Abstract
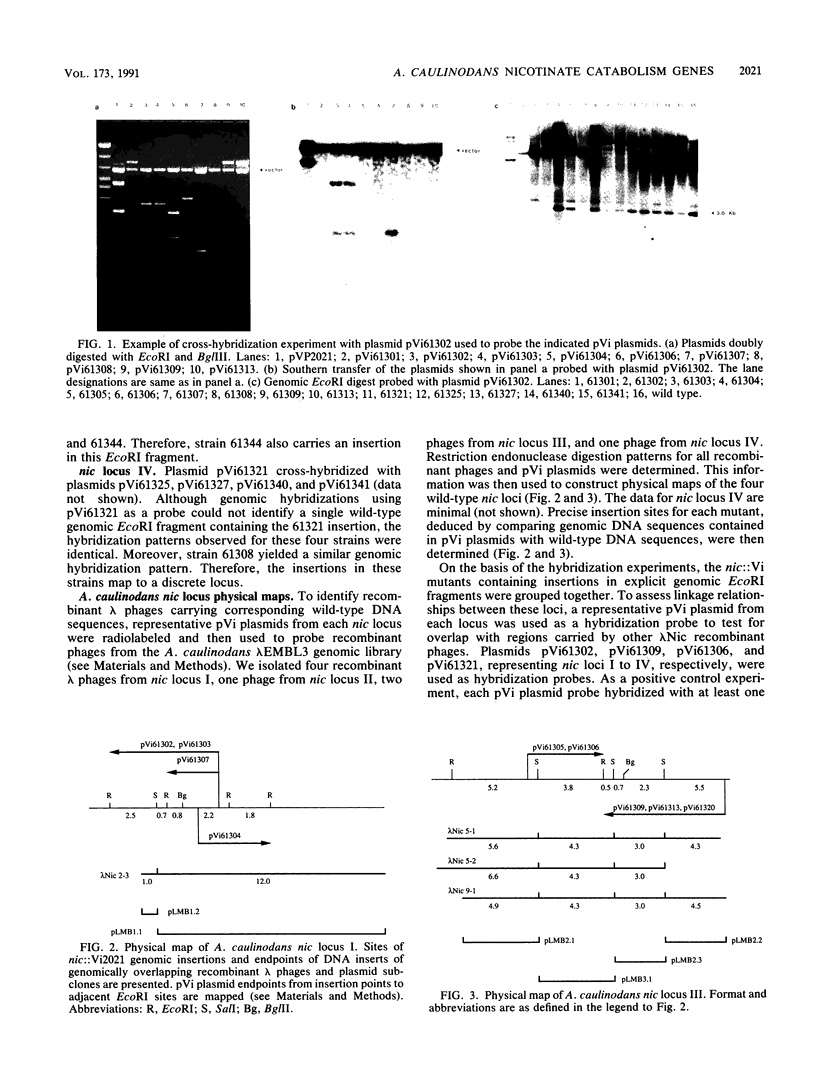
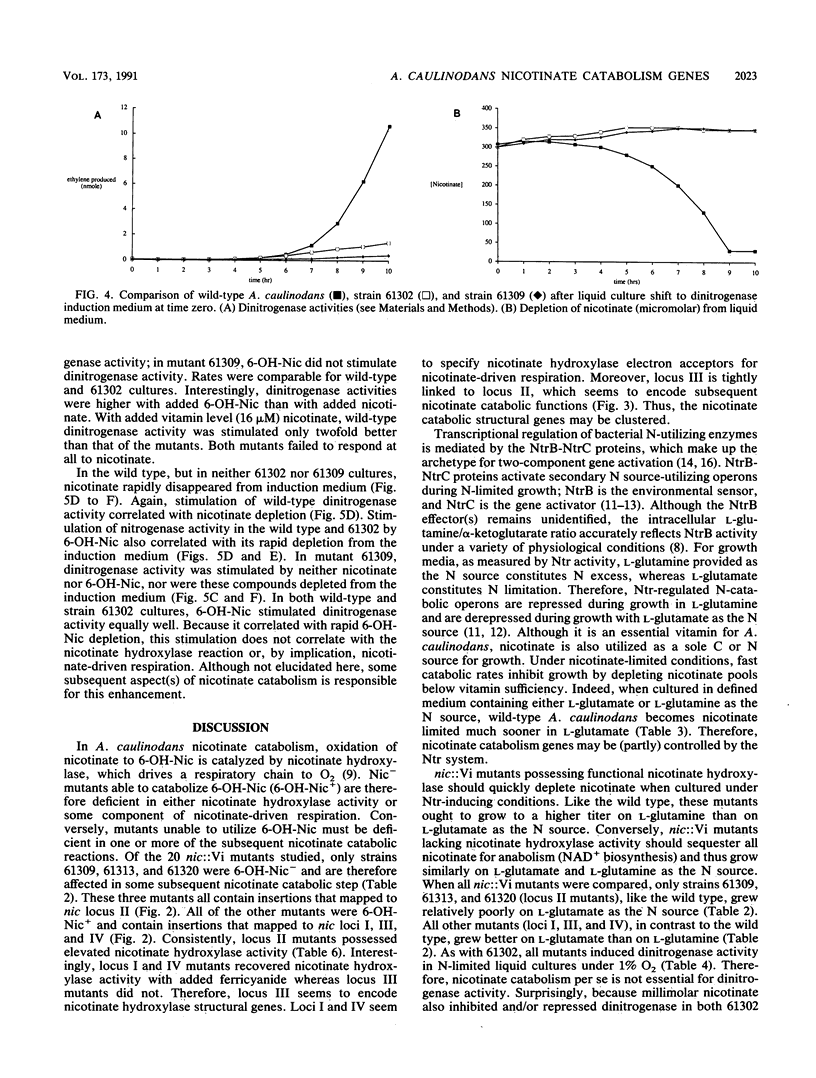
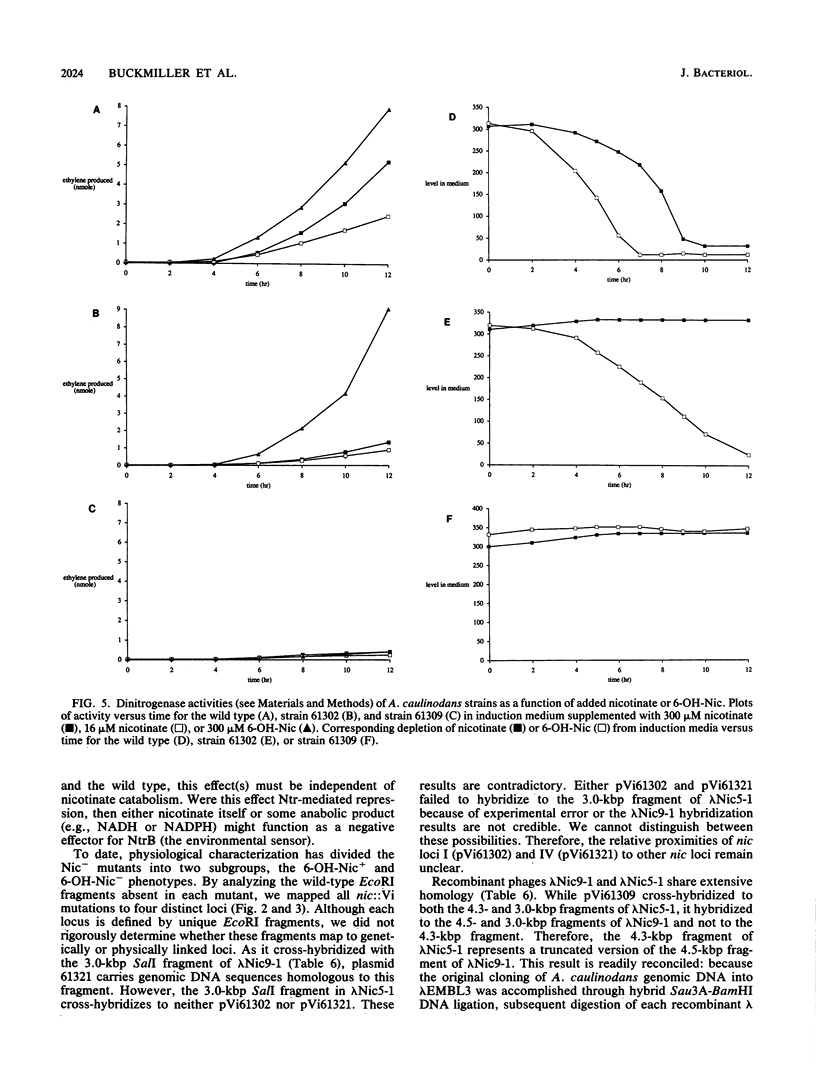
Twenty Azorhizobium caulinodans vector insertion (Vi) mutants unable to catabolize nicotinate (Nic- phenotype) were identified and directly cloned as pVi plasmids. These pVi plasmids were used as DNA hybridization probes to isolate homologous wild-type sequences. From subsequent physical mapping experiments, the nic::Vi mutants defined four distinct loci. Two, possibly three, of these loci are physically linked. A. caulinodans nic loci II and III encode the structural genes for nicotinate catabolism; nic loci I and IV encode nicotinate-driven respiratory chain components. Recombinant lambda bacteriophages corresponding to three of these loci were subcloned in pRK293; resulting plasmids were used for complementation tests with resolved nic::IS50 derivatives of the nic::Vi mutants. When wild-type A. caulinodans was cultured in defined liquid medium under 3% O2, nicotinate catabolism stimulated N2 fixation 10-fold. In these exponentially growing cultures, the entire (300 microM) nicotinate supplement was exhausted within 10 h. While nic::Vi mutants retained the ability to fix some N2, they did so at rates only 10% of that of the wild type: nitrogenase activity by nic::Vi mutants was not stimulated by 300 microM added nicotinate. Higher-level (5 mM) nicotinate supplementation inhibited N2 fixation. Because 5 mM nicotinate repressed nitrogenase induction in all nic::Vi mutants as well, this repression was independent of nicotinate catabolism. During catabolism, nicotinate is first oxidized to 6-OH-nicotinate by a membrane-bound nicotinate hydroxylase which drives a respiratory chain to O2. In A. caulinodans wild-type cultures, added 300 microM 6-OH-nicotinate stimulated N2 fixation twofold better than did added 300 microM nicotinate. Likewise, nic::Vi mutant 61302, defective in nicotinate hydroxylase, fixed N2 at wild-type levels when supplemented with 300 microM 6-OH-nicotinate. Therefore, nicotinate catabolism stimulates N2 fixation not by nicotinate hydroxylase-driven respiration but rather by some subsequent aspect(s) of nicotinate catabolism.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald R. G., Nees D. W., Raymond C. K., Loroch A. I., Ludwig R. A. Characterization of three genomic loci encoding Rhizobium sp. strain ORS571 N2 fixation genes. J Bacteriol. 1986 Jan;165(1):72–81. doi: 10.1128/jb.165.1.72-81.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald R. G., Raymond C. K., Ludwig R. A. Vector insertion mutagenesis of Rhizobium sp. strain ORS571: direct cloning of mutagenized DNA sequences. J Bacteriol. 1985 Apr;162(1):317–323. doi: 10.1128/jb.162.1.317-323.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus B. L., Elmerich C., Dommergues Y. R. Free-living Rhizobium strain able to grow on n(2) as the sole nitrogen source. Appl Environ Microbiol. 1983 Feb;45(2):711–713. doi: 10.1128/aem.45.2.711-713.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtel A., Merrick M. J. The Klebsiella pneumoniae PII protein (glnB gene product) is not absolutely required for nitrogen regulation and is not involved in NifL-mediated nif gene regulation. Mol Gen Genet. 1989 Jun;217(2-3):474–480. doi: 10.1007/BF02464920. [DOI] [PubMed] [Google Scholar]
- Kitts C. L., Schaechter L. E., Rabin R. S., Ludwig R. A. Identification of cyclic intermediates in Azorhizobium caulinodans nicotinate catabolism. J Bacteriol. 1989 Jun;171(6):3406–3411. doi: 10.1128/jb.171.6.3406-3411.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig R. A. Rhizobium sp. strain ORS571 grows synergistically on N2 and nicotinate as N sources. J Bacteriol. 1986 Jan;165(1):304–307. doi: 10.1128/jb.165.1.304-307.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B. Genetic control of nitrogen assimilation in bacteria. Annu Rev Genet. 1982;16:135–168. doi: 10.1146/annurev.ge.16.120182.001031. [DOI] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]



