Abstract
The GTPase dynamin I is essential for synaptic vesicle endocytosis in nerve terminals. It is a nerve terminal phosphoprotein that is dephosphorylated on nerve terminal stimulation by the calcium-dependent protein phosphatase calcineurin and then rephosphorylated by cyclin-dependent kinase 5 on termination of the stimulus. Because of its unusual phosphorylation profile, the phosphorylation status of dynamin I was assumed to be inexorably linked to synaptic vesicle endocytosis, however direct proof of this link has been elusive until very recently. This review will describe current knowledge regarding dynamin I phosphorylation in nerve terminals and how this regulates its biological function with respect to synaptic vesicle endocytosis.
Synaptic vesicle (SV) recycling in nerve terminals is essential for the maintenance of neurotransmission. On neuronal depolarization, Ca2+ influx through voltage-gated Ca2+ channels stimulates SV fusion with the plasma membrane (exocytosis) and thus neurotransmitter release. Following exocytosis, SVs are reformed from the plasma membrane in a process called endocytosis. SV endocytosis (SVE) is tightly coupled to SV exocytosis both temporally and spatially, since SVE is activated by the same Ca2+ influx that stimulates exocytosis [1]. After SVE, SVs are recycled locally within the nerve terminal where they are filled with neurotransmitter and prepared for the next round of exocytosis.
Dynamin I
Many proteins are essential for SVE, reflecting the complex nature of de novo generation of SVs from the plasma membrane. One of the most studied and best characterised is the presynaptic GTPase dynamin I. Dynamin I was initially discovered as a microtubule binding protein [2], however its essential role in SVE was highlighted when it was identified as the homologue of the shibre gene product in Drosophila [3]. Shibre mutants display a temperature-sensitive paralysis originating from a block at a late stage of SVE [4]. Electron micrographs of shibire nerve terminals show large infoldings of membrane or deeply invaginated pits, both originating from the plasma membrane with electron dense collars. These collars were identified as dynamin I, suggesting that dynamin I was required for SV fission from the plasma membrane.
Dynamin I is a member of a larger family of dynamin-like proteins [5-6] of which three mammalian isoforms exist. Dynamin I is solely expressed in the brain and enriched in nerve terminals where SVE occurs. Dynamin II is ubiquitously expressed and is essential for receptor-mediated endocytosis [6]. Dynamin III expression was thought originally to be restricted to testis, however it is also expressed in the dendritic spines of neurones, suggesting a role in postsynaptic receptor internalisation [7].
Dynamin I structure
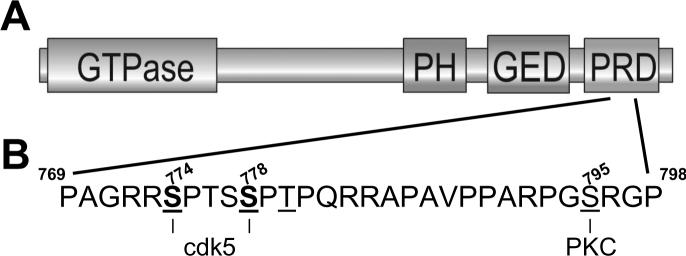
Dynamin I is a modular protein, with four identified domains (Fig 1): an N-terminal GTPase domain, pleckstrin homology (PH) domain, GTPase effector domain(GED), and C-terminal proline-rich domain (PRD). The GTPase activity of dynamin is essential for endocytosis. This was initially revealed by the shibire mutant, which has a defect in GTP binding as a result of a point mutation within the GTPase domain [3]. Subsequent studies using overexpression of similar dynamin point mutants abolish both receptor-mediated endocytosis and SVE [7-9]. The most likely role for dynamin GTPase activity in endocytosis is to produce a mechanical force to drive SV fission, either by constriction or expansion of the dynamin collar surrounding the neck of the invaginated vesicle [10-12].
Fig 1. Domain structure of dynamin I its phosphorylation sites.
(A) Dynamin is a modular protein. The GTPase domain is essential for SVE. The PH domain interacts with PI(4,5)P2 and is also essential for SVE. The GED controls both dynamin I assembly and activation of its GTPase domain. The C-terminal PRD interacts with many proteins including the endocytosis proteins amphiphysin 1 and 2, endophilin and syndapin, and signalling / cytoskeletal proteins such as P85 (regulatory subunit of PI3-kinase), Grb-2, phospholipase C γ, actin filament binding protein, profilin, cortactin, intersectin and calcineurin. (B) Location of the in vitro and in vivo phosphorlyation sites on the dynamin I PRD. The amino acid sequence corresponds to residues 769 to 798 of the Dynamin Iaa PRD. The bold underlined serines (774 and 778) indicate the endogenous cdk5 phosphorylation sites. The underlined serine (795) indicates the in vitro phosphorylation site for PKC, however this site is not phosphorylated in vivo. Amino acid sequences denoted using the single letter code.
The PH domain is principally for binding to phosphoinositides, specifically phosphatidylinositol (4,5) bisphosphate (PI(4,5)P2). PI(4,5)P2 binding to the PH domain greatly increases dynamin GTPase activity, and mutations that inhibit PI(4,5)P2 binding abolish receptor-mediated endocytosis [11,13].
When dynamin self-assembles into collars at the neck of an invaginating vesicle, its GTPase activity is activated. The GED controls this process since it regulates both dynamin I assembly and functions as a GTPase-activating domain. The role of these two functions in receptor-mediated endocytosis has been independently investigated, with overexpression of either assembly-deficient or GTPase inactive mutants increasing the amount of endocytosis [14]. This has lead to an alternative hypothesis for the role of dynamin GTPase activity in endocytosis, where activity is required to terminate interactions with downstream effectors [14-15].
The C-terminal PRD is the major protein interaction region of dynamin, encompassing multiple binding sites for proteins containing src-homology 3 (SH3) domains such as amphiphysin and endophilin [16-18]. Binding of several of these SH3 domain-containing proteins stimulate dynamin I GTPase activity, albeit to much lower levels than PI(4,5)P2 [19].
Dynamin I is a phosphoprotein that is dephosphorylated on nerve terminal stimulation
Dynamin I is a phosphoprotein and was originally discovered in nerve terminals due to its ability to undergo a rapid (< 5 sec) depolarisation-dependent dephosphorylation [20]. On termination of nerve terminal stimulation dynamin I is slowly (t1/2 approx. 40 sec) rephosphorylated by its endogenous kinase. Dynamin I is present in both the cytosol and membrane of nerve terminals, however phosphorylated dynamin I is restricted to the cytosol [21]. Thus the phosphorylation status of dynamin I would appear to regulate its localisation within the nerve terminal. It is tempting to speculate that the dephosphorylation of dynamin I facilitates its translocation to the plasma membrane for SVE and its rephosphorylation directs its return to the nerve terminal cytosol.
These initial observations on the physiology of dynamin I phosphorylation in nerve terminals were made approximately 10 years ago [20-21]. In the intervening time, many questions related to dynamin I phosphorylation and SVE have been answered, while many have still not been addressed or are contentious. The remainder of the review will summarise the current understanding of dynamin I phosphorlylation with respect to its biological activity and SVE.
Calcineurin is the in vivo dynamin I phosphatase
Dynamin I is dephosphorylated in a Ca2+-dependent manner on nerve terminal depolarisation. Thus the obvious candidate for the dynamin I phosphatase was calcineurin (PP2B), a Ca2+-dependent protein phosphatase that is enriched in nerve terminals. Calcineurin dephosphorylates purified phospho-dynamin I in vitro [22] and more importantly in vivo, since pharmacological calcineurin antagonists abolish KCl-evoked dynamin I dephosphorylation in nerve terminals [23-24]. Calcineurin co-ordinately dephosphorylates at least seven other nerve terminal proteins on nerve terminal stimulation, in addition to dynamin I. These proteins are termed the dephosphins and every member of this group of proteins (PIPK1(, AP180, epsin, eps15, amphiphysin 1, amphiphysin 2, synaptojanin and dynamin I) is essential for SVE in their own right [25].
Dynamin I binds calcineurin with high affinity within its C-terminal PRD [22]. Interestingly, calcineurin interacts with the PRD in a Ca2+-dependent manner, with binding stimulated by Ca2+ changes in the physiological range (EC50 between 100 - 400 nM) [26]. This adds another potential level of control to the dephosphoylation of dynamin I, with Ca2+-dependent recruitment of calcineurin preceding dynamin I dephosphorylation.
Cyclin-dependent kinase 5 (cdk5) is the in vivo dynamin I kinase
On termination of nerve terminal depolarisation dynamin I is slowly rephosphorylated. The identification of the endogenous dynamin I kinase was more problematic than its phosphatase, with a number of potential protein kinases identified within the past ten years. Protein kinases that phosphorylate dynamin I in vitro include - microtubule-associated protein kinase, casein kinase II, cyclin-dependent kinase 2, minibrain kinase and protein kinase C (PKC) [27-30]. PKC was assumed to be the dynamin I kinase for many years. This was because dynamin I was an excellent PKC substrate in vitro [21] and pharmacological PKC antagonists abolished dynamin I rephosphorylation in intact nerve terminals [31-32]. However there were always several inconsistencies with this assumption. For example, PKC substrates such as MARKS and GAP-43 were phosphorylated on nerve terminal stimulation, whereas dynamin I was dephosphorylated [33]. Also, dynamin I rephosphorylation only occurred after intracellular Ca2+ levels had returned to basal levels, thus after the removal of the stimulus for the Ca2+-dependent PKCs [20].
Recent evidence now proves that the endogenous dynamin I kinase is not PKC but cyclin-dependent kinase 5 (cdk5) [34]. Cdk5 is a nerve terminal enriched serine / threonine kinase that is active at rest and phosphorylates multiple proteins implicated in SV recycling in vitro [35-40]. Cdk5 phosphorylates either purified dynamin I or a GST fusion protein of the dynamin I PRD in vitro [34,39]. The in vitro phosphorylation sites for cdk5 on dynamin I were determined by mass spectrometry to be serine-774 and serine-778 [34]. Both of these residues are located in the dynamin I PRD (Fig 1) and were distinct from the previously determined in vitro PKC phosphorylation site (serine-795, [41]). Importantly, endogenous dynamin I purified from intact nerve terminals was phosphorylated on identical sites to that observed with in vitro cdk5-dependent phosphorylation [34]. Cdk5 also phosphorylates dynamin I in vivo, since dynamin I rephosphorylation is abolished by either cdk5 antagonists in intact nerve terminals or overexpression of dominant negative cdk5 in neuronal cultures [34]. The identified cdk5 phosphorylation sites are physiologically relevant, since blotting with phosphospecific antibodies raised against them showed decreases and increases in signal in depolarised and repolarised nerve terminal lysates respectively [34].
A recent in vitro study proposed that cdk5-dependent phosphorylation of dynamin I occurred on threonine-780 and not serine-774 / serine-778 [39]. However when this mass spectrometry data is re-evaluated it would seem that dynamin I is phosphorylated on serine-778 and not threonine-780. This was confirmed when the phosphorlyation sites on endogenous dynamin I were re-examined using a refined protocol employing graphite powder microcolumns [42]. There was no detectable phosphate on threonine-780, with all phosphate on serine-774 and serine-778. It was confirmed that dynamin I can be doubly phosphorylated, but in singly phosphorylated peptides phosphate was predominantly found on S-774 [42]. This suggests that S-774 may potentially be the most important site for the control of SVE.
Cdk5 rephosphoylates some, but not all, of the dephosphin group of proteins in addition to dynamin I. The lipid phosphatase synaptojanin is rephosphorylated by cdk5 in intact nerve terminals, whereas amphiphysin I and II and AP180 are not [32,34,40]. This highlights the difficulties in extrapolating in vitro phosphorlyation data to in vivo, since amphiphysin is an in vitro cdk5 substrate [38-39].
The identification of cdk5 as the dynamin I kinase has resolved some of the paradoxes raised by previous studies. For example, the inhibition of dynamin I rephosphorylation by PKC antagonists in nerve terminals was explained by the fact that the same antagonists potently inhibit cdk5 activity [34]. Likewise the reciprocal relationship between the Ca2+-dependence of the dynamin I phosphorylation cycle and PKC activation is now resolved. Since cdk5 is constitutively active in nerve terminals, there is no requirement for the activation of dynamin I phosphorylation after termination of stimulation. Thus the amount of phosphorylated or dephosphorylated dynamin I in nerve terminals is determined by the stimulation-dependent activity of calcineurin, and not cdk5.
How does dynamin I phosphorylation regulate its biological activity?
The identification of calcineurin and cdk5 as the in vivo dynamin I phosphatase and kinase allows the biological role of dynamin I phosphorylation to be investigated. Previous in vitro studies have shown that changes in dynamin I phosphorylation affect its GTPase activity and its interactions with both proteins and lipids.
Dynamin I phosphorylation and GTPase activity
In vitro phosphorylation of dynamin I by PKC stimulates its GTPase activity by approximately twelve-fold [27], with calcineurin reversing this activation [22]. However, these studies now have to be re-interpreted ([34], supplemental data). The conditions used to phosphorylate dynamin I with PKC in previous studies were found to stimulate dynamin self-assembly, resulting in an increase in GTPase activity that was independent of phosphorylation. When dynamin I self-assembly was disrupted after phosphorylation by PKC, no stimulation of GTPase activity was observed [34]. Using this modified protocol, the GTPase activity of dynamin I phosphorylated by cdk5 showed a modest three-fold stimulation. Thus phosphorylation of dynamin I by cdk5 may regulate its GTPase activity in nerve terminals. However phosphorylation cannot be the major activator, since nanotubules that incorporate PI(4,5)P2 can increase GTPase activity by approximately 1000 fold [11].
Dynamin I phosphorylation and protein-protein interactions
Dynamin I shares many interactions with other essential endocytosis proteins, however the understanding of how dynamin I phosphorylation regulates these interactions is still embryonic. Studies using rat brain detergent extracts suggested that phosphorylation of different endocytosis proteins may control their interactions [43]. The multiple protein kinase activities present in these extracts reduced the interaction of dynamin I with both amphiphysin and AP-2, an effect that was reversed by calcineurin antagonists. It is difficult to extrapolate these results to nerve terminals, since many of the protein kinases present in the lysates would not normally be able to phosphorlyate these proteins, since they would be in separate subcellular compartments.
Some in vitro studies have been performed to establish whether dynamin I phosphorylation regulates its protein interactions. For example, in vitro phosphorylation of purified dynamin I with either minibrain kinase or cdk5 inhibits its binding to the SH3 domain of amphiphysin [30,39]. However, in vitro phosphorylation of only the dynamin I PRD by cdk5 did not inhibit this interaction. Preincubation of the dynamin I PRD with the amphiphysin SH3 domain prevents its phosphorylation by cdk5 [34]. These studies suggest that other dynamin I regions may influence its putative phosphorylation-dependent interaction with amphiphysin and also indicate that dynamin I may have to be released from amphiphysin before it can be phosphorylated by cdk5, possibly on termination of SVE.
Dynamin I phosphorylation and protein-lipid interactions
Phosphorylated dynamin I is restricted to the cytosol of nerve terminals, suggesting a possible phosphorylation-dependent interaction with plasma membrane lipids [21]. In support, in vitro phosphorylation of dynamin I by either PKC or cdk5 inhibit its ability to bind to acidic phospholipids [34,41]. Dynamin I interacts with PI(4,5)P2 via its PH domain. This interaction is essential for dynamin I translocation to the plasma membrane, since point mutations within the PH domain that abolish PI(4,5)P2 binding restrict dynamin I to the cytosol [13]. If the PI(4,5)P2 - dynamin I interaction is regulated by phosphorylation, it should provide a mechanism for dynamin I movement between different subcellular compartments.
The physiological role of dynamin I phosphorylation in SVE
The phosphorylation of dynamin I regulates its biological activity, but does its phosphorylation status play any essential role in SVE? Studies in isolated nerve terminals and primary cell cultures have shown that the enzyme activities of both calcineurin and cdk5 are essential for SVE. Calcineurin is essential since the internalisation of the amphiphillic membrane marker FM2-10 was inhibited by calcineurin antagonists in isolated nerve terminals (synaptosomes) [24,32]. Cdk5 activity is also essential for SVE, with one important distinction. Inhibition of cdk5 activity does not affect SVE following nerve terminal stimulation, unlike calcineurin antagonists [32,34]. However if nerve terminals continue to be incubated with cdk5 antagonists, the next cycle of SVE is abolished. This is consistent with cdk5 being required to rephosphorylate dynamin I after SVE and thus reset the endocytic machinery for the next round of SV recycling. Morphological studies using electron microscopy also highlight the essential role for calcineurin and cdk5 in SVE. Synaptosomes preincubated with calcineurin antagonists display a severe SV depletion (indicative of a block in SVE) with no obvious changes in membrane appearance, showing SVE was arrested at an early stage (M.A.C., unpublished observations). Synaptosomes preincubated with cdk5 blockers had a normal appearance after the first stimulation of SVE, but displayed extensive SV depletion and multiple membrane invaginations after the second stimulation, indicative of a block in SVE at a specific step [34]. In addition, overexpression of dominant negative cdk5 in primary neuronal cultures greatly retarded the internalisation of the membrane marker FM4-64, highlighting its essential role in SVE [34].
The activities of calcineurin and cdk5 are essential for the activation and maintenance of SVE. However this does not reveal whether the phosphorylation status of dynamin I itself is essential, since both calcineurin and cdk5 have multiple substrates in nerve terminals. The specific role of dynamin I phosphorylation can now be addressed, since the physiological phosphorylation sites on dynamin I have been identified. We have conclusive evidence that a cycle of dynamin I phosphorylation at sites serine-774 and serine-778 is essential for SVE (KJS, unpublished observations). Thus both the initial dephosphorylation and subsequent rephosphorylation of dynamin I are required for SVE to proceed. The model in figure 2 proposes a possible mechanism of action for this process.
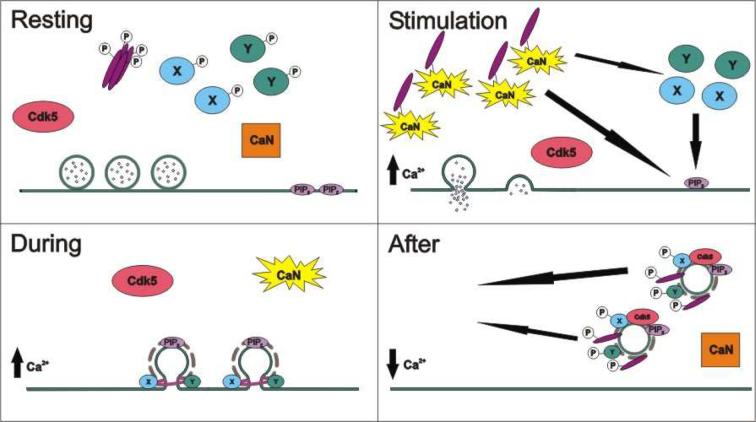
Fig 2. Model for the control of SVE by dynamin I phosphorylation.
(A) In a resting nerve terminal all phosphorylated dynamin I (purple bars) and its phosphorylation-dependent binding partner(s) (X & Y) are in the cytosol. Calcineurin (CaN) is inactive. (B) On nerve terminal depolarisation Ca2+ influx activates calcineurin, which dephosphorylates dynamin I and proteins X & Y. Dephosphorylation allows dynamin I to translocate to the plasma membrane via interactions with either or both phosphatidylinositol (4,5) bisphosphate (PIP2) or its phosphorylation-dependent binding partners (X & Y). (C) During depolarisation and SVE, dynamin I forms a collar round the neck of the retrieving SV and causes fission from the plasma membrane. (D) On termination of nerve terminal stimulation intracellular Ca2+ levels drop, inactivating CaN. This allows cdk5 to rephosphorylate dynamin I, X and Y. This rephosphorylation reduces its affinity for both PIP2 and proteins X & Y, facilitating the return of dynamin I to the cytosol for the next cycle of SVE.
Perspectives
Since the identification of dynamin I as a phosphoprotein that is dephosphorylated on nerve terminal stimulation, it has been hypothesised that its phosphorylation status may regulate its essential role in SVE. However it has taken many years for direct proof to emerge. This delay was mainly a result of identifying the physiological dynamin I kinase. However the proof of its essential role in SVE has rapidly followed the identification of its physiological phosphorylation sites. Now that calcineurin activity, cdk5 activity and the phosphorylation status of dynamin I itself have been demonstrated to be essential for SVE, this would seem to bring this story to a conclusion. However some questions regarding the phosphorylation status of dynamin I in SVE still remain unanswered.
For example, can dynamin I phosphorylation be modulated within the nerve terminal and does this control SVE? Cdk5 is activated by the proteins p39 or p35. P35 itself is a cdk5 substrate and is an unstable protein, with a half-life of 20 min. Inhibition of cdk5 activity greatly increases p35 stability, as does mutagenesis of its cdk5-dependent phosphorylation sites, suggesting a possible negative feedback mechanism [44]. Positive regulation of cdk5 activity could originate from stimulation of growth factor cascades. For example, nerve growth factor induces p35 expression in PC12 cells and brain-derived growth factor activates cdk5 activity in cultured primary neurones [45]. Also, the extent of dynamin I dephosphorylation may be controlled by the endogenous calcineurin inhibitor, cain [46]. Cain is expressed in nerve terminals, binds amphiphysin and its overexpression abolishes endocytosis in non-neuronal cells. Thus cain should be in the correct localisation to regulate dynamin I phosphorylation and possibly SVE. Further studies will determine whether this is the case.
Another question is, how does dynamin I phosphorylation control SVE? In vitro studies have hinted at three possible mechanisms, with the regulation of either or both protein or lipid interactions being most likely. However this question will not be definitively addressed until in vivo interaction studies are performed in real time, to establish whether the temporal and spatial properties of candidate interactions match their proposed role in SVE. The development of fluorescence resonance energy transfer-based interaction assays should facilitate these experiments.
We have shown that dual serine-774 and serine-778 mutants abolish SVE (KJS, unpublished observations), however do these sites control SVE independently of each other? Serine-774 may be the essential residue for SVE, since it is the predominately phosphorylated site on singly phosphorylated dynamin I [42]. This can be tested by overexpressing dynamin I mutants in neurones that encompass single substitutions at either serine-774 or serine-778 and examining the effect on SVE.
Now that the essential role for the phosphorylation status of dynamin I in SVE is established the remaining immediate questions will become more and more targeted. One final wider question is, do the phosphoryation status of all the other dephosphins also regulate SVE? This question will have to be addressed in a similar manner to dynamin I, via the identification of the physiologically relevant in vivo phosphorylation sites of the other dephosphins. When these questions are answered then we should be a long way to discovering the mechanism by which global dephosphorylation of the dephopshins controls SVE.
Acknowledgements
This work was supported by grants from The Wellcome Trust (Ref: GR070569) and a University of Edinburgh Medical Faculty Scholarship (to KJS).
Reference List
- 1.Cousin MA. Mol. Neurobiol. 2000;22:115–128. doi: 10.1385/MN:22:1-3:115. [DOI] [PubMed] [Google Scholar]
- 2.Shpetner HS, Vallee RB. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- 3.van der Bliek AM, Meyerowitz EM. Nature. 1991;351:411–414. doi: 10.1038/351411a0. [DOI] [PubMed] [Google Scholar]
- 4.Koenig JH, Ikeda K. J. Neurosci. 1989;9:3844–3860. doi: 10.1523/JNEUROSCI.09-11-03844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu JP, Robinson PJ. Endocr. Rev. 1995;16:590–607. doi: 10.1210/edrv-16-5-590. [DOI] [PubMed] [Google Scholar]
- 6.Urrutia R, Henley JR, Cook T, McNiven MA. Proc. Natl. Acad. Sci. U. S. A. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray NW, Fourgeaud L, Huang B, Chen J, Cao H, Oswald BJ, Hemar A, McNiven MA. Curr. Biol. 2003;13:510–515. doi: 10.1016/s0960-9822(03)00136-2. [DOI] [PubMed] [Google Scholar]
- 8.Herskovits JS, Burgess CC, Obar RA, Vallee RB. J. Cell Biol. 1993;122:565–578. doi: 10.1083/jcb.122.3.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL. J. Cell Biol. 1993;122:553–563. doi: 10.1083/jcb.122.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sweitzer SM, Hinshaw JE. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 11.Stowell MH, Marks B, Wigge P, McMahon HT. Nat. Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- 12.Marks B, Stowell MH, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 13.Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Curr. Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 14.Sever S, Muhlberg AB, Schmid SL. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- 15.Song BD, Schmid SL. Biochemistry. 2003;42:1369–1376. doi: 10.1021/bi027062h. [DOI] [PubMed] [Google Scholar]
- 16.David C, McPherson PS, Mundigl O, De Camilli P. Proc. Natl. Acad. Sci. U. S. A. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringstad N, Nemoto Y, De Camilli P. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8569–8574. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Heuvel E, Bell AW, Ramjaun AR, Wong K, Sossin WS, McPherson PS. J. Biol. Chem. 1997;272:8710–8716. doi: 10.1074/jbc.272.13.8710. [DOI] [PubMed] [Google Scholar]
- 19.Barylko B, Binns DD, Albanesi JP. Methods Enzymol. 2001;329:486–496. doi: 10.1016/s0076-6879(01)29110-1. [DOI] [PubMed] [Google Scholar]
- 20.Robinson PJ, Liu JP, Powell KA, Fykse EM, Südhof TC. Trends Neurosci. 1994;17:348–353. doi: 10.1016/0166-2236(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 21.Liu JP, Powell KA, Südhof TC, Robinson PJ. J. Biol. Chem. 1994;269:21043–21050. [PubMed] [Google Scholar]
- 22.Liu JP, Sim ATR, Robinson PJ. Science. 1994;265:970–973. doi: 10.1126/science.8052858. [DOI] [PubMed] [Google Scholar]
- 23.Bauerfeind R, Takei K, De Camilli P. J. Biol. Chem. 1997;272:30984–30992. doi: 10.1074/jbc.272.49.30984. [DOI] [PubMed] [Google Scholar]
- 24.Marks B, McMahon HT. Curr. Biol. 1998;8:740–749. doi: 10.1016/s0960-9822(98)70297-0. [DOI] [PubMed] [Google Scholar]
- 25.Cousin MA, Robinson PJ. Trends Neurosci. 2001;24:659–665. doi: 10.1016/s0166-2236(00)01930-5. [DOI] [PubMed] [Google Scholar]
- 26.Lai MM, Hong JJ, Ruggiero AM, Burnett PE, Slepnev VI, De Camilli P, Snyder SH. J. Biol. Chem. 1999;274:25963–25966. doi: 10.1074/jbc.274.37.25963. [DOI] [PubMed] [Google Scholar]
- 27.Robinson PJ, Sontag J-M, Liu JP, Fykse EM, Slaughter C, McMahon HT, Südhof TC. Nature. 1993;365:163–166. doi: 10.1038/365163a0. [DOI] [PubMed] [Google Scholar]
- 28.Hosoya H, Komatsu S, Shimizu T, Inagaki M, Ikegami M, Yazaki K. Biochem. Biophys. Res. Commun. 1994;202:1127–1133. doi: 10.1006/bbrc.1994.2045. [DOI] [PubMed] [Google Scholar]
- 29.Earnest S, Khokhlatchev A, Albanesi JP, Barylko B. FEBS Lett. 1996;396:62–66. doi: 10.1016/0014-5793(96)01074-5. [DOI] [PubMed] [Google Scholar]
- 30.Chen-Hwang M-C, Chen H-R, Elzinga M, Hwang Y-W. J. Biol. Chem. 2002;277:17597–17604. doi: 10.1074/jbc.M111101200. [DOI] [PubMed] [Google Scholar]
- 31.Robinson PJ. J. Biol. Chem. 1992;267:21637–21644. [PubMed] [Google Scholar]
- 32.Cousin MA, Tan TC, Robinson PJ. J. Neurochem. 2001;76:105–116. doi: 10.1046/j.1471-4159.2001.00049.x. [DOI] [PubMed] [Google Scholar]
- 33.Robinson PJ. Mol. Neurobiol. 1992;5:87–142. doi: 10.1007/BF02935541. [DOI] [PubMed] [Google Scholar]
- 34.Tan TC, Valova VA, Malladi CS, Graham ME, Berven LA, Jupp OJ, Hansra G, McClure SJ, Sarcevic B, Boadle RA, Larsen MR, Cousin MA, Robinson PJ. Nat. Cell Biol. 2003;5:701–710. doi: 10.1038/ncb1020. [DOI] [PubMed] [Google Scholar]
- 35.Matsubara M, Kusubata M, Ishiguro K, Uchida T, Titani K, Taniguchi H. J. Biol. Chem. 1996;271:21108–21113. doi: 10.1074/jbc.271.35.21108. [DOI] [PubMed] [Google Scholar]
- 36.Shuang RQ, Zhang L, Fletcher A, Groblewski GE, Pevsner J, Stuenkel EL. J. Biol. Chem. 1998;273:4957–4966. doi: 10.1074/jbc.273.9.4957. [DOI] [PubMed] [Google Scholar]
- 37.Rosales JL, Nodwell MJ, Johnston RN, Lee KY. J. Cell. Biochem. 2000;78:151–159. doi: 10.1002/(sici)1097-4644(20000701)78:1<151::aid-jcb14>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 38.Floyd SR, Porro EB, Slepnev VI, Ochoa GC, Tsai LH, De Camilli P. J. Biol. Chem. 2001;276:8104–8110. doi: 10.1074/jbc.M008932200. [DOI] [PubMed] [Google Scholar]
- 39.Tomizawa K, Sunada S, Lu YF, Oda Y, Kinuta M, Ohshima T, Saito T, Wei FY, Matsushita M, Li ST, Tsutsui K, Hisanaga SI, Mikoshiba K, Takei K, Matsui H. J. Cell Biol. 2003;163:813–824. doi: 10.1083/jcb.200308110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SY, Wenk MR, Kim Y, Nairn AC, De Camilli P. Proc. Natl. Acad. Sci. U. S. A. 2004;101:546–551. doi: 10.1073/pnas.0307813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Powell KA, Valova VA, Malladi CS, Jensen ON, Larsen MR, Robinson PJ. J. Biol. Chem. 2000;275:11610–11617. doi: 10.1074/jbc.275.16.11610. [DOI] [PubMed] [Google Scholar]
- 42.Larsen MR, Graham ME, Robinson PJ, Roepstorff P. Mol Cell Proteomics. 2004;3:456–465. doi: 10.1074/mcp.M300105-MCP200. [DOI] [PubMed] [Google Scholar]
- 43.Slepnev VI, Ochoa GC, Butler MH, Grabs D, De Camilli P. Science. 1998;281:821–824. doi: 10.1126/science.281.5378.821. [DOI] [PubMed] [Google Scholar]
- 44.Patrick GN, Zhou P, Kwon YT, Howley PM, Tsai LH. J. Biol. Chem. 1998;273:24057–24064. doi: 10.1074/jbc.273.37.24057. [DOI] [PubMed] [Google Scholar]
- 45.Dhavan R, Tsai LH. Nat. Rev. Mol. Cell Biol. 2001;2:749–759. doi: 10.1038/35096019. [DOI] [PubMed] [Google Scholar]
- 46.Lai MM, Luo HR, Burnett PE, Hong JJ, Snyder SH. J. Biol. Chem. 2000;275:34017–34020. doi: 10.1074/jbc.C000429200. [DOI] [PubMed] [Google Scholar]