Abstract
Specificity in the immune system is dictated and regulated by specific recognition of peptide/major histocompatibility complex (MHC) complexes by the T cell receptor. Such peptide/MHC complexes are a desirable target for novel approaches in immunotherapy because of their highly restricted fine specificity. Recently, phage display was used to isolate an antibody that has T cell receptor-like specificity. It recognizes mouse MHC class I H-2Kk molecules complexed with a H-2Kk-restricted influenza virus-derived hemagglutinin peptide (Ha255–262) but does not bind to class I H-2Kk alone, peptide alone, or H-2Kk complexed with other peptides. We have used this antibody to make a recombinant antibody–toxin fusion protein (immunotoxin) and show herein that it specifically kills antigen-presenting cells in a peptide-dependent manner and with T cell receptor-like specificity. We find a striking correlation between the fine specificity of binding of the antibody and the cytotoxic activity of the recombinant immunotoxin. We also show specific killing of influenza virus-infected target cells. The results suggest that it should be possible to develop novel immunotherapeutic strategies against human cancer by making recombinant antibodies that will recognize cancer-related peptides complexed with MHC class I molecules on the surface of cancer cells and using these to deliver toxins, radioisotopes, or cytotoxic drugs to the cancer cells.
Keywords: immunotoxin/targeted therapy
Expression of specific peptides in complex with major histocompatibility complex (MHC) class I molecules on cells was shown to be associated with cancer and autoimmune disorders (1–3). For example, human melanomas express tumor-specific peptides that are presented to the immune system in complex with class I HLA-A2 molecules (1). Specific targeting of drugs to these cells by using these specific peptide/MHC complexes will be a useful and promising therapeutic approach. To develop such a strategy, there is a need to isolate new targeting moieties such as recombinant antibodies that will recognize specifically peptide/MHC complexes.
Fab 13.4.1 was recently isolated from a phage display library. This recombinant antibody recognizes mouse H2-Kk class I molecules complexed with the Kk-restricted influenza virus-derived peptide of hemagglutinin (peptide Ha255–262) only and does not bind class I Kk molecules complexed with other peptides or class I molecules alone (4). Thus, this is a recombinant antibody with T cell receptor-like specificity and restriction. This antibody can bind to soluble Ha/H-2Kk complexes as well as complexes expressed on cells and it can inhibit specifically peptide-dependent T cell responses of T cell hybridoma lines specific for the Ha255–262/H-2Kk complex (4). More recently, it was also demonstrated that Fab 13.4.1 shares a striking similarity to the specificity of T cell hybridomas that recognize Ha255–262 and most of the peptide residues, which were found to be recognized by the T cells, could also be recognized by the antibody (5).
To determine whether such an antibody can be used for specific targeting of a drug or toxin to antigen-presenting cells (APCs), we have constructed a fusion protein composed of Fab 13.4.1 and a truncated form of Pseudomonas exotoxin (PE38) that contains the translocation and ADP ribosylation domains of whole PE but lacks the cell binding domain (6, 7). We describe herein the ability of the Fab 13.4.1–PE38 fusion protein to kill specifically APCs that express the class I MHC/peptide complexes recognized by Fab 13.4.1. Moreover, we show that the recombinant antibody–toxin fusion protein can kill specifically influenza virus-infected cells that present the MHC/peptide complex.
EXPERIMENTAL PROCEDURES
Construction, Expression, and Production of Fab 13.4.1–PE38.
Fab 13.4.1 was isolated previously from a phage display library as described (4). The Fd and Vκ were recloned by PCR using the oligonucleotides 5′-GGAAGCGTTGGCGCATATGCAGGTCCAGCTGCAGCAGTCT-3′ and 5′-CAGTAAAAGCTTTTTATCAACAATCCCTGGGCACAAT-3′ for the Fd and 5′-GGAAGCGTTGGCGCATATGGACATTCAGATGACACAGTCT-3′ and 5′-CAGTAAAAGCTTTACACTCATTCCTGTTGAAGCT-3′ for the Vκ. These oligonucleotides inserted also NdeI and HindIII cloning sites (underlined) and a stop codon after the Fd. The resulting Fd and Vκ PCR fragments were cloned into the expression vector pULI7 (8) and the resulting plasmids encode the Fab 13.4.1 Fd (pYR13.4.1/Fd) and Fab 13.4.1 Vκ fused to a gene encoding PE38 (pYR13.4.1/Vκ–PE38). The gene encoding PE38 that contains the translocation and ADP ribosylation domains of PE was fused to the C terminus of Fab 13.4.1 Lκ. The expression vectors for the Fab 13.4.1–PE38 fusion protein are driven by the T7 promoter. These constructs were expressed in Escherichia coli BL21lDE3 and accumulated in insoluble intracellular inclusion bodies. The recombinant Fab 13.4.1–PE38 and Fab 13.4.1 alone were produced from inclusion bodies by established protocols of solubilization and refolding (8, 9). Fab 13.4.1–PE38 was purified by ion-exchange chromatography on Q-Sepharose and Mono Q followed by size exclusion on TSK3000 column. Recombinant Fab 13.4.1 was purified from refolding solution by sequential chromatography on Q-Sepharose and S-Sepharose followed by chromatography on TSK3000 column.
Cytotoxicity Assays on RMA-S⋅Kk APCs.
RMA-S⋅Kk cells (107) were incubated overnight with 0.1 mM peptides at 26°C. Cells were then washed twice with medium and incubated for 20 hr with recombinant Fab 13.4.1–PE38. Protein synthesis inhibition is measured by incorporation of [3H]leucine into cell proteins. IC50 is the concentration of immunotoxin which causes 50% inhibition of protein synthesis.
Infection of Target Cells with Influenza Virus.
RMA-k or L929 cells (107) were incubated with various dilutions of influenza virus strain A2/Japan/305/57 and PR-8 in 1 ml PBS for 1 hr at 37°C. Serum-containing growth medium (RPMI 1640 plus 10% fetal calf serum) was than added and cells were further incubated for 12–16 hr at 37°C. Infection efficiency was monitored by fluorescence-activated cell sorter (FACS) analysis using an antinuclear protein mAb as described in the legend to Fig. 5.
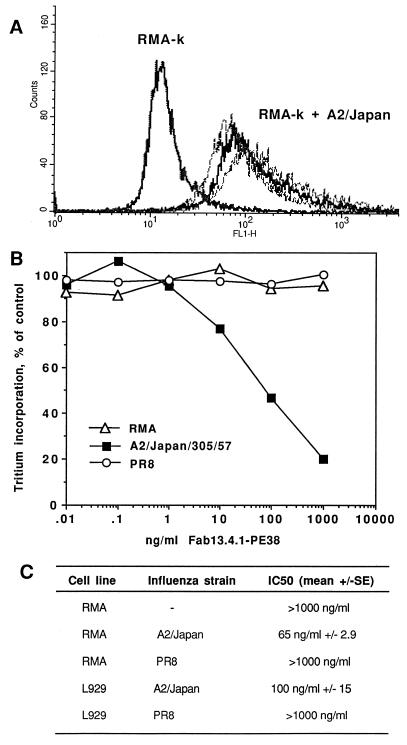
Figure 5.
Activity of Fab 13.4.1–PE38 on influenza virus-infected cells. (A) Detection of HA255–262 peptide/H-2Kk complexes on influenza virus infected-RMA-k cells by Fab 13.4.1. RMA-k cells were infected with various dilutions of influenza virus strain A2/Japan/305/57, and complexes were detected by FACS analysis using recombinant purified Fab 13.4.1 and FITC-labeled goat anti-mouse IgG or Fab. (B and C) Activity of Fab 13.4.1–PE38 on influenza virus-infected RMA-k cells. RMA-k cells were infected with influenza virus strains A2/Japan and PR-8. After infection cells were assayed for cytotoxicity by Fab 13.4.1–PE38.
FACS Analysis.
RMA-S⋅Kk cells (107) were incubated overnight with 0.1 mM peptide at 26°C. Cells were then washed twice with medium and incubated with fluorescein isothiocyanate (FITC)-labeled anti-mouse H-2Kk (0.5 mg/106 cells) for 90 min on ice. Cells were washed 3 times with PBS and analyzed by Becton Dickenson FACSort. Control RMA and RMA-S⋅Kk cells without peptide were incubated overnight at 37°C, washed, incubated with FITC-labeled antibody, and analyzed as described above. Analysis of viral infection and MHC/peptide complex expression on viral-infected cells was performed by staining with mouse antinuclear protein mAb (HB65) and FITC-labeled goat anti-mouse IgG. MHC/peptide complexes on influenza-infected cells were detected with purified recombinant Fab 13.4.1 and FITC-labeled goat anti-mouse IgG or Fab.
RESULTS AND DISCUSSION
Cytotoxic Activity of Fab 13.4.1-PE38 Toward RMA-S⋅Kk APCs.
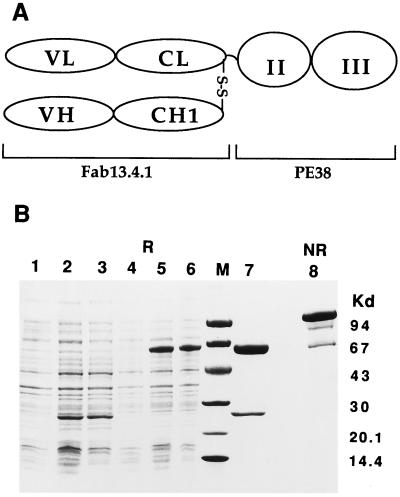
The recombinant antibody-toxin fusion was expressed in E. coli and isolated from intracellular inclusion bodies by refolding and purification (refs. 8 and 9 and as described in Experimental Procedures). Fig. 1 shows a highly purified homogeneous fusion protein with the expected size of ≈86 kDa on a nonreduced SDS/PAGE.
Figure 1.
SDS/PAGE analysis of recombinant purified Fab 13.4.1–PE38. (A) Schematics of the Fab 13.4.1–PE38 fusion protein. The gene encoding PE38 that contains the translocation and ADP ribosylation domains of PE was fused to the C terminus of Fab 13.4.1 (Lκ) κ chain. (B) SDS/gel analysis on 4–20% gradient gels. Lanes: 1, control noninduced cells; 2 and 3, expression of Fab 13.4.1 Fd; 4, control noninduced cells; 5 and 6, expression of Fab 13.4.1 Vκ–PE38; 7, Fab 13.4.1–PE38 reduced; 8, Fab 13.4.1–PE38 nonreduced.
To test the activity of Fab 13.4.1–PE38 on APCs that express the influenza hemagglutinin peptide in complex with class I Kk molecules, we used the temperature-sensitive TAP (transporter-associated with antigen processing) mutant mouse cell line RMA-S that was transfected with Kk (RMA-S⋅Kk cells) (10). The antigen-processing defective mutant RMA-S⋅Kk cells, when cultured at low temperature, express high amounts of MHC class I molecules that do not contain endogenously derived peptides. These empty MHC class I molecules can be stabilized by addition of MHC binding peptides. The RMA-S⋅Kk mutant cells were loaded at 26°C with the hemagglutinin Ha255–262 peptide and also with a control peptide, the influenza virus-derived nucleoprotein peptide NP50–57. FACS analysis with FITC-labeled anti-Kk antibody revealed expression of class I Kk molecules on the surface of peptide-loaded RMA-S⋅Kk cells but not on RMA-S⋅Kk cells that were not loaded with peptide (data not shown). Previous studies have shown that the Ha255–262 and NP50–57 peptides bind to a class I Kk molecule with an identical affinity (KD of 6.9 nM and 6.5 nM for the NP50–57 and Ha255–262 peptides, respectively) (11). The peptide-loaded RMA-S⋅Kk cells were then incubated with the recombinant Fab 13.4.1–PE38 fusion protein for 20 hr at 37°C. As shown in Fig. 2A, cytotoxicity by Fab 13.4.1–PE38 was observed only on RMA-S⋅Kk cells loaded with the hemagglutinin-specific peptide with an IC50 of 5–9 ng/ml. No cytotoxic activity was observed on RMA-S⋅Kk cells that were loaded with the influenza NP peptide, nor on RMA-S⋅Kk cells that were not incubated with peptide or on the parental cell line RMA. Cytotoxic activity by Fab 13.4.1–PE38 was also not observed on mouse cell lines L929 and NIH 3T3 (mouse fibroblast) nor on human tumor cell lines A431 (epidermoid carcinoma) or MCF7 (breast cancer) indicating that Fab 13.4.1–PE38 has no nonspecific cytotoxicity (results not shown).
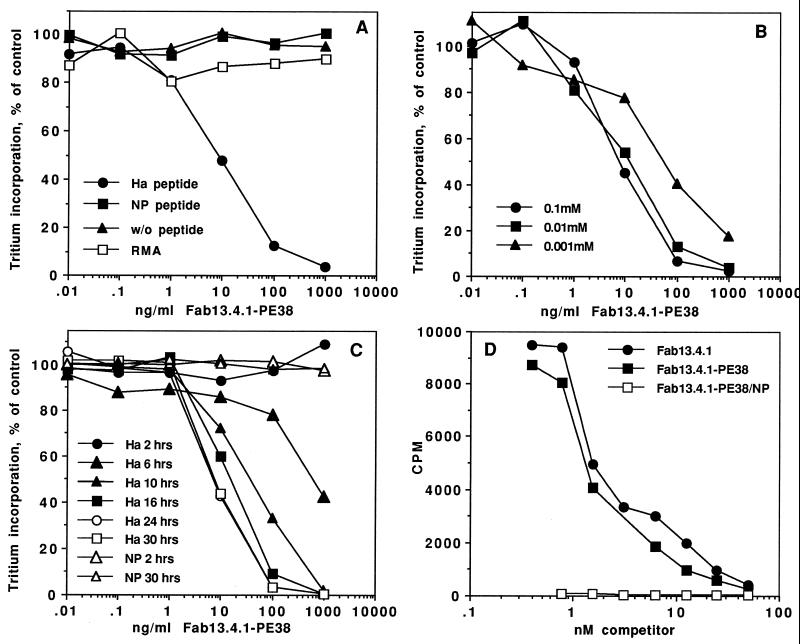
Figure 2.
Cytotoxicity and binding of Fab 13.4.1–PE38 to APCs. (A) Cytotoxic activity of recombinant Fab 13.4.1–PE38 to RMA-S⋅Kk cells with and without influenza peptides. RMA-S⋅Kk cells were incubated with 0.1 mM Ha and NP peptides at 26°C. Cells were then incubated for 20 hr with recombinant Fab 13.4.1–PE38. Protein synthesis is measured by incorporation of [3H]leucine into cell proteins. RMA cells are control cells that are the parental cells used to derive RMA-S⋅Kk. (B) Peptide titration for Fab 13.4.1–PE38 mediated cytotoxicity. RMA-S⋅Kk cells were incubated with various concentration of Ha peptide at 26°C. Cells were than exposed to Fab 13.4.1–PE38, and inhibition of protein synthesis was determined. (C) Kinetics of Fab 13.4.1–PE38 cytotoxic activity on RMA-S⋅Kk cells. RMA-S⋅Kk cells were incubated overnight with 0.1 mM Ha or NP peptide for the indicated times at 26°C. Cells were then exposed to Fab 13.4.1–PE38 and inhibition of protein synthesis was determined. (D) Binding of Fab 13.4.1 and Fab 13.4.1–PE38 to APCs. Competitive binding analysis of the ability of purified recombinant Fab 13.4.1 and Fab 13.4.1–PE38 to inhibit the binding of 125I-labeled Fab 13.4.1 to RMA-S⋅Kk cells loaded with 0.1 mM Ha and NP peptides. Apparent Kd is determined by the concentration of competitor which caused 50% inhibition of the binding of the iodinated Fab 13.4.1.
The cytotoxicity of Fab 13.4.1–PE38 was dependent on the concentration of the specific HA peptide used for loading onto RMA-S⋅Kk cells with a saturating Ha peptide concentration of ≈10 μM (Fig. 2B). The cytotoxic activity of Fab 13.4.1–PE38 on Ha peptide-loaded RMA-S⋅Kk cells increased with time (Fig. 2C) up to 24 hr at 26°C. This time-dependent increase in the cytotoxic activity of Fab 13.4.1–PE38 correlates with the kinetics of class I Kb and Db molecule expression on RMA-S cells as reported (10). Binding of Fab 13.4.1–PE38 to the antigen-presenting RMA-S⋅Kk cells loaded with the specific Ha peptide was performed by competitive binding analysis of the ability of purified recombinant Fab 13.4.1 and Fab 13.4.1–PE38 to inhibit the binding of 125I-labeled Fab 13.4.1.
As shown in Fig. 2D, the Fab 13.4.1–PE38 fusion protein and the purified Fab 13.4.1 bind with similar apparent Kd of ≈2–5 nM to Ha-loaded RMA-S⋅Kk cells as determined by competition binding analysis with purified radiolabeled Fab 13.4.1. No binding activity was observed on RMA-S⋅Kk cells that were loaded with the NP peptide (Fig. 2D). These results demonstrate that Fab 13.4.1–PE38 binds specifically and kills only cells that express the specific peptide in complex with class I Kk MHC molecules.
To demonstrate that the cytotoxic activity of Fab 13.4.1–PE38 is mediated through specific binding of the Fab, we performed a competition experiment in which Fab 13.4.1–PE38 cytotoxicity was competed by preincubation of Ha-loaded RMA-S⋅Kk cells with purified Fab 13.4.1. As shown in Fig. 3, the cytotoxic activity of Fab 13.4.1–PE38 was completely inhibited by preincubation with 50 μg/ml of purified Fab 13.4.1 but not with a control antibody. Fab 13.4.1–PE38 showed no cytotoxic activity on RMA-S⋅Kk cells that were loaded with the NP peptide.
Figure 3.
Specific cytotoxicity of Fab 13.4.1–PE38 to APCs. Competition of Fab 13.4.1–PE38 cytotoxicity by recombinant purified Fab 13.4.1. RMA-S⋅Kk cells were incubated overnight with 0.1 mM Ha and NP peptides at 26°C. Cells were then incubated for 20 hr with recombinant Fab 13.4.1–PE38. In competition experiments, peptide-loaded RMA-S⋅Kk cells were preincubated for 15 min with 50 μg/ml recombinant purified Fab 13.4.1 or control antibody HB21.
The Fine Specificity of the Activity of Fab 13.4.1-PE38 Toward APCs.
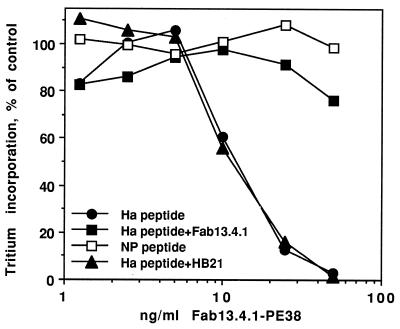
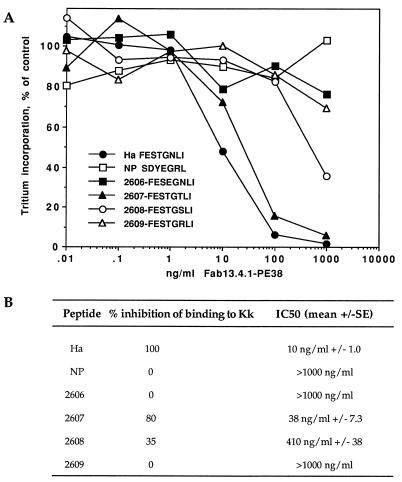
The fine specificity of peptide/MHC recognition by Fab 13.4.1 was recently analyzed and found to be very similar to the fine specificity of two T cell receptors with similar specificity for the Kk-restricted Ha255–262 peptide (5). This analysis was performed by using a complete set of singly substituted analogs of the Ha255–262 peptide. To further determine the killing specificity of Fab 13.4.1–PE38 to peptide/MHC complexes, we used several singly substituted mutants of the Ha peptide as listed in Fig. 4A. We chose two types of mutations that will affect the binding of Fab 13.4.1 to peptide/Kk complexes: mutations that will abolish binding of Fab 13.4.1 to the Ha peptide/Kk complex, or mutations that will maintain some binding activity. Mutation of threonine in position 4 of the Ha peptide to glutamate (peptide 2606) (same residue as in the NP peptide at position 4) will abolish completely the binding of Fab 13.4.1 to the peptide/Kk complex as determined previously by competition ELISA (5) (Fig. 4B). Mutation of asparagine to arginine in position 6 will also inhibit binding of Fab 13.4.1 (peptide 2609) while mutation of that position to threonine or serine (peptides 2607 and 2608, respectively) will restore 80% and 35% of the binding activity of Fab 13.4.1 to the peptide/MHC complex (compared with the binding to Ha255–262).
Figure 4.
Peptide fine specificity in the cytotoxic activity of Fab 13.4.1–PE38 on APCs. RMA-S⋅Kk cells (107) were incubated overnight at 26°C with 0.1 mM Ha, NP, and mutant peptides in which 1 amino acid was changed, as shown in A. Cells were then incubated for 20 hr with recombinant Fab 13.4.1–PE38. (A) Representative cytotoxicity experiment. (B) Summary of three independent experiments. Inhibition of peptide binding to purified H-2Kk by Fab 13.4.1 was determined by competition ELISA as described (5). Percent inhibition was calculated from the data in ref. 5.
To correlate these changes in fine specificity and binding of Fab 13.4.1 to the cytotoxic activity of Fab 13.4.1–PE38, we loaded RMA-S⋅Kk cells with the Ha, NP peptides, as well as with the four mutant peptides and determined the cytotoxic activity of Fab 13.4.1. As shown in Fig. 4, there is a striking correlation between the fine specificity and binding activity of Fab 13.4.1 to the mutant peptides and the cytotoxic activity of Fab 13.4.1–PE38 against the antigen presenting RMA-S⋅Kk cells that are loaded with the mutant peptides. The mutations that abolish binding (peptides 2606 and 2609) also abolish completely the cytotoxic activity of Fab 13.4.1–PE38. Mutations that allow weak binding activity are reflected by reduced cytotoxic activity. Peptide 2607, which retains 80% of the binding activity of Fab 13.4.1, when presented by RMA-S⋅Kk cells causes a mild reduction in cytotoxic activity of Fab 13.4.1–PE38 to an IC50 of 38 ng/ml compared with 10 ng/ml of wild-type Ha peptide (Fig. 4), while peptide 2608 which retains only 35% of the binding of Fab 13.4.1 causes a significant loss of cytotoxic activity to an IC50 of 400 ng/ml. The differences in cytotoxic activity by the mutant peptides are neither due to differences in the amount of MHC expression on RMA-S⋅Kk cells nor to different efficiencies of mutant peptides presentation by MHC class I Kk molecules, but rather reflect differences in specificity of binding because as analyzed by FACS with anti-mouse Kk antibodies, RMA-S⋅Kk cells loaded with the various peptides express the same number of MHC Kk molecules on the cell surface (data not shown). These results demonstrate that Fab 13.4.1–PE38 is a very specific agent that, like a T cell receptor, can differentiate between specific peptide/MHC complexes. Moreover, the cytotoxic activity of Fab 13.4.1–PE38 is fully correlated to the specific recognition of peptide/MHC complexes. Such specificity characteristics are necessary and highly desirable for new therapeutic agents that can target drugs or toxins to a defined population of cells that express a particular peptide/MHC class I complexes.
Characterization of the Sensitivity of RMA-S⋅Kk Cells to the Activity of Fab 13.4.1–PE38.
To study the sensitivity of antigen-presenting RMA-S⋅Kk cells to the cytotoxic activity of Fab 13.4.1–PE38, we measured the number of class I Kk molecules on the surface of peptide-loaded RMA-S⋅Kk cells. This indicates the number of MHC/peptide complexes on the cell surface, since the antigen-processing defective mutant RMA-S⋅Kk cells express class I Kk molecules at 37°C only when stabilized by a bound peptide. The number of MHC/peptide sites on the cell surface were determined by staining with FITC-labeled anti-Kk antibody microbeads that have varying capacities to bind mouse monoclonal IgG antibodies. The fluorescence intensity staining of the peptide-loaded RMA-S⋅Kk cells was corresponding to staining intensity of the microbeads that have ≈20,000 sites/bead (data not shown). The results indicate that ≈20,000 MHC/Ha peptide complexes are expressed on the surface of peptide-loaded RMA-S⋅Kk cells and that this number of sites is sufficient for efficient killing by Fab 13.4.1–PE38. Comparing the cytotoxic activity of Fab 13.4.1–PE38 with others recombinant toxins that have been constructed in our laboratory and directed to antigens that are highly expressed on the target cells (105–106 sites/cell) indicate that the efficient and specific binding of Fab 13.4.1–PE38 combined probably with efficient internalization and processing of the toxin in RMA-S⋅Kk cells that are loaded with the specific peptide leads to very good cytotoxic activity despite the relatively small number of MHC/peptide complex binding sites on the antigen-presenting target cells.
Activity of Fab 13.4.1–PE38 Toward Influenza Virus-Infected Cells.
To determine whether Fab 13.4.1–PE38 is also functional on virus-infected cells, we performed experiments in which RMA-k cells (RMA cells stably transfected with H2-Kk) were infected with influenza virus A2/Japan/305/57. T cell hybridomas that recognize the hemagglutinin peptide Ha255–262 of the A2/Japan strain and are restricted by class I H2-Kk were generated previously, and Fab 13.4.1 was shown to block specifically peptide-dependent stimulation of these hybridomas (4, 11).
Infection of RMA-k cells with the virus was efficient as detected by intracellular staining with an antinuclear protein (NP) antibody (HB65) (data not shown). All cells expressed influenza nuclear protein after ≈12 hr of incubation with virus. We also infected cells with a control influenza virus, PR-8, which presents a different HA peptide in complex with H2-Kk (Ha255–262 FEANGNLI) and is not recognized, when in complex with H2-Kk by Fab 13.4.1 (5). The infection efficiency with the two influenza strains, A2/Japan and PR-8, was similar as shown by staining with anti-NP antibody (data not shown). The RMA-k cells that are transfected with A2/Japan express on the cell surface Ha/H2-Kk complexes as evident by the specific staining with purified Fab 13.4.1 (Fig. 5A). However, cells infected by PR-8 are not recognized by Fab 13.4.1 (data not shown), demonstrating the fine specificity of the recombinant antibody recognition of the specific Ha/H2-Kk complex on the cell surface after the viral infection.
To test the cytotoxic activity of Fab 13.4.1–PE38 on virus-infected cells, RMA-k cells were infected with influenza virus strains A2/Japan and PR-8, washed, and than incubated with Fab 13.4.1–PE38. As shown in Fig. 5 B and C, Fab 13.4.1–PE38 was cytotoxic to RMA-k cells infected with A2/Japan but not with PR-8, which presents a different HA peptide in complex with H2-Kk that is not recognized by Fab 13.4.1. These results further indicate the highly restricted specificity of Fab 13.4.1–PE38 in killing only the cells that express the specific HA peptide/H2-Kk class I complex on their surface.
Summary and Concluding Remarks.
We have shown herein that a recombinant antibody with the antigen-specific, MHC-restricted specificity of a T cell receptor can be used very efficiently to deliver a cytotoxic drug or toxin to APCs that express the suitable peptide/MHC complex. These results demonstrate the feasibility of applying this approach to human diseases such as cancer and viral infections. For example, it is well established now that melanoma cells express tumor-associated peptides in complex with class I HLA-A2 molecules based on their recognition by melanoma-specific cytotoxic T lymphocyte (12–18). Accordingly, it is possible to use these specific MHC/peptide complexes to generate new antibodies that will recognize the complexes analogous to the melanoma-specific cytotoxic T lymphocytes. The generation of Fab 13.4.1 demonstrate that this is feasible, and the specific cytotoxic activity of Fab 13.4.1–PE38 described here suggests that such agents can be used to kill very specifically and in a peptide-dependent MHC-restricted manner APCs that express the correct peptide/MHC complex such as melanoma cells.
The generation of specific antibodies that recognize peptide/MHC complexes by conventional hybridoma technology is very limited, and only few reports exist on the successful generation of such reagents. This is probably because the peptides presented by MHC are buried deep inside the MHC molecule and only 100–300 Å2 of class I-bound peptide surface is facing outwards and is available for direct recognition (19–21). However, phage display antibodies (22, 23) makes it possible to generate libraries from immunized mice and screen tens of millions of individual clones by panning on antigen. Thus, it is possible to select specific and rare antibodies such as those directed to peptide/MHC complexes. Our results suggest the use and feasibility of this approach to develop new therapeutic agents against cancer or to block inappropriate T cell-mediated immune responses such as those leading to autoimmunity.
Acknowledgments
We thank Dr. Jonathan Yewdell (National Institute of Allergy and Infectious Diseases, National Institutes of Health) for providing the influenza virus strains and help with cell infections, Dr. George Vasmatzis for many helpful discussion, Inger Marguiles and Verity Fogg for valuable tissue culture expertise, and Jennie Evans and Althea Jackson for editorial assistance. A.D.C. is partially supported by the Leonardo Di Capua Association, Italy.
ABBREVIATIONS
- MHC
major histocompatibility complex
- APC
antigen-presenting cell
- PE
Pseudomonas exotoxin
- FACS
fluorescence-activated cell sorter
- FITC
fluorescein isothiocyanate
- HA
hemagglutinin
- NP
nucleoprotein
References
- 1.Rosenberg S A. Annu Rev Med. 1996;47:481–491. doi: 10.1146/annurev.med.47.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Goodnow C C. Proc Natl Acad Sci USA. 1996;93:2264–2271. doi: 10.1073/pnas.93.6.2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinman L. Proc Natl Acad Sci USA. 1996;93:2253–2256. doi: 10.1073/pnas.93.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andersen P S, Stryhn A, Hansen B E, Fugger L, Engberg J, Buus S. Proc Natl Acad Sci USA. 1996;93:1820–1824. doi: 10.1073/pnas.93.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stryhn A, Anderson P S, Pedersen L O, Svejgaard A, Holm A, Thorpe C J, Fugger L, Buus S, Engberg J. Proc Natl Acad Sci USA. 1996;93:10338–10342. doi: 10.1073/pnas.93.19.10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastan I, FitzGerald D. Science. 1991;254:1173–1137. doi: 10.1126/science.1683495. [DOI] [PubMed] [Google Scholar]
- 7.Pastan I, Chaudhary V, FitzGerald D. Annu Rev Biochem. 1992;61:331–354. doi: 10.1146/annurev.bi.61.070192.001555. [DOI] [PubMed] [Google Scholar]
- 8.Brinkmann U, Pai L H, FitzGerald D J, Pastan I. Proc Natl Acad Sci USA. 1991;88:8616–8620. doi: 10.1073/pnas.88.19.8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reiter Y, Brinkmann U, Lee B K, Pastan I. Nat Biotechnol. 1996;14:1239–1245. doi: 10.1038/nbt1096-1239. [DOI] [PubMed] [Google Scholar]
- 10.Ljunggren H-G, Stam N J, Ohlen C, Neefjes J J, Hoglund P, Heemels M T, Bustin J, Schumacher T N, Townsend A, Karre K, Ploegh H L. Nature (London) 1990;346:476–480. doi: 10.1038/346476a0. [DOI] [PubMed] [Google Scholar]
- 11.Stryhn A, Pedersen L O, Ortiz-Navarrete V, Buus S. Eur J Immunol. 1994;24:1404–1409. doi: 10.1002/eji.1830240625. [DOI] [PubMed] [Google Scholar]
- 12.Cox A L, Skipper J, Chen Y, Henderson R A, Darrow T L, Shabanowitz J, Engelhard V H, Hunt D F, Slingluff C L., Jr Science. 1994;264:716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 13.Van Der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 14.Gaugler B, van den Eynde B, van der Bruggen P, Romero P, Gaforio J J, De Plaen E, Lethe B, Brasseur F, Boon T. J Exp Med. 1994;179:921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brichard V, van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Cooli P, Boon T. J Exp Med. 1993;178:489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami Y, Eliyahu S, Delgano C H, Robbins P F, Rivoltini L, Topalian S L, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:3515–3519. doi: 10.1073/pnas.91.9.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawakami Y, Eliyahu S, Delgano C H, Robbins P F, Sakaguchi K, Appella E, Yannelli J R, Adema G J, Miki T, Rosenberg S A. Proc Natl Acad Sci USA. 1994;91:6458–6462. doi: 10.1073/pnas.91.14.6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawakami Y, Eliyahu S, Sakaguchi K, Robbins P F, Rivoltini L, Yannelli J R, Appella E, Rosenberg S A. J Exp Med. 1994;180:347–352. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fremont D H, Matsumura M, Stura E A, Peterson P A, Wilson I A. Science. 1992;257:919–927. doi: 10.1126/science.1323877. [DOI] [PubMed] [Google Scholar]
- 20.Young A C, Zhang W, Sacchettini J C, Nathenson S F. Cell. 1994;76:39–50. doi: 10.1016/0092-8674(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 21.Bjorkman P J, Saper M A, Samraoui B, Bennett W S, Strominger J L, Wiley D C. Nature (London) 1987;329:512–518. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 22.Winter G, Griffiths A D, Hawkins R E, Hoogenboom H R. Annu Rev Immunol. 1994;12:433–455. doi: 10.1146/annurev.iy.12.040194.002245. [DOI] [PubMed] [Google Scholar]
- 23.Barbas C F., III Nat Med. 1995;1:837–839. doi: 10.1038/nm0895-837. [DOI] [PubMed] [Google Scholar]