Abstract
Hereditary neuropathy with liability to pressure palsies (HNPP) is an autosomal dominant, demyelinating neuropathy. Point mutations in the PMP22 gene are a rare cause of HNPP. A novel PMP22 splice site mutation (c.179+1 G→C) is reported in an HNPP family. By reverse transcriptase‐polymerase chain reaction experiments, this mutation was shown to cause the synthesis of an abnormal mRNA in which a premature stop codon probably produces a truncated non‐functional protein.
Keywords: HNPP, splice site mutation, PMP22
Hereditary neuropathy with liability to pressure palsies (HNPP) (OMIM 162500) is an autosomal dominant disorder characterised by recurrent peripheral nerve palsies and electrophysiological and neuropathological evidence of widespread demyelination, with typical thickenings of the myelin sheaths called tomacula. The genetic defect in most HNPP patients is a 1.5 Mb deletion on chromosome 17p11.2 containing the PMP22 gene, thus producing a dose effect of this gene.1
A few HNPP patients harbouring point mutations in the PMP22 gene have been reported (http://molgen‐www.uia.ac.be/CMTMutations/), including two mutations involving splice sites.2,3 No RNA studies are available of these splice site mutations.
We report a novel splice site mutation (c.179+1G→C) which affects the 5′ donor splice site of PMP22 intron 2. Reverse transcriptase polymerase chain reaction (RT‐PCR) experiments showed that the c.179+1G→C mutation determines the formation, in the patient's sural nerve, of an abnormal mRNA in which a premature stop codon (E60fsX75) is likely to produce a truncated PMP22 protein.
Methods
Clinical and electrophysiological examination followed standard methods.
The duplication/deletion of 17p11.2 region was investigated by pulsed field gel electrophoresis (PFGE) and hybridisation with pNEA102 probe.4
The PMP22 coding sequence, including exon–intron boundaries, was amplified with the published primers5 and screened by single strand conformation polymorphism (SSCP) analysis.6 PCR products were sequenced in both strands using a 310 Genetic Analyser (PE Applied Biosystems, Foster City, California, USA) according to ABI Prism Dye Terminator chemistry.
RNA was isolated from the patient's sural nerve following a standard procedure. After DNAse treatment, cDNA was prepared using cMasterTM RT (Eppendorf AG, Germany) according to manufacturer's instructions. PMP22 cDNA was co‐amplified together with the glyceraldehyde‐3‐phosphate dehydrogenase (G3PDH) cDNA as an endogenous standard sequence.1
Two regions of the gene were amplified. A first set of primers (PMP22‐RT F 5′‐ TGTTCGTCTCCACGATCGTC‐3′and PMP22‐RT R 5′‐TCCCTTCCCTATGTACGCTC‐3′) was used to amplify the whole coding sequence (476 bp of cDNA, NCBI accession number NM_000304). A second reaction was carried out with primers PMP22‐RT F and PMP22‐intr2R (5′‐CCAATAAGCGTTTCCAGCTCT‐3′ in intron 2). Detailed protocols are available on request. Different sized PCR products were sequenced as described above.
Results
Family report
The proband (fig 1A, II.1), a 42 year old woman with 18 years of professional exposure to lead, had a 20 year history of paresthesiae, occasional weakness, and nocturnal cramps in the legs, recently involving the left arm as well. At 19 years of age she had an episode of right lateral popliteal nerve paralysis with complete recovery after two months. Neurological examination at the age of 40 years showed stocking‐glove hypoaesthesia, distal hypopallestesia, areflexia, and mild distal weakness in the lower limbs. All routine serum analysis were normal, including thyroid function, antinuclear antibodies, antiganglioside (GM1, GM2, GD1a, GD1b) and antisulphatide antibodies, serum lead level, aminolaevulinic acid deidrathase, zinc protoporphirin, cerebrospinal fluid (CSF) protein level, and CSF cell count. The neurophysiological study (table 1, II.1) revealed chronic denervation on distal muscles and a diffuse slowing of motor and sensory conduction velocities, more evident at the common entrapment sites.
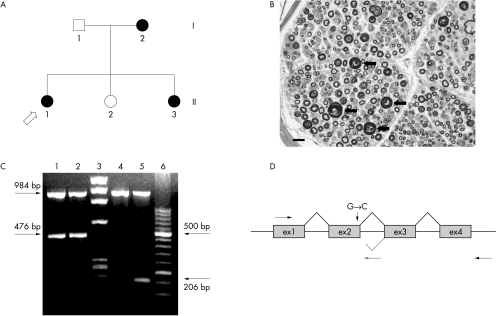
Figure 1 (A) Pedigree of the family with hereditary neuropathy with liability to pressure palsies (HNPP) with the splice site mutation. Black symbols indicate affected individuals; circles, females; squares, males; roman numerals, generations; arabic numbers, individuals. (B) Transverse semithin section from the sural nerve biopsy of patient II.1. The density of myelinated fibres is normal. Several fibres show focal thickenings of the myelin sheath (arrows), typical of HNPP. Stain, toluidine blue; bar = 20 µm. (C) Agarose gel electrophoresis of polymerase chain reaction (PCR) products obtained from the amplification of the cDNAs from the patient II.1 (lanes 2 and 5) and from normal control (lanes 1 and 4). The 984 bp PCR product corresponds to the control gene G3PDH (glyceraldeyde‐3‐phosphate dehydrogenase). Lane 3: DNA molecular weight marker IX (φX174). Lane 6: DNA molecular weight marker XIII (Roche). (D) Schematic figure representing the structure of the PMP22 gene and the position of the primers used; PMP22‐RT F (→), PMP22‐intr2R (←) and PMP22‐RT R (grey reverse arrow). Grey line shows a possible mechanism of splicing of the transcript from the mutated allele.
Table 1 Electrophysiological features of patients with the c.179+1G→C mutation.
| Motor nerve conduction velocities | Sensory nerve conduction velocities | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | Peroneal | Median | Sural | |||||||||
| DL (ms) (<4) | CMAP (mV) (>7) | MCV (m/s) (>50) | DL (ms) (<5) | CMAP (mV) (>5) | MCV (m/s) (>40) | DL (ms) | SAP (mV)(>14) | SCV (m/s)(>44) | DL (ms) | SAP (mV) (>4) | SCV (m/s) (>40) | |
| II.1 | 4.78* | 8.74 | 51.1 | 7.75* | 2.12* | 37.2* | 4.85 | 12 | 29.9* | 4.73 | 12.9 | 36.5* |
| II.3 | 6.87* | 7.45 | 54.7 | 7.35* | 3.34 | 42.6 | 4.5 | 10.4 | 30.2* | 4.19* | 14.8 | 33.4* |
| I.2 | 6.22* | 7.32 | 60.8 | 7.25* | 3.56* | 33.3* | 4.55 | 4.69* | 23.9* | 4.41* | 3.97 | 31.7* |
Normal reference values in brackets.
*Pathological values.
CMAP, compound muscle action potential amplitude (mV); DL, distal latencies; MCV, motor conduction velocity (m/s); SAP, sensory action potential (μV); SCV, sensory conduction velocity (m/s).
A sural nerve biopsy, done for diagnostic purposes at the age of 40 years, revealed a normal density of myelinated fibres. Several fibres showed thickening of the myelin sheath compared with the axon diameter, thus resembling tomacula. A teased fibre preparation confirmed the presence of focal swellings of the myelin sheath. Small onion bulbs were also occasionally observed (fig 1B).
Neurophysiological study of the 71 year old mother (I.2) and the 40 year old sister (II.3), both clinically asymptomatic, revealed a demyelinating neuropathy (table 1).
The duplication/deletion of the region 17p11.2‐12 in the proband was excluded by PFGE analysis.
SSCP screening of the PMP22 gene showed, in the proband, an altered fragment in exon 2. Sequence analysis of this fragment revealed a heterozygous G→C transversion at nucleotide c.179+1. This mutation is located at the 5′ donor splice site of intron 2. The same mutation was detected in the mother and in the sister of the proband but was not revealed in 208 control chromosomes.
To predict the influence of this mutation on RNA splicing, an RT‐PCR reaction using RNA from the sural nerve of the proband and of two control nerves was carried out. The amplification with PMP22‐RT F and PMP22‐RT R primers produced, in both patient and control nerves, an expected fragment of 476 bp (fig 1C). In order to check the presence of the retained intron, the cDNA was amplified with PMP22‐RT F and PMP22‐intr2R primers, the latest having its target sequence in intron 2. This reaction produced a 206 bp fragment in the proband, which was absent in control nerves (fig 1C). Sequencing analysis of these fragments showed that the longer product represents the full length wild type allele whereas the 206 bp product corresponds to the 5′ end of exon 1, exon 2, and a portion of intron 2 (∼82 bp) which is included in the transcript. The mutation of the 5′ donor splice site probably affects the correct splicing of the mRNA and introduces intron 2 in the abnormal mRNA. Therefore the first set of primers is unable to amplify the entire abnormal mRNA owing to the length of the intron (∼19.5 kb). The alteration of the sequence caused by c179+1G→C mutation predicts a premature stop codon at 45 nucleotides from the splice site (E60fsX75).
Discussion
We found a novel mutation at nucleotide c.179+1 that affects the 5′ donor splice site of the intron 2 of the PMP22 gene. Using RT‐PCR experiments we demonstrated that the c.179+1G→C mutation causes the synthesis of an abnormal mRNA which includes a portion of intron 2. Partial sequencing of this abnormal mRNA showed the formation of a stop codon at 45 nucleotides from the splice site. Translation of the mutant allele may produce a shorter protein unable to be processed and transported to the Schwann cell membrane and then to the myelin sheath.
The patients with c.179+1G→C mutation showed clinical, neurophysiological, and neuropathological features of a typical HNPP phenotype.1 Point mutations of PMP22 gene cause a wide variety of demyelinating neuropathies from the milder HNPP to the more severe Charcot‐Marie‐Tooth disease type 1A (CMT1A), Dejerine‐Sottas syndrome (DSS), and congenital hypomyelination (CH). The molecular mechanisms by which PMP22 mutations induce CMT1A, DSS, and CH probably include protein misfolding and abnormalities of the endocellular trafficking of wild type protein.7 To date, 11 mutations have been reported (http://molgen‐www.uia.ac.be/CMTMutations/) in HNPP patients and all of them are likely to cause a loss of function of the protein. Among them, only two splicing mutations were previously reported, for which no peripheral nerve RNA studies are available. The first mutation reported is a 5′‐splice site mutation which affects the splice donor site of intron 1 (c.78+1G→T). An ectopic mRNA analysis of leucocytes from the patient with this mutation suggested that exon 1 was skipped and the mutant allele acts as a null allele.2 The second one (c.179‐1 G→C) affects the splice acceptor site preceding exon 3 and most probably affects normal splicing of PMP22 mRNA by skipping of the exon 3 or by using a cryptic splice site.3
The c.179+1G→C mutation reported here affects a donor splice site and determines the production of an abnormal mRNA that contains a premature stop codon, probably resulting in a shorter protein. Thus the loss of function caused by a truncated protein that is likely to be rapidly degraded, resulting in a reduced dosage of PMP22, can be comparable to the 1.5 Mb deletion. This is in agreement with the fact that the “dosage sensitive” PMP22 gene accounts for two diseases—HNPP through monosomic underexpression, and CMT1A through trisomic overexpression.
Acknowledgements
This work was partly supported by Fondazione Mariani grant R‐05‐44 to PM, by Telethon grant GUP04009 to EB, and by Telethon grants GGP02169 and GUP04002 to AS. The Department of Neurosciences, Ophthalmology and Genetics participates to the European CMT Consortium with the Sections of Medical Genetics and Neurology. We are grateful to the patients for their collaboration.
Abbreviations
CMT1A - Charcot‐Marie‐Tooth disease type 1A
HNPP - hereditary neuropathy with liability to pressure palsies
PFGE - pulsed field gel electrophoresis
SSCP - single strand conformation polymorphism
Footnotes
Competing interests: none declared
References
- 1.Schenone A, Nobbio L, Mandich P.et al Underexpression of messenger RNA for peripheral myelin protein 22 in hereditary neuropathy with liability to pressure palsies. Neurology 199748445–449. [DOI] [PubMed] [Google Scholar]
- 2.Bort S, Nelis E, Timmerman V.et al Mutational analysis of the MPZ, PMP22 and Cx32 genes in patients of Spanish ancestry with Charcot‐Marie‐Tooth disease and hereditary neuropathy with liability to pressure palsies. Hum Genet 199799746–754. [DOI] [PubMed] [Google Scholar]
- 3.Meuleman J, Pou‐Serradell A, Lofgren A.et al A novel 3′‐splice site mutation in peripheral myelin protein 22 causing hereditary neuropathy with liability to pressure palsies. Neuromuscul Disord 200111400–403. [DOI] [PubMed] [Google Scholar]
- 4.Lorenzetti D, Pareyson D, Sghirlanzoni A.et al A 1.5 Mb deletion in 17p11.2‐p12 is frequently observed in Italian families with hereditary neuropathy with liability to pressure palsies. Am J Hum Genet 19955691–98. [PMC free article] [PubMed] [Google Scholar]
- 5.Roa B B, Garcia C A, Suter U.et al Charcot‐Marie‐Tooth disease type 1A. Association with a spontaneous point mutation in the PMP22 gene. N Engl J Med 199332996–101. [DOI] [PubMed] [Google Scholar]
- 6.Mandich P, Montera M, Bellone E.et al Three novel mutations in the Von Hippel‐Lindau tumour suppressor gene in Italian patients. Hum Mut 19981S268–S270. [DOI] [PubMed] [Google Scholar]
- 7.Sanders C R, Ismail‐Beigi F, McEnery M W. Mutations of peripheral myelin protein 22 result in defective trafficking through mechanisms which may be common to diseases involving tetraspan membrane proteins. Biochemistry 2001409453–9459. [DOI] [PubMed] [Google Scholar]