAIL+pDCHIV (Fig. 1D). We also observed data similar to those shown in Fig. 1 A–F using the infectious counterparts of both AT-2 HIV-1MN and HIV-1ADA (SI Fig. 5 A and D).
Generation of TRAIL+pDCHIV Occurs After Endosomal Acidification and Is Mediated by TLR.
Recently, pDC activation by AT-2 HIV-1 was shown to occur by TLR stimulation after endocytosis (6). To determine whether endocytosis is required for AT-2 HIV-1 induction of TRAIL+pDCHIV, we cultured pDC with AT-2 HIV-1 and the endosomal acidification inhibitor chloroquine (CQ) and looked for activation by staining for TRAIL. CQ inhibited TRAIL expression by 97 ± 8% (P = 0.005) (Fig. 1E) compared with cultures in the absence of CQ.
Because endocytosis is required for AT-2 HIV-1 activation of pDC, we investigated the intracellular receptors responsible for activation. TLR7 and TLR9 are intracellular receptors of the innate immune system that are present in pDC (23). To demonstrate the role of TLR in this system, we added the suppressive oligodinucleotide A151 to pDC cultured with AT-2 HIV-1. A151, also known as (TTAGGG)4, is a competitive inhibitor of TLR stimuli and has been shown to inhibit the activity of TLR7- or TLR9-transfected HEK293 cells (6). A151 significantly inhibited TRAIL expression on pDC cultured with AT-2 HIV-1 by 74 ± 15% (P = 0.02) compared with TRAIL+pDCHIV (Fig. 1F).
Because TLR stimulation is required for the generation of TRAIL+pDCHIV and TLR7 is used by human pDC for recognition of ssRNA (7, 24), we tested whether HIV-1 surface proteins or ssRNA were responsible for activation of pDC. We depleted ssRNA from HIV-1 particles by treatment with 80 mM β-cyclodextrin (HIVβCD), a chemical modification that induces a hole in the virion capsid, liberating ssRNA but retaining capsid proteins as previously shown (25). These RNA-free particles were used for the culture of pDC (SI Fig. 5G), as we had done with AT-2 HIV-1. RNA-free HIV-1 particles did not induce TRAIL expression on pDC (Fig. 1G). Similarly, HIVβCD did not induce CXCR4 or CCR5 down-regulation on pDC (data not shown). Taken together, these data indicate that TLR7 stimulation by viral RNA is required for the activation of pDC.
Role of TLR7 for Induction of TRAIL+pDC.
We tested whether TLR7 expression in pDC from 21 HIV-infected patients (HIV+) was different from pDC from 19 healthy control blood donors (HIV−). Peripheral blood mononuclear cells from HIV− and HIV+ individuals were tested for intracellular expression of TLR7 in pDC. We found that circulating pDC from both HIV− and HIV+ constitutively expressed TLR7. However, TLR7 expression was elevated in HIV+ patients compared with HIV− controls (MFI = 487 ± 58 and MFI = 99 ± 14 respectively; P = 0.001). This result establishes a parallel between TLR7 expression in our in vitro AT-2 HIV-1 data and in HIV+ patients.
To determine the effects of TLR7 stimulation in vitro, pDC isolated from peripheral blood mononuclear cells of healthy blood donors were cultured overnight with imiquimod (Imiq), an imidazoquinoline known to stimulate TLR7 but not TLR9 (26). TRAIL activation and migration markers were analyzed as above in the AT-2 HIV-1 experiments. TLR7 stimulation by Imiq induced TRAIL on 92 ± 6% of pDC (Fig. 2B). TRAIL+pDC generated by Imiq stimulation (TRAIL+pDCImiq) expressed high levels of CD83 and CCR7 (Fig. 2C), as well as HLA-DR, CD80, and CD86 (data not shown), compared with pDCmock. We also tested the effect of Imiq on HIV-1 coreceptors and observed that Imiq induced a marked down-regulation of both CXCR4 and CCR5 (Fig. 2D), similar to the findings above for AT-2 HIV-1. Therefore, the TRAIL+pDC generated by either AT-2 HIV-1 activation or a known TLR7 stimulator resulted in cells expressing identical surface markers.
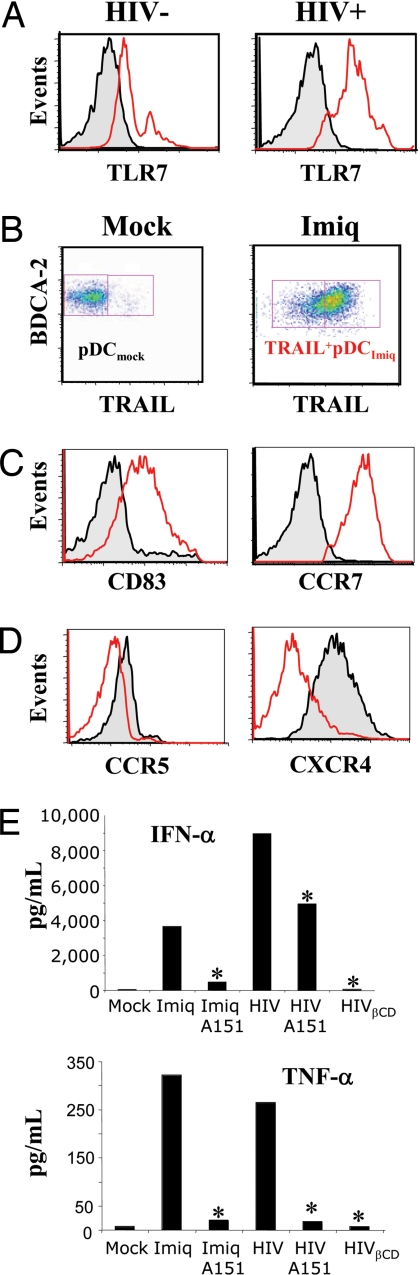
Fig. 2.
Role of TLR7 in IKpDC activation. (A) Increase of TLR7 expression in pDC of HIV+ patients (n = 21) compared with HIV− controls (n = 19) (MFI = 487 ± 58 and MFI = 99 ± 14, respectively; P = 0.001). (B) TRAIL expression on pDCmock (Left) compared with Imiq-stimulated pDC (pDCImiq) (Right). (C) Expression of CD83 (Left) and CCR7 (Right) by TRAIL+pDCImiq (red) compared with pDCmock (black). (D) Down-regulation of CCR5 (Left) and CXCR4 (Right) on TRAIL+pDCImiq (red) compared with pDCmock (black). (E) IFN-α and TNF-α production and inhibition with A151. *, P < 0.05, two-tailed Student's t test.
Finally, we tested whether IFN-α and TNF-α production (secretion of both are associated with activated pDC) were TLR-dependent. As shown in Fig. 2E, the TLR antagonist A151 inhibited >90% of IFN-α and TNF-α production by Imiq-stimulated pDC. TNF-α produced by AT-2 HIV-exposed pDC was also strongly reduced by A151 (95 ± 12% and 96 ± 7% inhibition; P = 0.002 and P = 0.01, respectively). However, IFN-α production by AT-2 HIV-activated pDC was partially reduced by A151 (46 ± 9% inhibition; P = 0.04).
TRAIL+pDC Induce Apoptosis of Target Cells.
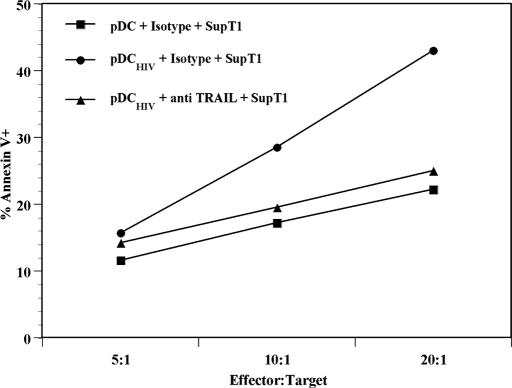
To determine whether the HIV-induced TRAIL expressed by pDC was functional, we performed cytotoxicity assays with purified pDC as effector cells and the TRAIL-sensitive SupT1 CD4+ T cell line as target cells (27, 28). Purified pDC were cultured with microvesicles (pDCmock) or AT-2 HIV-1 (pDCHIV) overnight. The supernatants were removed, and cells were washed. pDCmock or pDCHIV were then cocultured with the SupT1 cells at different effector:target cell ratios for 6 h, and apoptosis was tested by measuring annexin V on CD123−BDCA2−-gated cells (SI Fig. 6). The pDC did not express DR5, even when cultured with AT-2 HIV-1 (data not shown). pDCmock induced a baseline level of apoptosis in SupT1 cells at each effector:target combination (Fig. 3). In contrast, pDCHIV induced increased levels of annexin V expression in SupT1 cells that were proportional to increasing effector:target cell ratios. Furthermore, pDCHIV preincubated with a neutralizing anti-TRAIL antibody and cultured with SupT1 cells showed reduced expression of annexin V on SupT1 cells. These results demonstrate that AT-2 HIV-1 activation results in killer function of TRAIL+pDC and that the increase in the observed death is mediated by the TRAIL apoptotic pathway.
Fig. 3.
TRAIL+pDC kill DR5+ T cell targets. pDC stimulated with microvesicles (pDCMock) or AT-2 HIV-1 (pDCHIV) were used as effector cells against SupT1 target cells at various ratios in the absence or presence of a neutralizing anti-TRAIL antibody or matched isotype controls. Cytotoxicity was measured as the percentage of annexin V+ cells on the BDCA2−CD123− cell population. Data are presented as the mean of three experiments ± 95% CI.
IFN-α Regulates TRAIL, CXCR4, and CCR5 on TRAIL+pDC.
Because we and others previously demonstrated that AT-2 HIV-1 induced high levels of IFN-α by pDC (15–17), we tested whether IFN-α plays a role in the regulation of TRAIL on TRAIL+pDC. Additionally, because both CXCR4 and CCR5 are down-regulated on pDC of HIV+ individuals compared with healthy controls (22), and CXCR4 is down-modulated in response to cytokines (29), we tested whether IFN-α played a role in the down-regulation of CXCR4 and CCR5 observed on TRAIL+pDC. pDC were therefore cultured overnight with microvesicles, AT-2 HIV-1, or Imiq in the absence (−Anti-IFN) or presence (+Anti-IFN) of antibodies against IFN-α (Fig. 4A). TRAIL expression was significantly reduced when pDC were cultured with AT-2 HIV-1 or Imiq in the presence of IFN-α antibodies [67 ± 11% (P = 0.02) and 79 ± 19% inhibition (P = 0.003), respectively] compared with cultures without antibodies. Furthermore, the addition of neutralizing IFN-α antibodies abrogated the effect of AT-2 HIV-1 and Imiq on CXCR4 down-regulation (Fig. 4B Left). Similar results were obtained on CCR5 expression (Fig. 4B Right). To confirm that IFN-α was responsible for the down-regulation of CXCR4 and CCR5, we cultured pDC overnight with recombinant IFN-α. Both CXCR4 and CCR5 expression on pDC were significantly reduced by recombinant IFN-α compared with untreated pDC [82 ± 23% (P = 0.0001) and 71 ± 17% (P = 0.002) inhibition, respectively] (Fig. 4C).
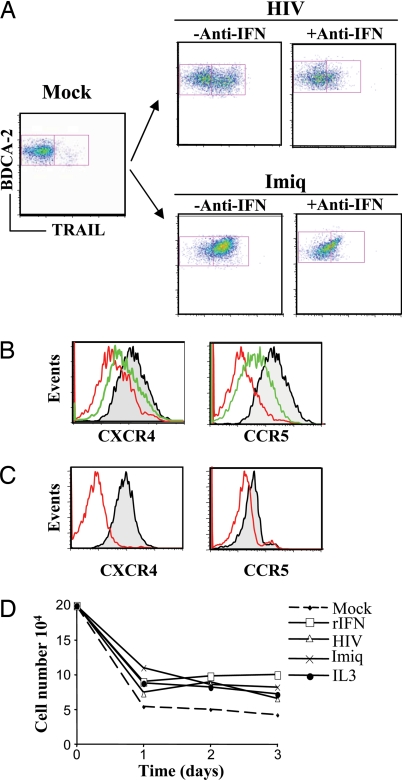
Fig. 4.
IFN-α regulation of TRAIL, CXCR4, and CCR5 by IKpDC. (A) TRAIL expression on pDC cultured overnight with AT-2 HIV-1 (Upper) or Imiq (Lower) in the absence (−Anti-IFN) or presence (+Anti-IFN) of antibodies against IFN-α. (B Left) Expression of CXCR4 on pDCMock (black), pDCHIV (red), and pDCHIV plus anti-IFN-α (green). (B Right) Expression of CCR5 on pDCMock (black), pDCHIV (red), and pDCHIV plus anti-IFN-α (green). (C) Down-regulation of CXCR4 (Left) and CCR5 (Right) by recombinant IFN-α. (D) Survival of isolated pDC in the presence of different stimuli during a 3-day period.
Because HIV-1 was associated with a decrease of pDC numbers in patients' blood (10), it was suggested that HIV-1 induced the death of pDC (12). Therefore, we tested the effect of IFN-α, AT-2 HIV-1, and Imiq on pDC survival. Isolated pDC from healthy blood donors were cultured with recombinant IFN-α, AT-2 HIV-1, Imiq, or IL-3 (positive control) for 3 days, and cells were counted at 24-h intervals. IFN-α, AT-2 HIV-1, Imiq, and IL-3 each increased the survival of pDC compared with mock-treated pDC after 2 and 3 days of culture (Fig. 4D). We also performed a cell proliferation assay. Isolated pDC were cultured overnight in the presence of infectious HIV, AT-2 HIV, IFN-α, and IL-3. We found that infectious HIV, AT-2 HIV, and IFN-α were not cytotoxic for pDC and induced a metabolic activation similar to IL-3 (SI Fig. 7). These data suggest that neither HIV-1 nor IFN-α exposure contributed to the death of pDC.
Discussion
The present article describes and identifies a unique subset of human pDC that possess killer activity. This killer activity was effected by TRAIL, a cytotoxic molecule that induces apoptosis of DR4+ and/or DR5+ target cells (30, 31). We show here that exposure of isolated pDC to either infectious or noninfectious HIV-1 induced TLR7-mediated IFN-α-dependent expression of TRAIL. These TRAIL-expressing pDC induced TRAIL-sensitive target cell apoptosis that was proportional to the pDC:target cell ratio. Therefore, we define this HIV-activated, TRAIL-expressing pDC as IKpDC.
We recently reported that the TRAIL/DR5 pathway contributed to HIV-induced selective apoptosis of CD4+ T cells in vitro (20) and that ex vivo apoptosis of HIV-infected patients' CD4+ T cells were blocked by an antibody against DR5 (20). Our earlier studies indicated that exposure of monocytes or CD4+ T cells from HIV− blood bank donors to infectious or noninfectious HIV-1 resulted in production of soluble TRAIL by monocytes and in membrane expression of TRAIL by both monocytes and CD4+ T cells (14, 17). Furthermore, the blood of HIV+ patients contained levels of plasma TRAIL and DR5-expressing CD4+ T cells that were directly (TRAIL) and inversely (DR5) associated with plasma viral load (20).
More than 99% of HIV-1 particles detected in the circulation are not productively infectious (32) but, as previously suggested, might contribute to HIV-induced immunopathogenesis (20, 33). For this reason, our investigations have included the effects of noninfectious AT-2 HIV-1 on TRAIL-mediated apoptosis of CD4+ T cells (20). The results of the present study that noninfectious HIV-1 particles induced IKpDC are consistent with those earlier reports, indicating that AT-2 HIV-1 activates resting pDC to produce IFN-α, resulting in TRAIL expression on CD4+ T cells in the absence of productive HIV infection (17). Our finding here that HIV-1 can induce IKpDC adds another cellular source of TRAIL to those mentioned above. However, it should be noted that IKpDC differ from HIV-induced, TRAIL-expressing CD4+ T cells in that T cells express both DR5 and TRAIL and therefore are themselves susceptible to TRAIL/DR5-mediated apoptosis. In contrast, because IKpDC express TRAIL but not DR5, this population of killer cells should not be susceptible to TRAIL/DR5-mediated apoptosis and could serve as an apoptosis-resistant population of killer cells.
Lymphoid tissues are major sites of CD4+ T cell depletion (34–36). Because CD4+ T cells expressing DR5 were elevated in lymphoid tissue (37), it is possible that IKpDC that express the migration marker CCR7 contribute to CD4+ T cell death by inducing apoptosis via the TRAIL pathway in lymphoid organs. Consistent with this hypothesis is the finding that AT-2 HIV-activated pDC expressed CCR7 and migrated to CCL19 in a transwell experiment (15). CCL19 is a chemokine that is found in lymphoid compartments and is the ligand of CCR7. These results can account for the fact that a high percentage of CD4+ T cells die in the lymphoid tissues during the early and late stages of HIV-1 disease (34, 38). In addition to CCR7, IKpDC also expressed the activation markers CD80, CD83, CD86, and HLA-DR, as was reported for activated pDC (15).
Interestingly, a novel TRAIL-expressing DC, IFN-producing killer DC, was recently described in a murine tumor model in which TRAIL expression was controlled by IFN-γ and the resulting DC expressed NK markers (39). In contrast, we did not detect the NK marker CD56 on IKpDC or IFN-γ production by these cells. It remains to be determined whether this difference is due to murine IFN-producing killer DC versus human IKpDC or whether it is due to a difference between tumor and viral stimuli. The answer to this question may reside in the nature of the stimulus, because influenza activated TRAIL expression by a human pDC cell line that is regulated by type I and not type II IFN (21).
TLR7 and TLR9 are sensor molecules of the innate immune system that recognize nucleic acid moieties of microbiological infectious agents (24, 40). Because TLR7 and TLR9 are not expressed on the surface of pDC, stimuli must gain access to intracellular compartments. A recent report showed that AT-2 HIV-1 enters pDC by endocytosis and stimulates TLR7 and possibly TLR9 to induce IFN-α (6). We found that CQ inhibited HIV-induced TRAIL expression on IKpDC and prevented HIV-induced loss of CXCR4 expression on IKpDC. These data indicate that endocytosis is required for these HIV-induced effects but do not distinguish whether the sensor molecule involved was TLR7 and/or TLR9. We used the TLR-stimulating agent Imiq, which is specific for TLR7, and demonstrated that it had the same effect as HIV-1 for inducing the up-regulation of TRAIL, CCR7, IFN-α, TNF-α, CD80, CD83, and CD86 and in the down-regulation of CXCR4 and CCR5 expression. In addition, the effect of Imiq on these parameters was abrogated by A151. Finally, we demonstrated that removal of viral RNA from HIV-1 particles with β-CD resulted in complete loss of the ability of these RNA-depleted HIV-1 particles to induce TRAIL expression by pDC (25). These results demonstrate that TLR7 is the only TLR that is required for HIV-induced transformation of pDC into IKpDC. Interestingly, pDC from HIV-1-infected patients express higher levels of TLR7 than pDC from healthy individuals, which is consistent with our in vitro findings. These results do not exclude the fact that TLR9 stimulation by CpG can induce activation of IKpDC (data not shown); however, in our model TLR9 is not responsible for pDC activation by HIV-1.
The major producers of IFN-α after viral infection are pDC (2, 3), and type I IFN has been shown to have antiviral activity against several viruses, including HIV-1 (41, 42). Furthermore, IFN-α induced TRAIL-mediated apoptosis of virus-infected cells in a murine model (43), and this mechanism was suggested to kill HIV-infected CD4+ T cells (44). However, the role of IFN-α in HIV-1 infection and pathogenesis is complex and controversial (45). Thus, high plasma titers of IFN-α are found during acute HIV-1 infection and reappear during late-stage disease as an indicator of poor clinical prognosis (13). Furthermore, HIV-1 disease progression was inhibited in AIDS patients immunized against IFN-α (46). In addition, we reported that a similar mechanism involving TRAIL/DR5-mediated apoptosis of CD4+ T cells is induced by IFN-α that is produced by AT-2 HIV-1-activated pDC (20). This immunopathogenic mechanism, which kills uninfected CD4+ T cells, would be favored in a setting in which most in situ HIV-1 is noninfectious, resulting in death of a disproportionately high percentage of uninfected T helper cells (20). Our finding here that infectious and noninfectious HIV-1 induces IFN-α, which activates TRAIL expression on IKpDC that kill DR5-expressing uninfected CD4+ T cells, provides another mechanistic example of IFN-α-induced immunopathogenesis.
We demonstrate here that IFN-α regulates TRAIL expression on IKpDC and is also responsible for HIV-1 coreceptor down-regulation and survival of IKpDC. The facts that both CXCR4 and CCR5 are down-regulated by IFN-α, coupled with the finding that IKpDC exhibited increased survival in an IFN-α-enriched environment, raise the possibility that IKpDC would not themselves be susceptible to HIV-1-induced death and could function as a persistent source of HIV-resistant killers of DR5-expressing target cells. Our data also highlight the ambiguous role of IFN-α. IFN-α up-regulates a cytotoxic molecule (TRAIL) and at the same time down-regulates HIV coreceptors, which may protect the cells from infection. This study provides a role for type I IFN in addition to those previously described for HIV-induced immunopathogenesis and disease progression (45, 47). The role, number, and function of IKpDC in vivo open a new area of DC research in HIV-1 immunopathogenesis and tumor cell biology.
Materials and Methods
Blood was collected from HIV+ patients who were enrolled in a U.S. Air Force natural history protocol at Wilford Hall Medical Center, Lackland Air Force Base (San Antonio, TX), and were not receiving antiretroviral therapy. Blood from HIV-1-seronegative blood bank donors was obtained from the Department of Transfusion Medicine at the National Institutes of Health (Bethesda, MD). All patients were studied under institutional review board-approved protocols and gave written, informed consent on forms from the National Cancer Institute and Wilford Hall Medical Center.
Isolation and Culture of Blood Leukocytes.
Peripheral blood mononuclear cells were isolated by density gradient centrifugation using lymphocyte separation medium (Cambrex, Gaithersburg, MD). The pDC were isolated from healthy donor peripheral blood mononuclear cells by using the BDCA-4 isolation kit (Miltenyi Biotec, Auburn, CA). Cells were cultured in RPMI medium 1640 (Invitrogen, Gaithersburg, MD) containing 10% FBS (HyClone, Logan, UT) and 1% Pen-Strep-Glut (Invitrogen) in the presence of cytokine, infectious HIV-1, or replication-incompetent HIV-1 preparations. A description of the preparation and concentration of stimuli used in this study is included in SI Experimental Methods.
Flow Cytometry.
After stimulation in culture, pDC phenotype was studied by flow cytometry. See SI Experimental Methods for a detailed description of staining procedures and antibodies used.
Cytotoxicity Experiment.
We used human lymphoid T cell line SupT1 (American Type Culture Collection, Manassas, VA) as target cells because SupT1 cells express DR5 but not TRAIL. In a 96-well plate, 5 × 103 target cells per well were incubated with pDC in different effector:target cell ratios of 5:1, 10:1, and 20:1. For inhibition of TRAIL-dependent lysis, 5 μg of monoclonal anti-TRAIL (clone RIK-2; eBioscience) or 5 μg/ml of an IgG1 isotype (eBioscience) was added to target cells 30 min before addition of effector cells. After 6 h of coculture, SupT1 cells were assayed for apoptosis by annexin V by flow cytometry. Briefly, cells were washed in annexin V buffer and stained with annexin V-FITC, CD123-phycoerythrin, and BDCA2-allophycocyanin. Because SupT1 cells do not express CD123 or BDCA2, an acquisition gate was placed on the CD123−BDCA2− population and 2,000 events were captured. Apoptosis was measured as the percentage of annexin V+ cells in the CD123−BDCA2− population. Data are presented as the mean of three independent experiments ± 95% confidence interval.
Statistical Analysis.
Experiments were repeated at least four times. P values were determined by using a two-tailed Student t test. P < 0.05 was considered statistically significant. Univariate distributions of flow cytometric data were performed by probability binning in 300 bins with FlowJo software (48).
Supplementary Material
Acknowledgments
We thank Dr. J. D. Lifson (SAIC-NCI, Frederick, MD) for providing the microvesicles, infectious HIV-1, and AT-2 HIV-1 particles. We thank Dr. Adriano Boasso for a critical reading of the manuscript. We thank the Agence Nationale de Recherches sur le SIDA and the Fondation pour la Recherche Médicale for financial support. This research was supported by the Intramural Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, and by the National Institutes of Health Intramural AIDS Targeted Antiviral Program.
Abbreviations
- TLR
Toll-like receptor
- DC
dendritic cell
- pDC
plasmacytoid DC
- IKpDC
IFN-producing killer pDC
- TRAIL
TNF-related apoptosis-inducing ligand
- CQ
chloroquine
- Imiq
imiquimod
- MFI
mean fluorescence intensity
- AT-2
aldrithiol-2.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0707244104/DC1.
References
- 1.Grouard G, Rissoan MC, Filgueira L, Durand I, Banchereau J, Liu YJ. J Exp Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegal FP, Kadowaki N, Shodell M, Fitzgerald-Bocarsly PA, Shah K, Ho S, Antonenko S, Liu YJ. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 3.Colonna M, Trinchieri G, Liu YJ. Nat Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 4.Kadowaki N, Antonenko S, Lau JY, Liu YJ. J Exp Med. 2000;192:219–226. doi: 10.1084/jem.192.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crozat K, Beutler B. Proc Natl Acad Sci USA. 2004;101:6835–6836. doi: 10.1073/pnas.0401347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beignon AS, McKenna K, Skoberne M, Manches O, DaSilva I, Kavanagh DG, Larsson M, Gorelick RJ, Lifson JD, Bhardwaj N. J Clin Invest. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 8.Coccia EM, Severa M, Giacomini E, Monneron D, Remoli ME, Julkunen I, Cella M, Lande R, Uze G. Eur J Immunol. 2004;34:796–805. doi: 10.1002/eji.200324610. [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto JK, Barre-Sinoussi F, Bolton V, Pedersen NC, Gardner MB. J Interferon Res. 1986;6:143–152. doi: 10.1089/jir.1986.6.143. [DOI] [PubMed] [Google Scholar]
- 10.Soumelis V, Scott I, Gheyas F, Bouhour D, Cozon G, Cotte L, Huang L, Levy JA, Liu YJ. Blood. 2001;98:906–912. doi: 10.1182/blood.v98.4.906. [DOI] [PubMed] [Google Scholar]
- 11.Muller-Trutwin M, Hosmalin A. Immunol Cell Biol. 2005;83:578–583. doi: 10.1111/j.1440-1711.2005.01394.x. [DOI] [PubMed] [Google Scholar]
- 12.Siegal F. Res Initiat Treat Action. 2003;8:10–13. [PubMed] [Google Scholar]
- 13.von Sydow M, Sonnerborg A, Gaines H, Strannegard O. AIDS Res Hum Retroviruses. 1991;7:375–380. doi: 10.1089/aid.1991.7.375. [DOI] [PubMed] [Google Scholar]
- 14.Herbeuval JP, Boasso A, Grivel JC, Hardy AW, Anderson SA, Dolan MJ, Chougnet C, Lifson JD, Shearer GM. Blood. 2005;105:2458–2464. doi: 10.1182/blood-2004-08-3058. [DOI] [PubMed] [Google Scholar]
- 15.Fonteneau JF, Larsson M, Beignon AS, McKenna K, Dasilva I, Amara A, Liu YJ, Lifson JD, Littman DR, Bhardwaj N. J Virol. 2004;78:5223–5232. doi: 10.1128/JVI.78.10.5223-5232.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yonezawa A, Morita R, Takaori-Kondo A, Kadowaki N, Kitawaki T, Hori T, Uchiyama T. J Virol. 2003;77:3777–3784. doi: 10.1128/JVI.77.6.3777-3784.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbeuval JP, Hardy AW, Boasso A, Anderson SA, Dolan MJ, Dy M, Shearer GM. Proc Natl Acad Sci USA. 2005;102:13974–13979. doi: 10.1073/pnas.0505251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lichtner M, Maranon C, Vidalain PO, Azocar O, Hanau D, Lebon P, Burgard M, Rouzioux C, Vullo V, Yagita H, et al. AIDS Res Hum Retroviruses. 2004;20:175–182. doi: 10.1089/088922204773004897. [DOI] [PubMed] [Google Scholar]
- 19.Miura Y, Misawa N, Maeda N, Inagaki Y, Tanaka Y, Ito M, Kayagaki N, Yamamoto N, Yagita H, Mizusawa H, Koyanagi Y. J Exp Med. 2001;193:651–660. doi: 10.1084/jem.193.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herbeuval JP, Grivel JC, Boasso A, Hardy AW, Chougnet C, Dolan MJ, Yagita H, Lifson JD, Shearer GM. Blood. 2005;106:3524–3531. doi: 10.1182/blood-2005-03-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaperot L, Blum A, Manches O, Lui G, Angel J, Molens JP, Plumas J. J Immunol. 2006;176:248–255. doi: 10.4049/jimmunol.176.1.248. [DOI] [PubMed] [Google Scholar]
- 22.Almeida M, Cordero M, Almeida J, Orfao A. AIDS. 2005;19:261–271. [PubMed] [Google Scholar]
- 23.Kadowaki N, Liu YJ. Hum Immunol. 2002;63:1126–1132. doi: 10.1016/s0198-8859(02)00751-6. [DOI] [PubMed] [Google Scholar]
- 24.Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 25.Graham DR, Chertova E, Hilburn JM, Arthur LO, Hildreth JE. J Virol. 2003;77:8237–8248. doi: 10.1128/JVI.77.15.8237-8248.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akira S, Hemmi H. Immunol Lett. 2003;85:85–95. doi: 10.1016/s0165-2478(02)00228-6. [DOI] [PubMed] [Google Scholar]
- 27.Secchiero P, Gonelli A, Celeghini C, Mirandola P, Guidotti L, Visani G, Capitani S, Zauli G. Blood. 2001;98:2220–2228. doi: 10.1182/blood.v98.7.2220. [DOI] [PubMed] [Google Scholar]
- 28.Stary G, Bangert C, Tauber M, Strohal R, Kopp T, Stingl G. J Exp Med. 2007;204:1441–1451. doi: 10.1084/jem.20070021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Busillo JM, Benovic JL. Biochim Biophys Acta. 2006;1768:952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herbeuval JP, Lambert C, Sabido O, Cottier M, Fournel P, Dy M, Genin C. J Natl Cancer Inst. 2003;95:611–621. doi: 10.1093/jnci/95.8.611. [DOI] [PubMed] [Google Scholar]
- 31.Wu GS, Burns TF, McDonald ER, III, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R, et al. Nat Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 32.Piatak M, Jr, Saag MS, Yang LC, Clark SJ, Kappes JC, Luk KC, Hahn BH, Shaw GM, Lifson JD. Science. 1993;259:1749–1754. doi: 10.1126/science.8096089. [DOI] [PubMed] [Google Scholar]
- 33.Esser MT, Bess JW, Jr, Suryanarayana K, Chertova E, Marti D, Carrington M, Arthur LO, Lifson JD. J Virol. 2001;75:1152–1164. doi: 10.1128/JVI.75.3.1152-1164.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, Douek DC. J Exp Med. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyrhol-Riise AM, Ohlsson M, Skarstein K, Nygaard SJ, Olofsson J, Jonsson R, Asjo B. Clin Immunol. 2001;101:180–191. doi: 10.1006/clim.2001.5102. [DOI] [PubMed] [Google Scholar]
- 36.Guadalupe M, Reay E, Sankaran S, Prindiville T, Flamm J, McNeil A, Dandekar S. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbeuval JP, Nilsson J, Boasso A, Hardy AW, Kruhlak MJ, Anderson SA, Dolan MJ, Dy M, Andersson J, Shearer GM. Proc Natl Acad Sci USA. 2006;103:7000–7005. doi: 10.1073/pnas.0600363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mehandru S, Poles MA, Tenner-Racz K, Horowitz A, Hurley A, Hogan C, Boden D, Racz P, Markowitz M. J Exp Med. 2004;200:761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taieb J, Chaput N, Menard C, Apetoh L, Ullrich E, Bonmort M, Pequignot M, Casares N, Terme M, Flament C, et al. Nat Med. 2006;12:214–219. doi: 10.1038/nm1356. [DOI] [PubMed] [Google Scholar]
- 40.O'Neill LA. Science. 2004;303:1481–1482. doi: 10.1126/science.1096113. [DOI] [PubMed] [Google Scholar]
- 41.Bekisz J, Schmeisser H, Hernandez J, Goldman ND, Zoon KC. Growth Factors. 2004;22:243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- 42.Pitha PM. Antiviral Res. 1994;24:205–219. doi: 10.1016/0166-3542(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 43.Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, Taniguchi T. Nature. 2003;424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- 44.Vilcek J. Nat Immunol. 2003;4:825–826. doi: 10.1038/ni0903-825. [DOI] [PubMed] [Google Scholar]
- 45.Herbeuval JP, Shearer GM. Clin Immunol. 2006;123:121–128. doi: 10.1016/j.clim.2006.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gringeri A, Musicco M, Hermans P, Bentwich Z, Cusini M, Bergamasco A, Santagostino E, Burny A, Bizzini B, Zagury D. J Acquired Immune Defic Syndr Hum Retrovirol. 1999;20:358–370. doi: 10.1097/00042560-199904010-00006. [DOI] [PubMed] [Google Scholar]
- 47.Zagury D, Lachgar A, Chams V, Fall LS, Bernard J, Zagury JF, Bizzini B, Gringeri A, Santagostino E, Rappaport J, et al. Proc Natl Acad Sci USA. 1998;95:3851–3856. doi: 10.1073/pnas.95.7.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roederer M, Treister A, Moore W, Herzenberg LA. Cytometry. 2001;45:37–46. doi: 10.1002/1097-0320(20010901)45:1<37::aid-cyto1142>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.