Abstract
β-Catenin (β-cat) is a key signaling component of the canonical Wnt pathway as well as an increasingly studied contributor to various pathways that regulate cell adhesion, proliferation, and differentiation. For its best known function, posttranslational stabilization of β-cat is required for T cell factor-dependent transcription of numerous downstream targets of Wnt, and this process is aberrantly active in a wide array of cancers. To enable direct monitoring of posttranslational stabilization of β-cat in cells and living animals, we constructed and characterized the bioluminescent fusion reporters β-cat firefly luciferase (β-cat-FLuc) and β-cat click beetle green luciferase (β-cat-CBG). These reporters provided real-time, noninvasive readout of modulators of β-cat stability in cellulo and, furthermore, enabled monitoring of changes in total β-cat levels in vivo in intact animals. In addition, using spectral unmixing, green β-cat-CBG was deconvoluted from a red TCF-dependent FLuc reporter (TOPFLASH), enabling analysis of β-cat processing and downstream transcriptional activation simultaneously. By using this system, the natural product epigallocatechin 3-gallate was found to block Wnt signaling, independent of β-cat processing. These β-cat reporters represent a powerful new strategy for identifying in cellulo and in vivo dynamic regulators and mechanism-based therapeutics of signaling pathways mediated by β-cat stabilization.
Keywords: bioluminescence, epigallocatechin, molecular imaging, signal transduction, Wnt
The highly conserved Wnt/β-catenin (β-cat) signal transduction pathway has been studied extensively for its key role in metazoan development (1) and the aberrant expression of its various vital components in colorectal tumors (2), gastric cancer, ovarian carcinomas, hepatocellular carcinoma, and endometrial carcinomas (3). In the absence of Wnt, β-cat, the key signaling molecule in the pathway, is phosphorylated while bound to a cytoplasmic destruction complex consisting of the tumor suppressor adenomatous polyposis coli (APC), a scaffold protein axin, and glycogen synthase kinase 3β (GSK3β) (1, 4). In this phosphorylated state, β-cat is ubiquitinated and consequently degraded via the proteasome (5). In the presence of Wnt, the destruction complex is inhibited (3, 6), hypophosphorylated β-cat is stabilized in the cytoplasm and translocates to the nucleus, where it forms a complex with T cell factor (TCF) or lymphoid enhancer-binding factor (LEF) (7) to activate transcription of >50 genes, including c-MYC, cyclin D1, gastrin, and matrilysin (8). A third population of β-cat resides in a membranous complex with E-cadherin and α-catenin and plays a pivotal role in intercellular adhesion (1, 4).
The role of β-cat in various signaling cascades continues to be pursued in model organisms by using genetic techniques that provide a phenotypic readout at the organismal level. In mammalian cells, a TCF-based reporter system (9), providing a firefly luciferase-based readout of Wnt pathway activation at the transcriptional level, is used in conjunction with Western blot analysis of β-cat protein levels or immunofluorescent microscopy to detect β-cat nuclear accumulation. However, these techniques lack dynamic resolution and are destructive in character. We present β-cat firefly luciferase (β-cat-FLuc) and β-cat click beetle green luciferase (β-cat-CBG) fusion reporters that allow direct readout of β-cat protein levels in real time. These reporters facilitate dynamic analysis of total β-cat levels in living cells in response to various ligand and drug stimuli. Using spectral deconvolution of multicolored luciferases (10), we demonstrate simultaneous imaging of posttranslational β-cat stabilization (β-cat-CBG) together with TCF-dependent transcriptional activity (pTOPFLASH) as a single-step live-cell assay. In addition, animal studies conducted with β-cat-FLuc now enable noninvasive monitoring of β-cat stabilization in intact animals. The β-cat reporters represent facile tools for the characterization and pharmacodynamic analysis of potential regulators and drugs that target the Wnt pathway via effects on β-cat stabilization.
Results
Monitoring Wnt-Induced Posttranslational Stabilization of β-Cat in Real Time in Cellulo.
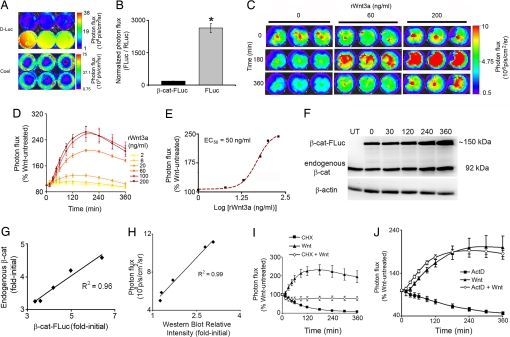
To enable direct real-time monitoring of β-cat protein levels, we created a bioluminescence reporter, β-cat-FLuc. To minimize any steric interference with known binding partners of β-cat, FLuc was fused to the C terminus of β-cat separated by an 11-aa flexible linker (11). Under basal Wnt unstimulated conditions, low β-cat-FLuc levels were maintained as evidenced by the ≈100-fold lower bioluminescence signal obtained from cells transfected with pβ-cat-FLuc as compared with cells transfected with an equal DNA amount of the control construct pFLuc (Fig. 1 A and B). In addition, only β-cat-FLuc cells showed a ligand-dependent increase in signal upon treatment with increasing concentrations of rWnt3a as would be expected because of the stabilizing effects of Wnt on β-cat protein levels (Fig. 1C). β-cat-FLuc stabilization was perceptible 30–45 min after Wnt treatment. Compared with treatment with vehicle, a 2.6-fold increase in signal intensity was observed with the highest concentration of Wnt tested, and peak stabilization was observed at ≈2–3 h (Fig. 1 C and D). Wnt-induced stabilization of β-cat-FLuc showed an apparent EC50 of 50 ng/ml (Fig. 1E), and consequently, 60 ng/ml rWnt3a was used in all subsequent experiments to activate the pathway. Control pFLuc-transfected cells did not respond to rWnt3a (data not shown), thus ruling out effects on either the CMV promoter or FLuc itself.
Fig. 1.
Validation of β-cat-FLuc as a reporter for posttranslation stabilization of total β-cat protein levels in cellulo. (A) Bioluminescence imaging of HEK293T cells transfected with equal DNA amounts of pβ-cat-FLuc (upper row) or pFLuc (lower row) and pRLuc each. Images show color-coded maps of photon flux produced by treatment with d-luciferin (D-Luc; Upper) or coelenterazine (Coel; Lower) superimposed on black and white photographs of the assay plates. (B) Quantification of bioluminescence signal from A normalized for transfection efficiency (FLuc/RLuc) and plotted as mean ± SEM (n = 3) for β-cat-FLuc (black bar) and FLuc (gray bar). *, P < 0.001 by an unpaired t test. (C) Bioluminescence images (n = 3 each) of β-cat-FLuc response to vehicle or representative concentrations (60 or 200 ng/ml) of rWnt3a at representative time points (0, 180, and 360 min after treatment). (D) Wnt-induced stabilization of β-cat-FLuc by increasing concentrations of rWnt3a plotted as percent of Wnt untreated versus time after treatment, calculated from fold-initial values (photon fluxx min/photon flux0). Each point represents mean ± SEM (n = 3). (E) Concentration–response curve of β-cat-FLuc to rWnt3a, generated by using percent Wnt untreated values from D at 180 min plotted versus log [rWnt3a concentrations]. The curve is fitted to a nonlinear variable slope sigmoidal concentration–response curve (EC50 = 50 ng/ml; R2 = 0.996). (F) Western blot analysis of total cell lysates of pβ-cat-FLuc-transfected HEK293T cells, probed with anti-β-cat antibody. An ≈150-kDa band corresponding to β-cat-FLuc reporter protein and a 92-kDa band corresponding to endogenous β-cat are seen in all lanes except the first lane, which contains control lysate from untransfected HEK293T cells. The second through sixth lanes contain lysates from cells treated with 60 ng/ml rWnt3A for 0, 30, 120, 240, and 360 min, respectively. Equal loading was confirmed by probing with anti-actin antibody. (G) Densitometric analysis of endogenous and reporter bands normalized to actin bands in F reveals excellent correlation between Wnt-induced β-cat-FLuc and endogenous β-cat protein levels. (H) Tight correlation between bioluminescence signal obtained from pβ-cat-FLuc-transfected HEK293T cells just before lysis for Western blot analysis, and densitometric analysis of the β-cat-FLuc reporter band from concordant Western blot probed with anti-β-cat antibody. (I and J) Inhibition of Wnt-induced stabilization of β-cat-FLuc by blocking translation with cycloheximide (100 μg/ml) (I) but not by blocking transcription with actinomycin D (ActD) (1 μM) (J). Graphs are represented as in D with treatments of drug plus rWnt3a plotted as percent of treatment with drug alone.
To demonstrate that β-cat-FLuc reporter levels were an accurate reflection of endogenous β-cat levels, we conducted Western blot analysis using an anti-β-cat antibody on total cell lysates of pβ-cat-FLuc-transfected cells cultured in DMEM and treated with rWnt3a for various periods of time (Fig. 1F). Semiquantitative densitometric analysis of Western blots showed excellent correlation between Wnt-stimulated endogenous β-cat and β-cat-FLuc protein levels (R2 = 0.96; Fig. 1G). Of note, Western blot analysis revealed that both endogenous and reporter β-cat protein levels continued to increase past 3 h—a stabilization profile that differed slightly from that seen with bioluminescence imaging (Fig. 1D). Because the conditions under which transfected cells were maintained for real-time imaging (37°C, non-CO2-buffered, nonhumidified chamber) differed from those under which cells were maintained before lysis for Western blot analysis (37°C, 5% CO2, humidified incubator), we performed a direct comparison of stabilization profiles over longer times, measured by bioluminescence imaging and by Western blot analysis of cells maintained under similar conditions in DMEM (37°C, 5% CO2, humidified incubator). For this comparison, bioluminescence images of pβ-cat-FLuc-transfected cells were taken immediately before lysis and correlations were determined between bioluminescence signal flux and semiquantitative densitometric analysis of the β-cat-FLuc protein levels by Western blot. Once again, an excellent correlation coefficient was observed (R2 = 0.99; Fig. 1H), suggesting that longer-term analysis of β-cat stabilization was feasible using the β-cat-FLuc reporter under modified imaging conditions.
Finally, to demonstrate that the increase in bioluminescence signal from β-cat-FLuc truly reflected posttranslational stabilization of β-cat-FLuc protein, we treated cells transfected with pβ-cat-FLuc with 100 μg/ml of the translation inhibitor cycloheximide either alone or together with rWnt3a. Inhibition of translation completely abrogated Wnt-induced stabilization of β-cat-FLuc (Fig. 1I), whereas treatment with 1 μM transcription inhibitor actinomycin D (ActD) did not (Fig. 1J). Together, these data suggested that our β-cat-FLuc reporter provided an accurate semiquantitative real-time readout of posttranslation stabilization of total β-cat protein levels upon Wnt stimulation in living cells.
Effects of Pharmacological Modulation of Wnt Pathway Components on β-Cat-FLuc in Cellulo.
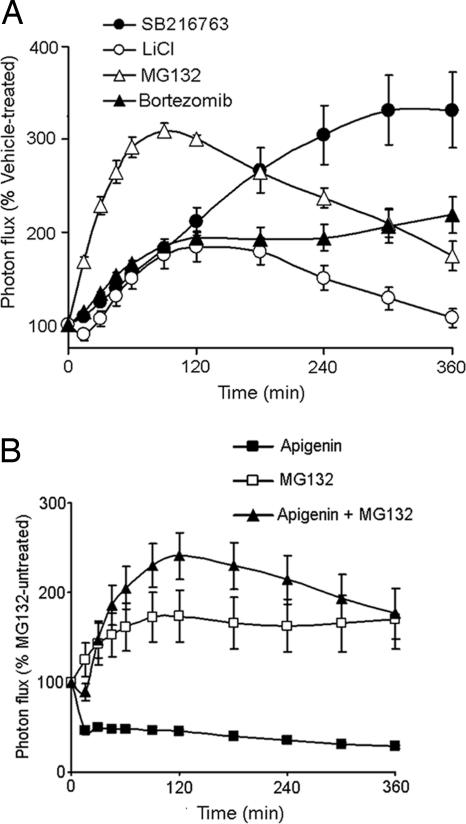
The availability of a direct readout of β-cat levels in intact cells may provide a new tool for discriminating whether drugs and compounds that are shown to modulate Wnt signaling by conventional TCF-dependent transcriptional readout actually act upstream or downstream of β-cat. To validate the approach, we tested four known stabilizers of β-cat: LiCl (12) (GSK3β inhibitor), SB216763 (13) (GSK3β inhibitor), MG132 (14, 15) (proteasome inhibitor), and bortezomib (14, 16) (proteasome inhibitor) (Fig. 2A). We also tested a known negative regulator of the Wnt/β-cat pathway, apigenin (17) (a CKII inhibitor) (Fig. 2B). Treatment with 50 mM LiCl or 5 μM SB216763 resulted in 2- to 3-fold increases in the β-cat-FLuc signal, whereas the FLuc signal did not show a concordant increase (data not shown). Similar increases in β-cat-FLuc levels, but not FLuc levels, were evident upon treatment with 25 μM MG132 or 1 μM bortezomib, with effects observed as early as 15 min after treatment. Conversely, exposure of pβ-cat-FLuc-transfected cells to 80 μM apigenin caused an immediate decrease in β-cat-FLuc levels, an effect that was completely reversible by treatment with MG132 (Fig. 2B). Thus, these data validate the β-cat-FLuc reporter for rapid molecular-specific readout of Wnt/β-cat pathway modulators in living cells.
Fig. 2.
β-cat-FLuc response to known modulators of β-cat stability. Changes in bioluminescence (0–360 min) of HEK293T cells transfected with pβ-cat-FLuc (A) upon treatment with GSK3β inhibitors [SB216763 (5 μM) and LiCl (50 mM)] or proteasome inhibitors [MG132 (25 μM) and bortezomib (1 μm)]. (B) Effect of CKII inhibitor apigenin (80 μM) alone, or together with MG132, on β-cat-FLuc dynamics. Graphs are plotted as percent untreated cells.
Simultaneous Monitoring of Real-Time and Transcriptionally Coupled Events in the Wnt Pathway.
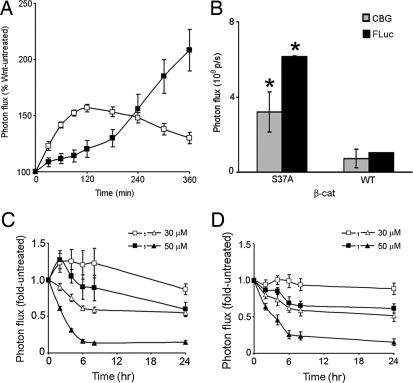
The recent development of a technique for spectral unmixing of multicolored luciferases (10) allows for monitoring of bioluminescent signal from two different reporters simultaneously. We used this technique to monitor concurrent stabilization of β-cat levels and consequent activation of TCF-dependent transcription. Herein, β-cat fused to a green-emitting luciferase CBG (pβ-cat-CBG) was cotransfected with the TCF-dependent red-emitting FLuc reporter (pTOPFLASH). Spectral deconvolution of signal from the two luciferase sources allowed us to determine the relative contribution of each of the two reporters to the total bioluminescence emitted. Upon treatment with rWnt3a, an early stabilization of β-cat-CBG levels and a delayed transcription-dependent FLuc activation from pTOPFLASH (Fig. 3A), but not from pFOPFLASH (data not shown), were evident. We further used this technique to demonstrate that the β-cat reporters were physiologically active in this system. Either wild-type (WT) β-cat or a constitutively active β-cat mutant (S37A) (18), fused to CBG, was cotransfected with pTOPFLASH. Spectral unmixing of signal from the two luciferase sources showed that both β-cat levels (CBG) and TCF-induced FLuc activity (FLuc) were higher in cells cotransfected with S37A as compared with WT (Fig. 3B).
Fig. 3.
Simultaneous imaging of real-time β-cat-FLuc stabilization and consequent pTOPFLASH induction in cellulo. (A) Spectrally unmixed values of bioluminescent signal from HEK293T cells cotransfected with both green-emitting pβ-cat-CBG (open squares) and red-emitting pTOPFLASH (filled squares) and treated with rWnt3a (60 ng/ml). (B) Both β-cat levels (CBG, gray bars) and TCF-dependent transcription (FLuc, black bars) are higher in HEK293T cells cotransfected with pTOPFLASH and a stable β-cat mutant pS37A-CBG, as compared with those cotransfected with pTOPFLASH and WT pβ-cat-CBG. Spectrally unmixed values are represented as mean ± SEM (n = 6). *, significance at P < 0.01 by unpaired t tests. In the presence of either WT β-cat-CBG (C) or a nondegradable β-cat mutant, S37A-CBG (D), EGCG (30 and 50 μM) exerts a time- and concentration-dependent inhibitory effect on β-cat levels (CBG, open and filled squares) and TCF-dependent transcription (FLuc, open and filled triangles). Spectrally unmixed values are plotted as fold-untreated as normalized from fold-initial values and represented as mean ± SEM (n = 3).
Characterization of Epigallocatechin 3-gallate (EGCG).
Next, we used the β-cat reporters to examine the mechanism of action of EGCG, an antioxidant constituent of green tea extract. Various studies have documented the inhibitory effect of EGCG on TCF-dependent activity, but there have been conflicting reports as to whether this occurs in a β-cat-dependent manner (19–22). In HEK293T cells, inhibition of pTOPFLASH expression (FLuc) in both WT and S37A β-cat-expressing cells was noticeable within 2 h of EGCG treatment and continued to decline over the ensuing 24 h (Fig. 3 C and D). By comparison, at 2 h, EGCG transiently stabilized WT β-cat by ≈25% but not mutant S37A β-cat. However, treatment for up to 24 h with EGCG resulted in a concentration-dependent decrease of β-cat levels (CBG) in cells expressing WT β-cat (Fig. 3C) and, remarkably, a proportionally identical decrease in cells expressing the oncogenic constitutively active S37A β-cat mutant (Fig. 3D). Thus, continuous treatment with EGCG inhibited the Wnt signaling machinery, potentially at the level of nuclear import or by β-cat transcriptional repression, independent of long-term β-cat protein processing.
Noninvasive Imaging of Drug Effects on β-Cat-FLuc Stabilization in Vivo in Mice.
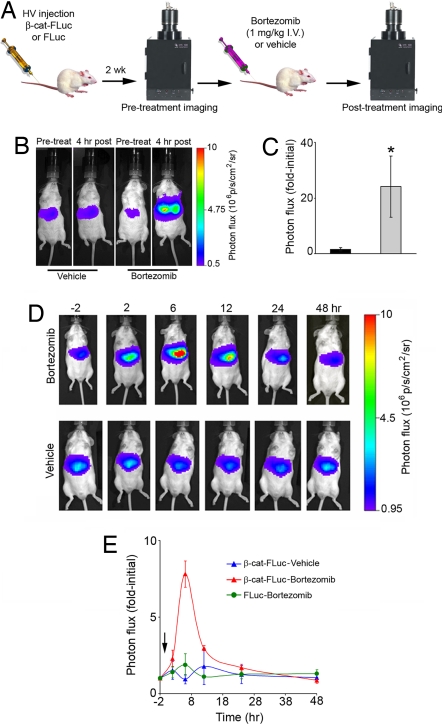
A reporter that allowed noninvasive in vivo imaging would enable pharmacodynamic analysis of β-cat-targeted drugs in vivo. To demonstrate this capability of the β-cat-FLuc reporter, we performed somatic gene transfer to hepatocytes in vivo by i.v. injection of naked pβ-cat-FLuc or control pFLuc DNA (23, 24). Steady levels of bioluminescence signal intensity were observed 2 weeks after DNA injection from livers of injected mice. On the day of treatment, a pretreatment image was taken 10 min after i.p. injection of d-luciferin (150 μg/g of body weight), followed by i.v. administration of either bortezomib (1 μg/g of body weight) or vehicle (Fig. 4A). Four hours later, a posttreatment image was obtained 10 min after i.p. injection of d-luciferin, as before. The bortezomib-treated cohort (n = 5) revealed >20-fold stabilization in β-cat-FLuc as compared with vehicle-treated mice (n = 5) (Fig. 4 B and C), demonstrating the effectiveness of the reporter as a readout of β-cat stabilization upon proteasomal inhibition in vivo. To further characterize the temporal profile of the effects of bortezomib on β-cat-FLuc stabilization in vivo, we performed repetitive imaging at −2, 2, 6, 12, 24, and 48 h after i.v. administration of a single dose of bortezomib (1 μg/g of body weight) or vehicle. β-cat-FLuc, but not FLuc control, was stabilized in vivo between 2 and 6 h, and signal intensity was completely restored to pretreatment levels by 48 h after bortezomib treatment (Fig. 4 D and E). This demonstrated the utility of the reporter as a tool for screening drugs effective on β-cat processing as well as determining their in vivo molecular-specific pharmacodynamic profiles.
Fig. 4.
Imaging pharmacological modulation of β-cat in living mice. (A) Schematic representation of the high-volume (HV) somatic gene transfer and imaging experimental timeline. (B) Representative images of β-cat-FLuc bioluminescence signal from livers of mice treated with vehicle (saline) or bortezomib (1 μg/g) taken before (first and third photographs) or 4 h after treatment (second and fourth photographs). (C) Photon flux (fold-initial; mean ± SEM) for the 4 h vehicle-treated (black bar) and bortezomib-treated (gray bar) cohorts (n = 5 mice each). Regions of interest were manually drawn over the liver of mice, and signal intensities were calculated as fold-initial pretreatment images. *, significance at P < 0.01 by a Mann–Whitney test. (D) Representative β-cat-FLuc images of a mouse treated with bortezomib (1 μg/g) and a mouse treated with vehicle (saline) at −2, 2, 6, 12, 24, and 48 h after treatment. (E) Kinetics of bortezomib-induced stabilization of β-cat in living mice. Regions of interest were manually drawn over the liver of mice, and signal intensities were calculated at each time point. Signal intensities are displayed as fold-initial for each time point for β-cat-FLuc-transfected bortezomib-treated (red triangles) (n = 3) and vehicle-treated (blue triangles) (n = 2) mice as well as control FLuc-transfected bortezomib-treated (green circles) (n = 2) mice (mean ± SEM). The arrow indicates bortezomib administration at time 0.
Discussion
Activation of the Wnt signaling pathway by stabilization of β-cat, an essential event for normal growth and pro-proliferative processes, when dysregulated, also causes aberrant growth in various types of cancer. The ability to monitor β-cat processing in real time in both intact cells and animals provides a unique means to study one node of normal and abnormal Wnt signaling, as well as to monitor additional signal transduction cascades that relay through β-cat. To this end, we have developed bioluminescence-based β-cat reporters (β-cat-FLuc and β-cat-CBG) that facilitate in cellulo and in vivo analysis of signal transduction via regulation of posttranscriptional stabilization of β-cat.
Quantitative analysis of β-cat-FLuc signal intensity under basal and Wnt-stimulated conditions characterized the biochemical behavior of the reporter. In addition, correlation with densitometry from Western blot analysis, the traditional assay used to measure β-cat stabilization, showed that β-cat-FLuc responded identically to endogenous β-cat, despite the fact that the endogenous and reporter proteins were transcribed from different promoters. β-cat-FLuc profiles in response to various known modulators of Wnt signaling closely matched the overall predicted outcome—i.e., stabilization or destabilization—but also provided detailed temporal information about the effect of each compound. These assays illustrated several properties of our β-cat-FLuc reporter assay that distinguished it from current methods used to interrogate the canonical Wnt pathway, including recently published techniques for monitoring β-cat accumulation such as a luminometric antibody-based assay (25) and an automated fluorescence microscopy assay to monitor nuclear translocation of β-cat (26): (i) the ability to monitor events in real time instead of a delayed response due to the transcription dependence of TCF-based reporters, (ii) live cell assays instead of analysis of lysates or fixed cells, (iii) direct readout of β-cat levels instead of indirect monitoring of Wnt pathway activation, and (iv) independence from the use of additional reagents that require extensive optimization, such as antibodies. In addition, this assay could have widespread uses beyond studying details of canonical Wnt pathway activation, as evidenced by ever-increasing reports of β-cat involvement in cross-talk between Wnt and numerous signaling cascades (27), often mediated by GSK3β (28).
An attribute of assays that distinguish between various subcellular populations of β-cat, which is lacking in our reporter, is the ability to monitor compartment shifts in β-cat populations that do not result in an increase in total β-cat levels or to study just the “active” β-cat population in isolation. A next generation of β-cat reporters can be engineered to represent only a specific β-cat population—i.e., membrane-tethered β-cat-FLuc—for conditions in which tracking subcellular localization is essential. However, reporters such as β-cat-FLuc that provide an online readout of the total β-cat population will continue to offer a physiologically relevant holistic view, because long-term effects of stimuli are rarely restricted to a single subcellular population of a protein, even if the immediate effect may appear to be.
To date, the Wnt pathway could only be interrogated singularly, either at the level of β-cat stabilization or transcription of target genes of the TCF-β-cat complex. Using spectral unmixing of multicolored luciferases, we were able to simultaneously monitor both β-cat accumulation and TCF transactivation in the same cells in real time. In addition to providing a means of simultaneously capturing both an upstream and a downstream event in the Wnt signaling cascade, use of dual reporters in this way also allowed determination of whether a given stimulus impacted transcriptional activity in a β-cat-dependent manner. We used this technique to examine the mechanism of action of EGCG, a natural antioxidant that is under investigation as a chemopreventive agent in a variety of cancers (29). Studies suggesting EGCG-mediated effects on β-cat are currently under debate (19–22). Our data showed that long-term exposure to EGCG inhibited both β-cat levels and TCF-dependent transcription but only after a 24-h treatment regimen. Although the inhibitory effects on pTOPFLASH were observed within a few hours, the length of treatment required for both WT and stabilized β-cat (S37A-CBG) levels to be significantly diminished provided support for the hypothesis that EGCG down-regulated β-cat expression levels in the long term (21) and thus suggested that the dominant effect of EGCG inhibition on the Wnt pathway is at the transcriptional level. An ideal therapeutic candidate for targeting an aberrantly active Wnt pathway may be one that is able to inhibit constitutively active TCF transcription caused by the presence of nondegradable oncogenic forms of β-cat. As captured by spectral unmixing, EGCG treatment was capable of inhibiting oncogenic β-cat levels as well as the ensuing constitutively active β-cat-dependent transcription, thereby providing a natural product lead for novel anticancer agents.
Finally, the utility of the β-cat-FLuc reporter as a tool for pharmacological characterization of drugs in vivo was demonstrated for the proteasome inhibitor bortezomib. Noninvasive imaging of intact mice whose hepatocytes were transfected with the reporter was used for single-time-point pharmacodynamic analysis as well as for temporal profiling of targeted drug action. The robustness and noninvasive nature of the assay allowed a remarkably small number of mice (n = 20) to be used to perform all of the drug studies described. This reporter strategy now provides quantitative monitoring of β-cat levels in live animals and a unique tool that may be used for investigating potential therapeutics acting on β-cat in vivo.
Signaling via β-cat continues to be pursued in a host of model organisms and cell-based studies even as additional functions, newer contexts, alternate pathways, and aberrant activities are discovered for this multifaceted protein. Our novel β-cat reporters provide versatile tools for mechanism-based studies of β-cat signaling, in vivo noninvasive pharmacodynamic analysis of therapeutic candidates in living animals, and a complementary tool to established Wnt reporters for potential cell-based high-throughput screening of small molecule libraries that target normal and abnormal Wnt pathway activation.
Methods
Plasmids and Cells.
HEK293T cells (American Type Culture Collection) were cultured in DMEM supplemented with 10% heat-inactivated FBS and 1% glutamine. β-catS37A plasmid was kindly provided by Bert Vogelstein (Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, Baltimore, MD). To generate pβ-cat-FLuc, we first PCR-amplified the β-cat ORF from β-catS37A and then performed site-directed mutagenesis of A37→S with a QuikChange kit (Stratagene). Primers 5′-GTACCGCGGGCCACCATGGCTACTCAAGCTGATTTGATGGAGTTGGAC and 3′-GTAGGATCCCAGGTCAGTATCAAACCAGGCCAGCTGATT were used for PCR amplification, and 5′-CCTGGACTCTGGAATCCATTCTGGTGCCACTACCACAGCTCC and 3′-GGAGCTGTGGTAGTGGCACCAGAATGGATTCCAGAGTCCAGG were used for site-directed mutagenesis. The WT cDNA was then fused to codon-optimized firefly (Photinus pyralis) luciferase (FLuc) from the pGL-3 control plasmid (Promega) by using a strategy described in ref. 30. Construction of the pFLuc control plasmid is described in ref. 30. In the final constructs, β-cat-FLuc and FLuc expression were driven by an immediate-early CMV promoter. To generate pβ-cat-CBG and pS37A-CBG, FLuc was replaced by the click beetle (Pyrophorus plagiophthalmus) green luciferase pCBG99 (Promega) in the WT and mutant β-cat constructs, respectively. TCF-FLuc reporter plasmid (pTOPFLASH) and its corresponding negative control (pFOPFLASH) were purchased from Upstate, and pRLuc-N3, expressing codon-optimized Renilla luciferase (RLuc), was purchased from BioSignal Packard.
In Cellulo Bioluminescence Assays.
HEK293T cells were batch-transfected in suspension by using FuGENE 6 reagent (Roche) and immediately plated. For assays conducted in 24-well plates, 105 cells were plated per well, and plasmid quantities were as follows: pβ-cat-FLuc, 200 ng per 105 cells; mixture of pFLuc and pcDNA3.1, 50 ng and 150 ng, respectively, per 105 cells. For the initial comparison between the two constructs, 1 μg each of pβ-cat-FLuc or pFLuc per 106 cells together with 0.2 μg of pRluc-N3 per 106 cells was used, and cells were plated in six-well plates. Twenty-four hours later, medium was replaced with colorless modified Earle's balanced salt solution (MEBSS) buffer (23) supplemented with 10% FBS, 1% glutamine, and 50 μg/ml d-luciferin (BIOSYNTH International) for FLuc imaging with an IVIS 50 imaging system (Xenogen). Acquisition parameters were as follows: exposure time, 30–60 sec; binning, 4 or 8; no filter; f-stop, 1; field of view (FOV), 10 or 15 cm. For RLuc imaging, d-luciferin-containing buffer was replaced with colorless MEBSS buffer supplemented with 10% heat-inactivated FBS, 1% glutamine, and 1 μg/ml coelenterazine (Biotium). Acquisition parameters were as follows: exposure time, 60 sec; binning, 8; <510 filter; f-stop, 1; FOV, 10 cm. FLuc assays were conducted in the absence or presence of increasing concentrations of rWnt3A (R&D Systems), proteasome inhibitor MG-132, GSK3β inhibitors LiCl or SB216763, or CKII inhibitor apigenin (Sigma–Aldrich). rWnt3a was reconstituted in PBS containing 0.2% human serum albumin, and MG-132, SB216763, and apigenin were dissolved in DMSO. Bioluminescence imaging was performed before, and at the indicated time points after, addition of various reagents. Data were normalized as FLuc reporter/RLuc reporter or first calculated as fold-initial (photon fluxtime x/photon fluxtime 0) and then expressed as percentage of photon counts of Wnt pathway modulator-treated over corresponding vehicle-treated cells. To calculate basal versus ligand-induced reporter turnover, cells were imaged (0–360 min at various intervals, acquisition parameters as above) after addition of cycloheximide (100 μg/ml) or actinomycin D (ActD) (1 μM) (Sigma–Aldrich), with or without Wnt stimulation.
Spectral Unmixing of Multicolored Luciferases.
HEK293T cells were cotransfected with 100 ng per 105 cells each of pTOPFLASH and pβ-cat-CBG, and plated in black 24-well plates. In addition, each plate had two control wells—one transfected with pFLuc alone and the other with pβ-cat-CBG alone. Twenty-four hours after transfection, medium was replaced with colorless MEBSS buffer supplemented with 10% heat-inactivated FBS, 1% glutamine, and 50 μg/ml d-luciferin. Images were taken pretreatment and at indicated time points after treatment with rWnt3a (60 ng/ml) or vehicle. Three sequential images were acquired on an IVIS 100 imaging system using no filter, a 540AF20 filter, and a >650 filter, respectively, with the following acquisition parameters (identical for all three images): exposure time, 10–60 sec; binning, 4 or 8; f-stop, 1; FOV, 10 cm. Acquired images were analyzed by using plug-ins created for ImageJ software as described in ref. 10, which allow the mixed luciferase signal from each well to be spectrally resolved to calculate the contribution of each of the two luciferases to the total bioluminescence signal. To study β-cat-mediated transcriptional activation by either WT or mutant β-cat, 100 ng per 105 cells of pTOPFLASH was cotransfected with an equal amount of either pβ-cat-CBG or pS37A-CBG. Twenty-four hours after transfection, imaging and analysis was performed as above, with and without EGCG (Sigma–Aldrich; reconstituted in sterile water).
Western Blot Analysis.
HEK293T cells were batch-transfected with 1 μg per 106 cells of pβ-cat-FLuc and plated in 35-mm, tissue-culture-treated dishes. Twenty-four hours later, cells were treated with 60 ng/ml rWnt3A or vehicle for the indicated times and lysed in sample buffer [62.5 mM Tris·HCl (pH 6.8), 2% (wt/vol) SDS, 10% glycerol, 50 mM DTT, 0.01% bromophenol blue]. Total β-cat and fusion reporter protein levels were detected by using a rabbit polyclonal antibody against human β-cat (Santa Cruz Biotechnology). Uniform loading was confirmed with anti-β-actin antibody (Sigma–Aldrich). Immune complexes were detected by horseradish-peroxidase-labeled antibodies and enhanced chemiluminescence reagent (Amersham Biosciences). Exposure and densitometric analysis were performed by using the IVIS imaging system and Living Image (Xenogen) and Igor (WaveMetrics) software. For the correlation study between bioluminescence signal and reporter protein levels, cells were transfected as above. Twenty-four hours later, an image was acquired by adding 50 μg/ml d-luciferin directly into the medium (exposure time, 60 sec; binning, 8; f-stop, 1; FOV, 10 cm), followed by immediate lysis into sample buffer.
Bioluminescent Imaging of Mice.
Animal protocols were approved by the Animal Studies Committee of Washington University School of Medicine. Bortezomib was a gift from Millennium Pharmaceuticals and was diluted in saline. In vivo transfection of mouse hepatocytes was performed by using the hydrodynamic method as described in refs. 23 and 24. Briefly, either pβ-cat-FLuc (30 μg) or pFLuc (5 μg) were diluted in PBS in a volume of 1 ml per 10 g of body weight and injected rapidly (5–7 sec) into tail veins of mice (FVB mice, 6-week-old males; Taconic) by using a 3-ml syringe fitted with a 27-gauge needle. Two weeks later, mice were anesthetized (isoflurane) and imaged for liver FLuc expression after i.p. injection of d-luciferin (150 μg per g of body weight) by using the IVIS imaging system (exposure time, 10–60 sec; binning, 2–8; no filter; f-stop, 1; FOV, 10 cm). Mice were treated with bortezomib (1 μg per g of body weight, i.v.) or its corresponding vehicle, saline. For single-time-point analysis, immediately after pretreatment imaging, mice were treated with bortezomib, and posttreatment imaging was performed 4 h later (as above). To obtain a temporal response profile, 2 h after pretreatment imaging, mice were injected with bortezomib, and posttreatment imaging was performed at 2, 6, 12, and 24 h after treatment (as above) after i.p. injection of d-luciferin at each of those times. Images were taken before injection of d-luciferin for all time points, except pretreatment, to visualize any residual signal, which was then used to calculate corrected posttreatment signal intensity. Regions of interest were defined manually over the liver for determining total photon flux (photons per second). Corrected posttreatment photon flux was calculated as posttreatment minus residual signal for each time point. Data were expressed as mean fold-initial (corrected posttreatment/pretreatment) across all mice receiving the same treatment.
Statistical Analyses.
Curve fitting, correlation coefficients, linear regression, Student's t test, and Mann–Whitney tests were performed by using Prism software (GraphPad).
Acknowledgments
We thank Dr. A. Pichler-Wallace for performing hydrodynamic injections, Dr. S. Gross for discussion, and Dr. S. Gammon for assistance with spectral unmixing of multicolored luciferases. This work was supported by National Institutes of Health Grant P50 CA94056 and the Cancer Biology Pathway training program through the Alvin J. Siteman Cancer Center at Barnes-Jewish Hospital–Washington University School of Medicine.
Abbreviations
- EGCG
epigallocatechin 3-gallate
- TCF
T cell factor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Clevers H. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Korinek V, Barker N, Morin PJ, van Wichen D, de Weger R, Kinzler KW, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 3.Giles RH, van Es JH, Clevers H. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 4.Riggleman B, Schedl P, Wieschaus E. Cell. 1990;63:549–560. doi: 10.1016/0092-8674(90)90451-j. [DOI] [PubMed] [Google Scholar]
- 5.Kimelman D, Xu W. Oncogene. 2006;25:7482–7491. doi: 10.1038/sj.onc.1210055. [DOI] [PubMed] [Google Scholar]
- 6.Willert K, Brink M, Wodarz A, Varmus H, Nusse R. EMBO J. 1997;16:3089–3096. doi: 10.1093/emboj/16.11.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Townsley FM, Cliffe A, Bienz M. Nat Cell Biol. 2004;6:626–633. doi: 10.1038/ncb1141. [DOI] [PubMed] [Google Scholar]
- 8.Willert J, Epping M, Pollack JR, Brown PO, Nusse R. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gammon ST, Leevy WM, Gross S, Gokel GW, Piwnica-Worms D. Anal Chem. 2006;78:1520–1527. doi: 10.1021/ac051999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Proc Natl Acad Sci USA. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein PS, Melton DA. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cross DA, Culbert AA, Chalmers KA, Facci L, Skaper SD, Reith AD. J Neurochem. 2001;77:94–102. doi: 10.1046/j.1471-4159.2001.t01-1-00251.x. [DOI] [PubMed] [Google Scholar]
- 14.Luker G, Pica C, Song J, Luker K, Piwnica-Worms D. Nat Med. 2003;9:969–973. doi: 10.1038/nm894. [DOI] [PubMed] [Google Scholar]
- 15.Lee DH, Goldberg AL. Mol Cell Biol. 1998;18:30–38. doi: 10.1128/mcb.18.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan C, Waldmann TA. Cancer Res. 2002;62:1083–1086. [PubMed] [Google Scholar]
- 17.Song DH, Sussman DJ, Seldin DC. J Biol Chem. 2000;275:23790–23797. doi: 10.1074/jbc.M909107199. [DOI] [PubMed] [Google Scholar]
- 18.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. J Biol Chem. 1997;272:24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 19.Dashwood WM, Orner GA, Dashwood RH. Biochem Biophys Res Commun. 2002;296:584–588. doi: 10.1016/s0006-291x(02)00914-2. [DOI] [PubMed] [Google Scholar]
- 20.Dashwood WM, Carter O, Al-Fageeh M, Li Q, Dashwood RH. Mutat Res. 2005;591:161–172. doi: 10.1016/j.mrfmmm.2005.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pahlke G, Ngiewih Y, Kern M, Jakobs S, Marko D, Eisenbrand G. J Agric Food Chem. 2006;54:7075–7082. doi: 10.1021/jf0612530. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Zhang X, Rieger-Christ KM, Summerhayes IC, Wazer DE, Paulson KE, Yee AS. J Biol Chem. 2006;281:10865–10875. doi: 10.1074/jbc.M513378200. [DOI] [PubMed] [Google Scholar]
- 23.Pichler A, Zelcer N, Prior JL, Kuil AJ, Piwnica-Worms D. Clin Cancer Res. 2005;11:4487–4494. doi: 10.1158/1078-0432.CCR-05-0038. [DOI] [PubMed] [Google Scholar]
- 24.Liu F, Song Y, Liu D. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 25.Yeow K, Novo-Perez L, Gaillard P, Page P, Gotteland JP, Scheer A, Lang P. Assay Drug Dev Technol. 2006;4:451–460. doi: 10.1089/adt.2006.4.451. [DOI] [PubMed] [Google Scholar]
- 26.Borchert KM, Galvin RJ, Frolik CA, Hale LV, Halladay DL, Gonyier RJ, Trask OJ, Nickischer DR, Houck KA. Assay Drug Dev Technol. 2005;3:133–141. doi: 10.1089/adt.2005.3.133. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Pamukcu R, Thompson WJ. Cancer Biol Ther. 2002;1:621–625. doi: 10.4161/cbt.309. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Rath O, Kolch W, Cho KH. Oncogene. 2007 doi: 10.1038/sj.onc.1210230. [DOI] [PubMed] [Google Scholar]
- 29.Khan N, Afaq F, Saleem M, Ahmad N, Mukhtar H. Cancer Res. 2006;66:2500–2505. doi: 10.1158/0008-5472.CAN-05-3636. [DOI] [PubMed] [Google Scholar]
- 30.Gross S, Piwnica-Worms D. Nat Methods. 2005;2:607–614. doi: 10.1038/nmeth779. [DOI] [PubMed] [Google Scholar]