Abstract
Endogenous retroviruses (ERVs) are the remnants of ancient retroviral infections of germ cells and have been maintained in whole or part as heritable genomic elements. The last known endogenization events occurred several million years ago, and therefore stepwise analysis of retroviral endogenization has not been possible. A unique opportunity to study this process became available when a full-length ERV isolated from koalas (KoRV) was shown to have integrated into their germ line within the past 100 years. Even though KoRV shares 78% nucleotide identity with the exogenous and highly infectious gibbon ape leukemia virus (GALV), the infectivity of KoRV, like that of other ERVs, is substantially lower than that of GALV. Differences in the protein coding regions of KoRV that distinguish it from GALV were introduced into the GALV genome, and their functional consequences were assessed. We identified a KoRV gagpol L domain mutation as well as five residues present in the KoRV envelope (env) that, when substituted for the corresponding residues of GALV, resulted in vectors exhibiting substantially reduced titers similar to those observed with KoRV vectors. In addition, KoRV env protein lacks an intact CETTG motif that we have identified as invariant among highly infectious gammaretroviruses. Disruption of this motif in GALV results in vectors with reduced syncytia forming capabilities. Functional assessment of specific sequences that contribute to KoRV's attenuation from a highly infectious GALV-like progenitor virus has allowed the identification of specific modifications in the KoRV genome that correlate with its endogenization.
Keywords: adaptation, endogenous retrovirus, koala
Koala retrovirus (KoRV) infection is widespread among koalas of mainland Australia. In the early 1920s, a founder population of koalas from the southeastern state of Victoria was established on Kangaroo Island. The Kangaroo Island koalas were recently reported free of KoRV. The discovery of a KoRV-free population of koalas, together with the observations that KoRV remains actively transcribed in its host and that KoRV has integrated into germ line tissue, suggests that this retrovirus is a recently introduced endogenous retrovirus (ERV) (1). KoRV's closest genomic relative is the exogenous gibbon ape leukemia virus (GALV). The only recorded GALV outbreak was confined to gibbon apes originating from an animal holding facility in Thailand in the late 1960s through the 1970s (2). In contrast, since the initial observation of leukemias and neoplasias in koalas in the 1960s (3) and the description of a gammaretrovirus as the possible cause (4), KoRV is now recognized as endemic among Australian mainland koalas (1, 5).
Although ERVs are for the most part dormant, there is an increasing amount of evidence to suggest that mobile retroelements contribute to genomic evolution (6, 7). KoRV is unusual in that it coexists as both an exogenous and endogenizing viral agent, providing a very rare real-time model for the role, if any, of viral endogenization in species evolution (1). Based on the hypothesis that the close similarity of GALV and KoRV implies their descent from a common retrovirus progenitor (1, 8), this similarity can in turn be exploited to evaluate viral elements altered on stable integration into the host germ line and their functional importance in the endogenization process.
Results
Retroviral Vectors Containing KoRV Group-specific Antigen and Polymerase (gagpol) Components Show Reduced Titer When Compared with GALV.
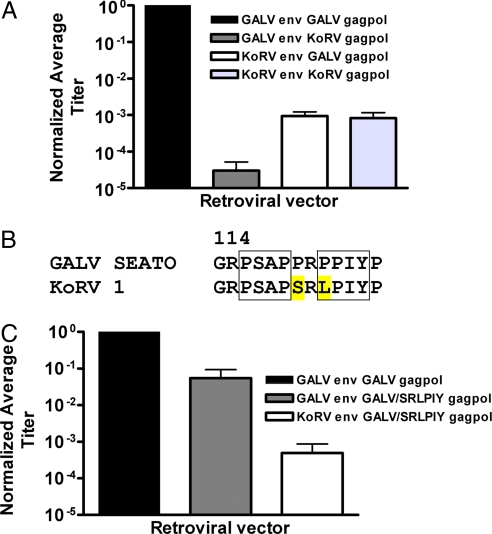
Gammaretroviruses are simple in composition, containing three ORFs encoding internal structural proteins (gag), viral enzymes (pol), and envelope (env) proteins. The relative contribution of these proteins to the diminished infectivity of ERVs such as KoRV can be directly assessed by using retroviral vectors. We have previously described the construction of retroviral vectors containing GALV§ envelope and gagpol proteins (9). Vectors composed of KoRV gagpol and envelope proteins have a greatly diminished titer compared with their GALV counterparts. The gagpol or env proteins encoded by KoRV can be selectively incorporated into infectious retroviral vectors and their effect on infectivity can be directly measured. We compared vectors containing GALV env with GALV or KoRV gagpol and KoRV env with GALV or KoRV gagpol to determine which of the KoRV proteins contribute to the diminished infectivity of KoRV vectors and which may accompany early stages of viral attenuation. All vectors contained the same murine leukemia virus (MLV)-based genome encoding β-galactosidase as a marker for infection. Retroviral vectors containing either KoRV gagpol or env proteins are substantially less infectious than are vectors comprised exclusively of GALV proteins (Fig. 1A). As expected, vectors containing GALV env and GALV gagpol efficiently infected murine cells expressing inorganic phosphate transporter 1 (PiT1) (MDTFPiT1), the receptor for both GALV and KoRV (10) (Fig. 1A). In comparison, vectors composed of KoRV env and either GALV or KoRV gagpol displayed a 1,000-fold reduced titer compared with GALV env with GALV gagpol vectors, whereas vectors composed of GALV env with KoRV gagpol exhibited a 30,000-fold reduced titer. Thus, residues in the KoRV gagpol proteins that differ from GALV residues have a considerable effect on GALV enveloped vector titer.
Fig. 1.
Infectivity studies of GALV and KoRV retroviral vectors. (A) Titers of GALV and KoRV enveloped vectors containing different gagpol proteins. MDTFPiT1 cells were exposed to GALV or KoRV enveloped vectors assembled with either GALV or KoRV gagpol proteins. Titers were normalized to GALV env with GALV gagpol vectors (titer, average ± SEM, 2.6 × 106 ± 9.3 × 105) and were determined by averaging the number of blue-forming units (bfu) per milliliter of vector supernatant from at least three independent experiments ± SEM. (B) Alignment of part of GALV gagpol and KoRV gagpol with two L domains shown in boxes. The two amino acids as encoded by KoRV gagpol and targeted for mutagenesis in GALV gagpol are highlighted in yellow. Residue numbers for GALV gagpol. (C) MDTFPiT1 cells were exposed to GALV or KoRV enveloped vectors assembled with mutant GALV/SRLPIY gagpol proteins. Titers were normalized as described in A.
A Disrupted L Domain in gagpol Contributes to Reduced Vector Titer.
We aligned the gagpol protein sequences of KoRV and GALV to assess what modifications of KoRV gagpol might be responsible for reduced infectivity. Retroviral gagpol proteins contain late domain consensus sequences (L domains). These motifs promote viral release from the infected cell membrane after viral assembly and budding (11). It has been hypothesized that L domain defects impair budding of enveloped viruses causing the accumulation of viral components within producer cells and reducing viral infectivity (12). Many groups have also characterized the importance of residues surrounding L domains in affecting virus infectivity (11). A PSAP L domain precedes a second L domain PPIY with residues PR between the two L domains in GALV gagpol (residues 116–125). Alignment revealed that the corresponding residues in KoRV are PSAP followed by SRLPIY (Fig. 1B). We constructed a mutant GALV gagpol (GALV/SRLPIY gagpol) within which the GALV residues were mutated to their KoRV equivalents, and compared the titers of GALV enveloped vectors assembled with either GALV or GALV/SRLPIY gagpol. GALV vectors containing GALV/SRLPIY gagpol displayed a 10-fold lower titer than vectors containing the GALV gagpol (Fig. 1C). KoRV enveloped vectors containing the GALV/SRLPIY gagpol had a ≈1,800-fold lower titer than GALV enveloped vectors containing GALV gagpol. Thus, disruption of an L domain can lower retroviral vectors titers and this mutation could aid in attenuating the infectivity of endogenous KoRV retroviruses.
Disruption of the CETTG Motif in the GALV Envelope Results in Reduced Cytopathology.
We have reported that substitution of the KoRV receptor binding domain (RBD) for the corresponding region of GALV env diminishes the titer of GALV vectors 1,000-fold (10). Substitution of GALV env for KoRV env in vectors containing the GALV/SRLPIY gagpol improved infectivity 100-fold (Fig. 1C), indicating that the KoRV env also contributes to the reduced infectivity of KoRV retroviral vectors. Alignment of the RBDs of GALV and KoRV envs revealed residue mismatches in the highly conserved CETTG motif. We made the observation that the CETTG motif (residues 132–136 of the mature GALV envelope) is invariably present in all known mammalian gammaretroviral env sequences derived from infectious MLVs, feline leukemia viruses (FLVs), GALV, and woolly monkey virus (WMV) as determined by Blast of virus sequences deposited in GenBank (Table 1). The presence and the conservation of the CETTG motif among exogenous and highly infectious inducible endogenous murine gammaretroviruses has not previously been described. This KoRV env region has been sequenced from 17 independent KoRV isolates (accession numbers ABH05071–ABH0585, AAF15099, and AAZ99990). Of these isolates, 15 have CETAG and two have CGTAG in place of the CETTG present in the four GALV isolate sequences (GALV SEATO, GALV Brain, GALV Hall's Island, and GALV SF). As shown in Table 1, these two KoRV quasispecies, KoRV 1 (CETAG) and KoRV 2 (CGTAG), as well as the ERVs PERV B, PERV C, HERV K, and BAEV contain variations of the CETTG motifs. To investigate the functional consequence of changes within this motif on GALV infectivity, we compared the titers of vectors bearing wild-type GALV env to those with either mutant GALV/CETAG or GALV/CGTAG envs. All three vectors displayed similar titers based on the number of blue-forming nuclei per milliliter of viral supernatant (data not shown). However, the cell-to-cell fusion induced after exposure to vectors bearing wild-type GALV env was diminished by 47 and 64%, respectively, when compared with vectors bearing either of the two mutant GALV envelopes (Fig. 2A). To gain insight into how these modifications of the CETTG motif reduce GALV syncytia-inducing capability, we imposed these residue changes on a crystal structure model of the Friend MLV RBD (13) (Fig. 2B). The E133G mutation results in a smaller side chain and the loss of a carboxyl group (Fig. 2C). A separate mutation, T135A, leads to the loss of two hydrogen bonds (Fig. 2C). Neither of these mutations is likely to result in markedly different protein structure. Possible loss of posttranslational carbohydrate modifications could account for the significant reduction in cytopathology. Virus envelopes are heavily glycosylated, bearing both N-linked and O-linked glycosylated moieties. N-Linked glycosylation can occur on asparagine residues present in the motif NXS/T. Thus, the CETTG motif disruption as seen in KoRV does not lead to the loss of an N-linked glycosylation site. O-Linked glycosylation sites, on the other hand, are found on serine and threonine residues. Thus, the CETTG to CETAG mutation could lead to loss of an O-linked glycosylation site. The further loss of in vitro cytopathology with two mutations in the CETTG motif, CGTAG, suggests that additional mutations in this motif have a cumulative effect on reducing syncytia formation.
Table 1.
Conservation of the CETTG motif among gammaretroviral envelope proteins
| Virus | Residues | Sequence | Virus category |
|---|---|---|---|
| 4070A MLV | 104–108 | CETTG | Infectious |
| Moloney MLV | 144–148 | CETTG | Infectious |
| M813 MLV | 120–124 | CETTG | Infectious |
| AKR MLV | 146–150 | CETTG | Infectious |
| Cas-Br-E MLV | 146–150 | CETTG | Infectious |
| DG-75 MLV | 104–108 | CETTG | Infectious |
| HoMLV | 145–149 | CETTG | Infectious |
| Mink cell focus-forming MLV | 131–135 | CETTG | Infectious |
| Mus dunni MLV | 126–130 | CETTG | Infectious |
| Radiation MLV | 146–150 | CETTG | Infectious |
| Xenotropic MLV | 104–108 | CETTG | Infectious |
| HEMV | 119–123 | CETTG | Infectious |
| FeLV A | 119–123 | CETTG | Infectious |
| FeLV B | 104–108 | CETTG | Infectious |
| FeLV C | 116–120 | CETTG | Infectious |
| GALV SEATO | 132–136 | CETTG | Infectious |
| WMV | 132–136 | CETTG | Infectious |
| KoRV 1 | 132–136 | CETAG | Endogenizing |
| KoRV 2 | 132–136 | CGTAG | Endogenizing |
| BAEV | 102–107 | CYTSY | Endogenous |
| PERV B | 91–95 | CVTSN | Endogenous |
| PERV C | 91–95 | CVTSN | Endogenous |
Residue numbers are given for the mature envelope proteins. Accession numbers for the envelope sequences are given in Materials and Methods.
Fig. 2.
Disruption of the conserved CETTG motif in GALV envelope results in reduced cytopathogenicity in target cells. (A) MDTFPiT1 cells were exposed to vectors composed of GALV gagpol and GALV env or mutant GALV/CETAG or GALV/CGTAG env. Seventy-two hours after the cells were exposed to vectors, syncytia were determined by counting the number of cells containing four or more nuclei. Data were derived from five independent experiments and are normalized to the number of syncytia observed on MDTFPiT1 cells exposed to GALV enveloped particles ± SEM. (B) Structural model of Friend MLV RBD (Protein Data Bank ID code 1AOL). The CETTG motif is shown as spheres. (C) CETTG and mutant CETAG and CGTAG motifs shown in spheres. The CETAG T135A and CGTAG E133G and T135A mutations are shown element color mode. Mutations are indicated with arrowheads. Depictions of structural models were made by using the PyMol (DeLano Scientific, Palo Alto, CA) software program.
Identification of the Residues in the KoRV Envelope That Reduce Vector Titer.
To identify candidate envelope residues that reduce KoRV titer, we aligned residues within the RBD of KoRV to the corresponding regions of four closely related, highly infectious GALV isolates (GALV SEATO, GALV Brain, GALV Hall's Island, and GALV SF) with the intent of identifying residues within this region that were conserved among the different GALV isolates but not between the GALVs and KoRV (Fig. 3A). GenBank contains 33 env sequences derived from primary KoRV isolates: two correspond to the complete env sequences, 15 span parts of the C-terminal RBD region, and 15 different partial sequences span the N terminus of the RBD. The 15 partial N terminus KoRV RBD sequences all contain the residues T86 and L87 (Fig. 3B), whereas all four GALV isolates contain A86 and I87 residues at these positions (Fig. 3A). Other residue differences between KoRV and GALV are shaded orange, yellow, blue, purple, and pink (Fig. 3B). Fourteen of 15 partial C terminus RBD sequences contain QPR residues (pink) at positions 141–143; one of the partial C terminus RBD sequence encodes HPR instead of QPR at this position. The QPR residues are not conserved between GALV SEATO and KoRV (Fig. 3B) but other GALV isolates share either one (e.g., GALV Brain and GALV Hall's Island) or two of the residues in this motif (GALV SF) (Fig. 3A). Substitution of the two GALV residues A86 and I87 for the analogous KoRV residues (T86 and L87) had the greatest effect on KoRV enveloped vectors. Mutant KoRV/AI vectors (Fig. 3C, white bar) exhibited titers within two orders of magnitude of GALV vector titers (Fig. 3C, black bar). We next assessed whether the titer of mutant KoRV/AI could be enhanced by further substitution of GALV RBD-specific residues into KoRV RBD (Fig. 3C). Of the additional substitutions, the Q141L, P142S, R143K (pink) mutant showed the most prominent increase in KoRV enveloped vector titer (Fig. 3C). Thus, the substitution of as few as five GALV residues for the corresponding residues of KoRV allowed for a substantive enhancement in titer of KoRV enveloped vectors to within one order of magnitude of wild-type GALV enveloped vectors (3C).
Fig. 3.
Infectivity studies of mutated KoRV and GALV envelope retroviral vectors. (A) Partial alignment of individual GALV envelope proteins GALV SEATO, GALV SF, GALV Brain, and GALV HI (Hall's Island). Residues highlighted in yellow are those that vary among the four proteins. (B) Partial alignment of KoRV and GALV SEATO mature env proteins. Schematic representation of the mature envelope protein is shown above alignment to designate where the aligned residues are found. Residue numbers are for GALV mature envelope. Highlighted residues represent groups of mutations made in the respective wild-type envs. The mutants are color coded as follows: GTDV residues 59–62 (orange); SKRV residues 64, 66–68 (yellow); DTAK residues 74, 76, 77, 80 (blue); AI residues 86, 87 (black); MST residues 98, 100, 102 (purple); LSK residues 141–142 (pink). (C) Titers on MDTFPiT1 cells exposed to GALV (black), KoRV (gray), or KoRV/AI (white) enveloped vectors. Normalized titers of KoRV/AI enveloped vectors containing one additional group of mutations are color coded to correspond to the mutations shown in Fig. 3B. All vectors were assembled with GALV gagpol. Vectors bearing KoRV/AI envelopes wherein the GALV DTAK (blue) residues were substituted failed to assemble into infectious particles (data not shown). Titers were normalized to that of GALV env with GALV gagpol and determined as described in Fig. 1. (D) Vector titers of GALV, mutant GALV/QPR, or GALV/TL/QPR env assembled with GALV gagpol. Vectors bearing GALV/TL+QPR env and GALV gagpol failed to assemble into infectious particles. (E) Vector titers on MDTFPiT1 cells exposed to GALV env or GALV/TL env with GALV gagpol or mutant GALV/PRLPIY gagpol vectors. Titers were normalized to that of GALV env with GALV gagpol and determined as described for Fig. 1. (F) Alignment of KoRV and GALV envelope residues 51–151 with residues in the KoRV envelope that have been shown to be critical for the attenuation of envelope functions are highlighted in yellow.
To validate the importance of these five residues in envelope function, we made and tested GALV vectors bearing envelopes containing the reciprocal mutations. Vectors assembled with mutant GALV/TL env showed a 70-fold and those assembled with GALV/QPR env showed a 10,000-fold reduction in titer compared with GALV enveloped vectors (Fig. 3D). The combination mutant GALV/TL/QPR env failed to infect MDTFPiT1 cells.
We then assessed the combinatorial effects of vectors containing both GALV/SRLPIY gagpol and GALV A86T-I87L mutant env on vector titers. GALV/TL vectors containing GALV/SRLPIY mutant gag proteins exhibited a 100-fold reduction in titer relative to GALV vector titers (Fig. 3E). Thus, mutations in the GALV env protein in combination with mutations in GALV gagpol that correspond to those found in KoRV result in a reduction in GALV vector infectivity and suggests that the presence of these residues in the KoRV env accompanied by the loss of an L domain both contribute to a decrease in KoRV's infectivity.
Discussion
Much of the investigation of mammalian retroviruses that took place in the latter part of the 20th century focused on how exogenous retroviruses came into being and what elements within each of these viruses accounted for their unique pathology. These findings shed light on how the endogenous viral elements could, either directly or after a series of recombination events, be mobilized to generate infectious viruses. The discovery and cloning of an endogenizing retrovirus, KoRV, and the determination of its relatedness to the highly infectious retrovirus GALV provides a unique opportunity to study how viruses endogenize.
It is important to note that the sequence of KoRV is truly that of the endogenized virus, and not representative of a single viral quasispecies, and that those residues divergent from GALV are not derived from mutations accrued in the virus during viral manipulation ex vivo. The endogenous (integrated) viruses were sequenced directly from the various host koala genomes. The possibility of further vector adaptation in culture, before assessment of parameters of infectivity or cytopathology, was eliminated by sequence reconfirmation. Residue differences that distinguish KoRV from GALV and result in a reduction in GALV titer when incorporated into replication-incompetent GALV-based vectors, are residues present in 13 of the 15 envelope sequences.
We have identified three regions within KoRV that affect viral infectivity or cytopathology. The first region identified was an L domain in KoRV gagpol that contains a SRLPIY in place of the PRPPIY motif found in GALV. The abrogation of this L domain results in the assembly of vector particles with reduced titers (Fig. 1C). This defect in KoRV could provide an explanation for the pleomorphic particle-budding properties of KoRV observed in infected koala bone marrow and lymphoma cells, which is similar to that of viruses containing mutant L domains (14, 15). In contrast, this pleomorphic phenotype is not observed in cells productively infected with GALV (16).
The second motif that appears to be modified early in the process of endogenization is the CETTG motif present in GALV envelope but absent from the envelope of KoRV; to date, its presence among all known exogenous gammaretroviruses and infectious murine retroviruses and its absence from all known ERVs has not been noted (Table 1). This intriguing observation together with the discovery that this motif is disrupted in one of two ways in the KoRV env led us to evaluate the effects of its disruption on GALV env function. Our findings show that as few as two residue changes (CETTG to CGTAG) in the GALV env results in a 64% reduction in syncytia formation. This region of the retroviral env protein has not been implicated in exogenous virus-induced cell fusion nor is it present in the human endogenous virus env protein, syncytin, which has been shown to be involved in hominoid placental morphogenesis (17). It is not readily evident how these subtle alterations in the proposed protein structure of the GALV RBD exert their effect. However, it has been reported that one or two residue changes in the RBD of murine retroviruses can have profound effects on viral-induced cytopathology and host range. For example, a W102G mutation in MLV envelope is required to induce neurological disease and syncytia formation of cerebral vessel endothelial cells (18). Additionally, Jinno-Oue et al. (19) have reported that two residue changes in the RBD of the murine retrovirus PVC-211 (E116G and E129K) are sufficient for conferring brain capillary endothelial cell tropism to this virus, a neuropathogenic variant of the leukemia-inducing Friend MLV.
Sequence alignment of only four GALV isolates showed a high degree of variability in the RBD region of their envelope proteins (Fig. 3A). In contrast, protein sequences of the 17 independent isolates obtained from the same region of the KoRV envelope is invariant with two exceptions: two of the 17 isolates have a CGTAG compared with 15 having a CETAG at residues 132–136, and one of the 17 isolates contains an H-to-Q substitution at position 141. The relative invariance among the 17 KoRV isolates compared with the high degree of variability between GALV isolates (Fig. 3A) is surprising, considering the widespread infection of koalas that significantly predates the limited 10-year infection of captive gibbons. Alignment of KoRV and GALV allowed us to anticipate key residue differences that may play a role in the diminished titer of KoRV when compared with the four GALV isolates (Fig. 3B). Key residue differences between KoRV and GALV env proteins that result in KoRV's diminished infectivity include T86, L87, Q141, P142, and R143 (Fig. 3F). These five residue differences may be reflective of the adaptations preceding complete inactivation of most ERVs caused by multiple in-frame stop codons and reading frame shifts. We hypothesize that these mutations correspond to key adaptive changes that made KoRV capable of entering and persisting in the koala germ line. Regions within the 100-residue stretch of envelope that contribute to KoRV attenutation by either reducing the vector titer or reducing the ability of viral particles to form syncytia are highlighted in Fig. 3F. The coexistence of conspecific exogenous and endogenous gammaretroviruses has allowed a deliberate inspection of the retroviral genome for adaptive mutations acquired during endogenization. This adaptive process may involve the attenuation of the viral env and gagpol proteins to render the virus less infectious, while at the same time allowing continued expression of env protein that may function to block challenge infection by a similar virus.
Koalas are not considered an endangered species; however, they are protected in all seven Australian states and territories. Survival of some koala populations is compromised by factors such as disease and encroachment by human development, whereas other areas, such as Kangaroo Island, are experiencing overpopulation. The koalas found on Kangaroo Island have reduced genetic variability owing to their descent from a smaller number of animals in the founder population. As an active ERV that has been etiologically linked to neoplasias in its host (20), KoRV can be considered an agent that, while in the process of adapting to its host and self-inactivating, is also providing selective pressure on mainland koalas. The founder populations of the island koalas together with the selection of a small number of mainland lineages that survive KoRV infection and subsequent endogenization could result in an overall loss of genetic diversity in the koala population that imposes a grave genetic bottleneck. KoRV may eventually provide an opportunity to study the impact of endogenization on its host.
Materials and Methods
Cell Culture and Retrovirus Infection.
MDTFPiT1 and 293T cells were maintained in DMEM supplemented with 10% FBS and 100 units of penicillin and 100 μg of streptomycin per milliliter, and grown at 37°C and 5% CO2. Retroviral vectors were produced by a three-plasmid transfection (21) with the calcium phosphate method using Profection Mammalian Transfection System (Promega, Madison, WI). Supernatants containing vector particles were harvested at 48 and 72 h after transfection and filtered through a 0.45-μm filter before use. Histochemical assays using 5-bromo-4-chloro-3-indolyl-B-d-galactopyranoside were performed to determine the transduction efficiency of the vectors by measuring expression of β-galactosidase. Vector titers were determined by averaging the number of blue foci obtained on each of the cell lines tested in three or more independent experiments. Stained cells were photographed by using the Q Image and iVision software (Biovision, Exton, PA) with a ×10 objective lens.
Plasmids.
Mutations in the KoRV env were made in the pcDNAZeo3.1 (Invitrogen, Carlsbad, CA) KoRV env expression plasmid and GALV env mutations were made in the pCIneoGALV env expression vector by using the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). Oligonucleotide primers were designed to introduce each group or individual amino acid mutation. The KoRV gagpol expression vector was made by PCR amplification of the gagpol sequence from the pGEMTKoRV provirus plasmid obtained from Jeff McKee (Genomics Research Centre, Griffith University, Gold Coast, Australia), which was cloned into the pCR2.1 plasmid (Invitrogen). The gagpol sequence was then subcloned into the pIK.GALVgagpol expression plasmid between the SphI and DraIII restriction enzyme sites. The mutant GALV/SRLPIY (R123S/P125L) gagpol expression plasmid was produced by site-directed mutagenesis of the pIK.GALVgagpol plasmid by using the QuikChange II XL Site-Directed Mutagenesis kit. All mutant plasmids were sequenced to confirm the presence of desired mutations and to check for unscheduled mutations. Primer sequences are available on request.
Alignments and Accession Numbers.
Alignments were performed by using Align X, part of the Vector NTI 10 software suite (Invitrogen), using default settings. Accession numbers of amino acid sequences included in the retroviral env alignments are as follows: 4070A MLV env, AAA46515; Moloney MLV env, NP_955587; M813 MLV env, AAK16156; AKR MLV env, P03386; Cas-Br-E MLV env, P08360; DG-75 MLV env, AAG29094; HoMLV env, P21436; SP2 MLV env, CAA63866; Mink cell focus-forming MLV env, P03388; Mus dunni MLV env, AAC31806; Radiation MLV env, P31794; Xenotropic MLV env, ABA54271; HEMV env, AAV68489; FeLV A env, P08359; FeLV B env, AAA43048; FeLV C env, A46165; GALV SEATO env, AAC96083; GALV SF, AAC96086; GALV Brain, AAC96085; GALV Hall's Island, AAC96084; WMV env, AAC96087; KoRV 1 env, AAF15099; KoRV 2 env, ABH05084; BAEV env, P10269; HERV K env, O42043; PERV B env, CAB86211; and PERV C env, AAK94075. KoRV 1 and GALV SEATO were also used in the RBD alignment. Accession numbers of amino acid sequences included in the gag alignments are as follows: GALV, SEATO AAA46809; and KoRV 1, Q9TTC2.
Statistical Analysis.
Nonnormalized titer data of GALV gagpol and GALV/SRLPIY gagpol and GALV enveloped versus GALV/TL enveloped vectors were subjected to a one-tailed, two-factor model I ANOVA with a significance level of 0.05. Nonnormalized syncytia counts of MDTFPiT1 cells infected with GALV/CETTG, GALV/CETAG, and GALV/CGTAG enveloped vectors were subjected to a one-tailed, one-factor model I ANOVA with significance level of 0.05.
Acknowledgments
We thank Carolyn Wilson and Karen Farrell for extensive review of the manuscript. We thank Rachel Carr for preparing DNA constructs. In addition, we express our thanks to James Nagle and Debbie Kauffman of the National Institute of Neurological Disorders and Stroke DNA sequencing facility for technical assistance.
Abbreviations
- KoRV
Koala retrovirus
- GALV
gibbon ape leukemia virus
- MLV
murine leukemia virus
- FeLV
feline leukemia virus
- WMV
woolly monkey virus
- ERV
endogenous retrovirus
- gagpol
group-specific antigen and polymerase
- env
envelope
- RBD
receptor binding domain
- PiT1
inorganic phosphate transporter 1.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
GALV SEATO was the source of GALV vector components used in these experiments.
References
- 1.Tarlinton RE, Meers J, Young PR. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 2.Johnsen DO, Wooding WL, Tanticharoenyos P, Bourgeois CH., Jr J Am Vet Med Assoc. 1971;159:563–566. [PubMed] [Google Scholar]
- 3.Heuschele WP, Hayes JR. Cancer Res. 1961;21:1394–1395. [PubMed] [Google Scholar]
- 4.Worley M, Rideout B, Shima A, Jansen D. Proc Annu Meet Am Assoc Zoo Vet. 1993;1:181–182. [Google Scholar]
- 5.Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. J Virol. 2000;74:4264–4272. doi: 10.1128/jvi.74.9.4264-4272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 7.Kazazian HH., Jr Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 8.Benveniste RE, Callahan R, Scherr CJ, Chapman V, Todaro GJ. J Virol. 1977;21:849–862. doi: 10.1128/jvi.21.3.849-862.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eglitis MA, Schneiderman RD, Rice PM, Eiden MV. Gene Ther. 1995;2:486–492. [PubMed] [Google Scholar]
- 10.Oliveira NM, Farrell KB, Eiden MV. J Virol. 2006;80:3104–3107. doi: 10.1128/JVI.80.6.3104-3107.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Demirov DG, Freed EO. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Bieniasz PD. Virology. 2006;344:55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 13.Fass D, Davey RA, Hamson CA, Kim PS, Cunningham JM, Berger JM. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 14.Fiebig U, Hartmann MG, Bannert N, Kurth R, Denner J. J Virol. 2006;80:5651–5654. doi: 10.1128/JVI.02597-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarlinton R, Meers J, Hanger J, Young P. J Gen Virol. 2005;86:783–787. doi: 10.1099/vir.0.80547-0. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami TG, Buckley P, Huff S. In: Goldsmith EI, Moor-Jankowski J, editors. Medical Primatology: Proceedings of the Third Conference on Experimental Medicine and Surgery in Primates; 1972; Lyon. Basel: Karger; 1972. pp. 163–168. [Google Scholar]
- 17.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, Lavallie E, Tang XY, Edouard P, Howes S, et al. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 18.Chung M, Kizhatil K, Albritton LM, Gaulton GN. J Virol. 1999;73:9377–9385. doi: 10.1128/jvi.73.11.9377-9385.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinno-Oue A, Oue M, Ruscetti SK. J Virol. 2001;75:12439–12445. doi: 10.1128/JVI.75.24.12439-12445.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masters P, Duka T, Berris S, Moss G. Wildl Res. 2004;31:267–272. [Google Scholar]
- 21.Farrell KB, Ting YT, Eiden MV. J Virol. 2002;76:4267–4274. doi: 10.1128/JVI.76.9.4267-4274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]