Abstract
Genetic fine mapping of the first locus identified for genetically complex forms of stroke, STRK1 (which has been mapped to chromosome 5q12 in Icelandic families), has identified the phosphodiesterase 4D gene (PDE4D) gene as a good candidate gene. Association analysis of single nucleotide polymorphisms (SNPs) in the PDE4D gene in an Icelandic stroke cohort demonstrated genetic association between six SNPs in the 5′ region of PDE4D and ischaemic stroke. The present study aimed to test whether the same six SNPs in PDE4D were also associated with stroke in a large stroke cohort from northern Germany (stroke patients with acute completed ischaemic stroke: n = 1181; population based controls: n = 1569). None of the six SNPs showed significant association with ischaemic stroke in the whole stroke sample before and after adjustment for conventional stroke risk factors (age, sex, hypertension, diabetes, and hypercholesterolaemia). Haplotype analysis did also not reveal any significant association. Marginally positive statistical measures of association in the subgroup with cardioembolic stroke did not remain significant after correction for multiple testing. In conclusion, this study was unable to demonstrate an association between the six SNPs which had showed significant single marker association with stroke in the Icelandic stroke cohort and ischaemic stroke in a large German cohort.
Keywords: ischaemic stroke; STRK1, PDE4D; association study
Stroke is the third commonest cause of mortality and a major cause of functional impairment among older people worldwide.1 Twin studies have shown that genetic components play a role in the pathogenesis of ischaemic stroke.2 Aetiologically, ischaemic stroke can be subdivided into the three major categories based on the Trial of Org 10172 in Acute Stroke Treatment (TOAST) classification: (1) large artery atherosclerosis, (2) cardioembolism, and (3) small vessel occlusion with lacunar infarction.3 Hypertension, diabetes, and hypercholesterolaemia are important known risk factors for ischaemic stroke. However, they are also partially determined by genetic factors and should be taken into account in genetic studies of ischaemic stroke.
A number of genetic defects causing some rare, monogenetically inherited forms of stroke have been identified in recent years, but the genetic determinants of common, genetically complex ischaemic stroke types remain largely unknown. The first chromosomal locus for common ischaemic stroke has been mapped to a candidate region on chromosome 5q12 in Icelandic families.4 Fine mapping of the locus pinpointed phosphodiesterase 4D (PDE4D) as one possible candidate gene and subsequent association analysis in an Icelandic stroke cohort showed association of six single nucleotide polymorphisms (SNPs) and a combined SNP and short tandem repeat (STR) marker haplotype in the 5′ region of PDE4D with ischaemic stroke.5
Here, we aimed to examine whether these six SNPs in PDE4D were also associated with ischaemic stroke in a large German case–control study.
Methods
Patients
We recruited ischaemic stroke patients from hospitals in northwest Germany through the regional Westphalian Stroke Register and from the University Hospital of Greifswald in northeast Germany. The treating physicians used standardised patient assessment forms to collect sociodemographic and disease related data as well as blood samples. For this study we included all available patients with completed ischaemic stroke, proved by computed tomography (CT) or magnetic resonance imaging (MRI) and classified as: large artery atherosclerosis (TOAST 1), cardioembolism (TOAST 2), or small vessel occlusion (TOAST 3). We excluded patients who had experienced a transient ischaemic attack (TIA) or haemorrhagic stroke, as well as patients falling into other TOAST categories, to maximise aetiologic homogeneity.
Individuals of similar age and sex (table 1) without a history of stroke served as controls and were drawn from the cross‐sectional, prospective, population based Dortmund Health Study for the Westphalian cases and the Study of Health in Pomerania (SHIP)6,7 for the Greifswald cases. The participants of the Dortmund Health Study were randomly selected from the registration office. They took part in a face‐to‐face health interview for assessment of their cardiovascular risk factors and comorbidities.
Table 1 Demographic and medical characteristics of the stroke patient and the control groups.
| Stroke patients | Controls | p value* | |
|---|---|---|---|
| Number | 1181 | 1569 | |
| Male (n (%)) | 633 (54) | 765 (49) | 0.012 |
| Age in years (SD) | 66.9 (14.6) | 55.9 (13.7) | 0.0001 |
| Hypertension (n (%)) | 907 (77) | 649 (41) | 0.0001 |
| Diabetes mellitus (n (%)) | 350 (30) | 143 (9) | 0.0001 |
| Hypercholesterolaemia (n (%)) | 334 (28) | 385 (25) | 0.03 |
| TOAST 1 (n (%)) | 515 (44) | – | – |
| TOAST 2 (n (%)) | 411 (35) | – | – |
| TOAST 3 (n (%)) | 255 (21) | – | – |
*p values for the difference between stroke patients and controls calculated using χ2 tests for dichotomous variables and two sided Student's t test for the continuous variable age.
TOAST 1, large artery atherosclerosis; TOAST 2, cardioembolism; TOAST 3, small vessel occlusion.
The ethics committee of the University of Münster approved the stroke genetic study and the respective committees of the Universities of Münster and Greifswald approved the two population studies. All participants gave their informed consent.
Genotyping
We obtained the DNA sequences of the SNPs, the surrounding genomic DNA, and the genomic region containing the PDE4D gene from the dbSNP database (www.ncbi.nlm.nih.gov/SNP/), Human Genome Browser (http://genome.ucsc.edu/), and electronically linked databases. Genomic DNA was prepared from fresh or frozen venous EDTA anticoagulated blood samples. The SNPs were genotyped by TaqMan assays, custom designed and synthesised by Applied Biosystems (Foster City, CA). Primer and probe sequences are available upon request. The amplification reactions were performed in a 5 μl volume containing 10–25 ng of genomic DNA, 0.125 μl of SNP genotyping assay mix (Applied Biosystems, Foster City, CA) containing the primers and probes for the SNP, and 2.5 μl of qPCR master mix (Eurogentec, Seraing, Belgium). Amplification and plate read were performed in a 7900HT real time polymerase chain reaction (PCR) system (Applied Biosystems, Foster City, CA).
Statistical analyses
We chose the six PDE4D SNPs on the basis of their previous association with stroke in the Icelandic study population.5 The study had 85% power to detect an association with an odds ratio (OR) of 1.4 with 95% confidence. We used a one degree of freedom goodness‐of‐fit test in the control population to calculate the Hardy–Weinberg equilibrium. Odds ratios and 95% confidence intervals were first calculated as a measure of association using logistic regression and case–control status as the dependent variables and each SNP as the independent variable. This was done for each of the three model assumptions (dominant, recessive, additive). A p value of less than 0.05 was considered statistically significant in this step. Crude odds ratios and p values were subsequently adjusted for confounding factors such as sex, age, hypertension, diabetes and hypercholesterolaemia. For these analyses the SPSS statistical package (version 11.0) was used. We chose to test first for single marker association followed by adjustment for confounding factors and correction for multiple testing. Permutation testing has become an accepted procedure to address the issue of multiple testing as the Bonferroni adjustment is highly conservative and low in power. We performed robust permutation testing using 10.000 bootstrap replicates and calculated an empirical p value for each marker according to Westfall and Young's8 step‐down procedure. Subsequently we performed a haplotype analysis using the program Haploview 3.2.9
Results
The stroke cases were on average 10 years older and had higher prevalence of hypertension and diabetes in comparison with the controls (see table 1).
We first calculated crude p values and odds ratios in the stroke cohort without adjustment for confounding factors (table 2). In this analysis we did not find a significant association between any of the six tested SNPs and ischaemic stroke. Subsequent adjustment for sex, age, hypertension, diabetes, and hypercholesterolaemia did not change this lack of association. All statistical measures of association remained non‐significant in the whole stroke cohort (table 2). In the next step we stratified the stroke cohort according to the TOAST classification into the following subgroups: large artery atherosclerosis (TOAST 1), cardioembolism (TOAST 2), and a combined group (TOAST 1 and 2). Crude and adjusted measures of association were again clearly non‐significant for five of the SNPs (table 2). Only SNP 87 (rs2910829) showed a p value of 0.0345 before adjustment and 0.0411 after adjustment for sex, age, hypertension, diabetes, and hypercholesterolaemia in the subgroup of patients with cardioembolic stroke, but correction for multiple testing resulted in a non‐significant p value of 0.0985. Calculation of the major allele frequencies in the controls and comparison with the three other studies exploring the relation between SNPs in PDE4D and stroke showed considerable differences between the allele frequencies in our control sample and the Icelandic study by Gretarsdottir et al.5 The remaining two studies reported allele frequencies for only one of the SNPs, revealing that the allele frequencies in our northern German control cohort were similar to the ones found in a southern German cohort and relatively close to a Swedish cohort (table 3).10,11
Table 2 Statistical measures of association between SNPs in PDE4D and ischaemic stroke before correction for multiple testing; p values are two sided.
| db SNP accession | rs152312 (HWpVal: 0.985) | rs12188950 (HWpVal: 0.481) | rs702553 (HWpVal: 0.099) | rs966221 (HWpVal: 0.121) | rs2910829 (HWpVal: 0.946) | rs1396476 (HWpVal: 0.507) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Designation by Gretarsdottir et al5 | SNP 41 | SNP 45 | SNP 56 | SNP 83 | SNP 87 | SNP 89 | ||||||||||||
| p | OR | 95% CI | p | OR | 95% CI | p | OR | 95% CI | p | OR | CI | p | OR | CI | p | OR | CI | |
| Complete sample, crude | 0.0949 | 0.85 | 0.70 to 1.03 | 0.5548 | 1.05 | 0.89 to 1.26 | 0.6075 | 0.96 | 0.81 to 1.13 | 0.4687 | 1.06 | 0.90 to 1.26 | 0.4181 | 1.09 | 0.89 to 1.33 | 0.5073 | 0.85 | 0.52 to 1.38 |
| Complete sample, adjusted | 0.1463 | 0.85 | 0.68 to 1.06 | 0.2158 | 1.14 | 0.93 to 1.39 | 0.4651 | 1.07 | 0.89 to 1.29 | 0.3586 | 1.09 | 0.90 to 1.33 | 0.5215 | 1.08 | 0.85 to 1.36 | 0.4944 | 0.82 | 0.46 to 1.45 |
| Subgroup TOAST 1, crude | 0.1279 | 0.82 | 0.63 to 1.06 | 0.6105 | 1.06 | 0.84 to 1.33 | 0.7182 | 1.04 | 0.84 to 1.29 | 0.2003 | 1.16 | 0.93 to 1.44 | 0.8657 | 1.02 | 0.78 to 1.34 | 0.9960 | 1.00 | 0.51 to 1.96 |
| Subgroup TOAST 1, adjusted | 0.1310 | 0.80 | 0.60 to 1.07 | 0.1517 | 1.20 | 0.93 to 1.55 | 0.2286 | 1.16 | 0.91 to 1.47 | 0.1646 | 1.19 | 0.93 to 1.53 | 0.9530 | 1.01 | 0.75 to 1.36 | 0.8090 | 1.10 | 0.51 to 2.36 |
| Subgroup TOAST 2, crude | 0.7370 | 0.95 | 0.73 to 1.25 | 0.8539 | 0.98 | 0.76 to 1.26 | 0.3549 | 0.89 | 0.71 to 1.13 | 0.4489 | 0.88 | 0.63 to 1.23 | 0.0345 | 1.38 | 1.02 to 1.85 | 0.8279 | 0.93 | 0.46 to 1.86 |
| Subgroup TOAST 2, adjusted | 0.6959 | 0.94 | 0.69 to 1.28 | 0.7509 | 1.05 | 0.79 to 1.40 | 0.9587 | 1.01 | 0.77 to 1.31 | 0.3005 | 0.82 | 0.57 to 1.19 | 0.0411 | 1.41 | 1.01 to 1.97 | 0.7342 | 0.87 | 0.39 to 1.94 |
| Subgroup TOAST 1+2, crude | 0.1250 | 0.88 | 0.72 to 1.08 | 0.6718 | 1.04 | 0.87 to 1.25 | 0.8699 | 0.99 | 0.84 to 1.16 | 0.7058 | 1.03 | 0.87 to 1.22 | 0.2373 | 1.13 | 0.92 to 1.40 | 0.9391 | 0.99 | 0.83 to 1.19 |
| Subgroup TOAST 1+2, adjusted | 0.2482 | 0.87 | 0.69 to 1.10 | 0.2582 | 1.13 | 0.92 to 1.39 | 0.5586 | 1.06 | 0.88 to 1.28 | 0.8224 | 1.02 | 0.84 to 1.24 | 0.2480 | 1.15 | 0.91 to 1.46 | 0.7669 | 1.03 | 0.84 to 1.26 |
All values are shown for the additive model before correction for multiple testing only. HWpVal: p value for deviation from the Hardy–Weinberg equilibrium. Except for SNP 87 (rs2910829) which also showed weakly significant p values in the dominant and recessive model in the subgroup TOAST 2, no other SNP yielded significant p values.
TOAST 1, large artery atherosclerosis; TOAST 2, cardioembolism.
Table 3 Major allele frequencies (%) in the control groups of different studies examining the role of PDE4D in stroke.
| SNP | Allele | Münster | Iceland | Munich | Sweden |
|---|---|---|---|---|---|
| SNP 45 (rs12188950) | G | 85.5 | 78.0 | 86.3 | 88.0 |
| SNP 56 (rs702553) | T | 68.7 | 65.5 | NR | NR |
| SNP 83 (rs966221) | C | 61.0 | 52.0 | NR | NR |
| SNP 87 (rs2910829) | T | 53.1 | 48.4 | NR | NR |
| SNP 89 (rs1396476) | A | 83.7 | 81.8 | NR | NR |
Major allele frequencies are derived from our own study (Münster) and the studies by Gretarsdottir et al (Iceland),5 Lohmussaar et al (Munich),10 and Nilsson‐Ardnor et al (Sweden).11 Bevan et al12 did not report allele frequencies of the SNPs. SNP41 is not reported because it received the wrong designation in the original paper by Gretarsdottir et al (rs152312) which was subsequently corrected to rs121537989. As only the study by Nilsson‐Ardnor typed SNP rs121537989 but the other studies typed SNP rs152312 we have not shown the allele frequencies of this SNP.
Allele: A, adenine; C, cytosine; G, guanine; T, thymidine.
NR, not reported; SNP, single nucleotide polymorphism designation.
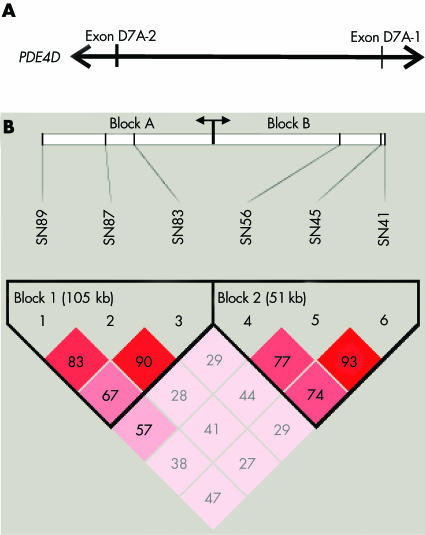
Haplotype analysis revealed two haplotype blocks with a block structure comparable with that found in the Icelandic study (fig 1). Haplotype association was performed using all six SNPs (data not shown) as well as without SNP 41 (rs152312) and SNP 89 (rs1396476) to enable comparison with the Icelandic study (table 4). SNP 41 had to be excluded because it was erroneously reported to be rs152312 in the Icelandic study but this SNP had not been typed (corrigendum to reference 5 and personal communication, Gretarsdottir, 2005) whereas SNP 89 is not included in the Icelandic haplotype analysis. In our study, both haplotype analyses did not reveal a significant association with stroke. In comparison with the Icelandic study we found the same three major two marker haplotypes for block A and block B but sizeable differences in some haplotype frequencies in patients as well as controls (table 4).
Figure 1 Haplotype block structure. (A) Schematic drawing of the 5′ portion of PDE4D spanning the six SNPs which contain the exons D7A‐2 and D7A‐1.5 (B) Haplotype block structure showing two haplotype blocks in the our German population (block A and block B) which correspond exactly to the haplotype blocks described in the Icelandic population.5 The D′ values (×100) are given in the blocks.
Table 4 Comparison of haplotypes and haplotype association between the Münster and Icelandic studies.
| SNP | 87 | 83 | p | Aff% | Ctrl% | 56 | 45 | p | Aff% | Ctrl% |
|---|---|---|---|---|---|---|---|---|---|---|
| HT block | Block A | Block B | ||||||||
| Iceland | ||||||||||
| 1 | T | C | – | 49.4 | 41.8 | T | G | – | 61.5 | 53.4 |
| 2 | C | T | – | 31.6 | 39.3 | A | A | – | 7.7 | 13.9 |
| 3 | C | C | – | 2.1 | 2.9 | A | G | – | 6.2 | 5.6 |
| Münster | ||||||||||
| 1 | T | C | 0.71 | 51.8 | 51.3 | T | G | 0.99 | 66.4 | 66.4 |
| 2 | C | T | 0.73 | 36.8 | 37.3 | A | G | 0.24 | 17.9 | 19.2 |
| 3 | C | C | 0.67 | 9.3 | 9.6 | A | A | 0.15 | 13.4 | 12.1 |
The three commonest haplotypes from the Icelandic study and our study are shown. The Icelandic haplotypes are summed haplotypes formed of all multimarker haplotypes described by Gretarsdottir et al5 for the respective blocks which contained the two marker haplotypes shown in this table, making it impossible to calculate p values for the Icelandic haplotypes.
SNP, single nucleotide polymorphism designation; p, p value for association between the haplotype block and stroke; Aff%, frequency of the haplotype in affected stroke patients in per cent; Ctrl%, frequency of the haplotype in controls in percent; HT block, haplotype block; Block A, haplotype block A according to fig 1; Block B, haplotype block B according to fig 1.
Discussion
Our goal was to evaluate the association between SNPs in the PDE4D gene and ischaemic stroke in well powered, large German case–control study. Association between six SNPs and a combined SNP and STR marker haplotype was previously found in an Icelandic stroke cohort. We chose to genotype the six SNPs showing strong association with ischaemic stroke in the Icelandic cohort and not the extended stroke associated marker haplotype for three main reasons:
haplotype structure and haplotypes could be different in an Icelandic and a German population, rendering the uncritical use of haplotype tagging SNPs impossible
nearly all SNPs in the genomic region of the PDE4D gene that were not highly correlated with each other were screened in the Icelandic study, making it likely that one or more of the significantly associated SNPs are the causative SNPs
to avoid the genetic and statistical problems inherent in combined SNP and STR haplotypes due to multiple alleles and different mutation rates of STR markers compared with SNPs.
Our results did not show any significant association before or after correction for the major conventional risk factors of age, hypertension, diabetes, and hypercholesterolaemia in the complete ischaemic stroke cohort, either for single SNPs or for SNP haplotypes. As in the Icelandic sample the strongest association was found in the combined sample of patients with large artery atherosclerosis (TOAST 1) and cardioembolism (TOAST 2) we also decided to stratify our sample into these groups of stroke subtypes. In these analyses SNP 87 (rs2910829) was the only SNP yielding a significant p value in the logistic regression analysis in the subgroup with cardioembolic stroke. However, the resulting p value after correction for multiple testing by permutation analysis was not significant (p = 0.0985), the odds ratio was moderate (1.41 after adjustment for conventional risk factors) and the lower boundary of the 95% confidence interval was very close to 1 (1.01) indicating that this association has to be treated with caution. We did not investigate the subgroup with stroke caused by small vessel occlusion (TOAST 3) any further because this subgroup was comparatively small (255 patients) and therefore lacked statistical power. In addition, no association with SNPs in PDE4D and small vessel disease was found in the Icelandic study.
Three other investigations have recently studied the PDE4D gene in German, British, and Swedish stroke cohorts. All three groups did not find a significant association of SNPs in PDE4D with ischaemic stroke in the respective study populations.10,11,12 However, in Swedish families with ischaemic stroke suggestive linkage with the chromosome 5q12 locus containing PDE4D was found.11 The German study by Lohmussaar et al, with regard to the study design, is complementary to our investigation because they employed the haplotype tagging approach using tagging of SNPs from the Icelandic study.10 Comparison of allele frequencies revealed close concordance—for the one common SNP—between the controls in the two German studies as well as the Swedish study but considerable differences from the Icelandic study. Nevertheless, the haplotype block structure in our population was the same as in the Icelandic population.
In summary, we did not find a significant association in our German stroke cohort between the six SNPs in PDE4D that had showed a significant association with ischaemic stroke in an Icelandic population. Our data in conjunction with the data previously reported by others suggest the presence of genetic heterogeneity of PDE4D within European populations, which may explain the discrepant association results in the various subpopulations.10,11,12 Further studies—that is, a meta‐analysis of all the study populations may help to elucidate further the role of genetic variation in PDE4D and the pathogenesis of stroke.
Acknowledgements
The contributions of the departments of neurology of following hospitals are gratefully acknowledged: Evangelisches Krankenhaus Castrop‐Rauxel, St Elisabeth‐Stift Damme, Klinikum Dortmund, Hans‐Susemihl‐Krankenhaus Emden, Evangelisches Krankenhaus Gelsenkirchen, Universitätsklinik Greifswald, St Johannes Hospital Hagen, Marienhospital Hamm, Friederikenstift Hannover, Klinikum Minden, Universitätsklinik Münster, Klinikum Neubrandenburg, Klinikum Osnabrück, St Vincenz‐Krankenhaus Paderborn, Ammerlandklinik Westerstede.
Abbreviations
PDE4D - phosphodiesterase 4D gene
SNP - single nucleotide polymorphism
STRK1 - Stroke 1 genetic locus
TOAST - Trial of Org 10172 in Acute Stroke Treatment
Footnotes
This project is in part sponsored by the German ‘Competence Net Stroke' which is supported by the German Federal Ministry of Education and Research (01GI9909/3). The “Dortmund Health Study” is supported by the German Migraine and Headache Society. The Study of Health in Pomerania (SHIP) is funded by grants from the German Federal Ministry of Education and Research (BMBF,01ZZ96030), and from the Ministry for Education, Research and Cultural Affairs and the Ministry for Social Affairs of the Federal State of Mecklenburg‐Vorpommern.
Competing interests: none declared
References
- 1.Feigin V L, Lawes C M, Bennett D A.et al Stroke epidemiology: a review of population‐based studies of incidence, prevalence, and case‐fatality in the late 20th century. Lancet Neurol 2003243–53. [DOI] [PubMed] [Google Scholar]
- 2.Flossmann E, Schulz U G, Rothwell P M. Systematic review of methods and results of studies of the genetic epidemiology of ischemic stroke. Stroke 200435212–227. [DOI] [PubMed] [Google Scholar]
- 3.Adams H P, Jr, Woolson R F, Biller J.et al Studies of Org 10172 in patients with acute ischemic stroke. TOAST Study Group. Haemostasis 19922299–103. [DOI] [PubMed] [Google Scholar]
- 4.Gretarsdottir S, Sveinbjornsdottir S, Jonsson H H.et al Localization of a susceptibility gene for common forms of stroke to 5q12. Am J Hum Genet 200270593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gretarsdottir S, Thorleifsson G, Reynisdottir S T.et al The gene encoding phosphodiesterase 4D confers risk of ischemic stroke. Nat Genet 200335131–138. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt W P, Berger K, Taeger D.et al Influence of institutional factors in neurological, medical and geriatric departments on length of stay in patients with stroke. Dtsch Med Wochenschr 2003128979–983. [DOI] [PubMed] [Google Scholar]
- 7.John U, Greiner B, Hensel E.et al Study of Health In Pomerania (SHIP): a health examination survey in an east German region: objectives and design. Soz Praventivmed 200146186–194. [DOI] [PubMed] [Google Scholar]
- 8.Westfall P H, Young S S. P value adjustments for multiple tests in binomial models. J Am Soc Assoc 198984780–786. [Google Scholar]
- 9.Barrett J C, Fry B, Maller J.et al Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 200521263–265. [DOI] [PubMed] [Google Scholar]
- 10.Lohmussaar E, Gschwendtner A, Mueller J C.et al ALOX5AP gene and the PDE4D gene in a central European population of stroke patients. Stroke 200536731–736. [DOI] [PubMed] [Google Scholar]
- 11.Nilsson‐Ardnor S, Wiklund P G, Lindgren P.et al Linkage of ischemic stroke to the PDE4D region on 5q in a Swedish population. Stroke 2005361666–1671. [DOI] [PubMed] [Google Scholar]
- 12.Bevan S, Porteous L, Sitzer M.et al Phosphodiesterase 4D gene, ischemic stroke, and asymptomatic carotid atherosclerosis. Stroke 200536949–953. [DOI] [PubMed] [Google Scholar]