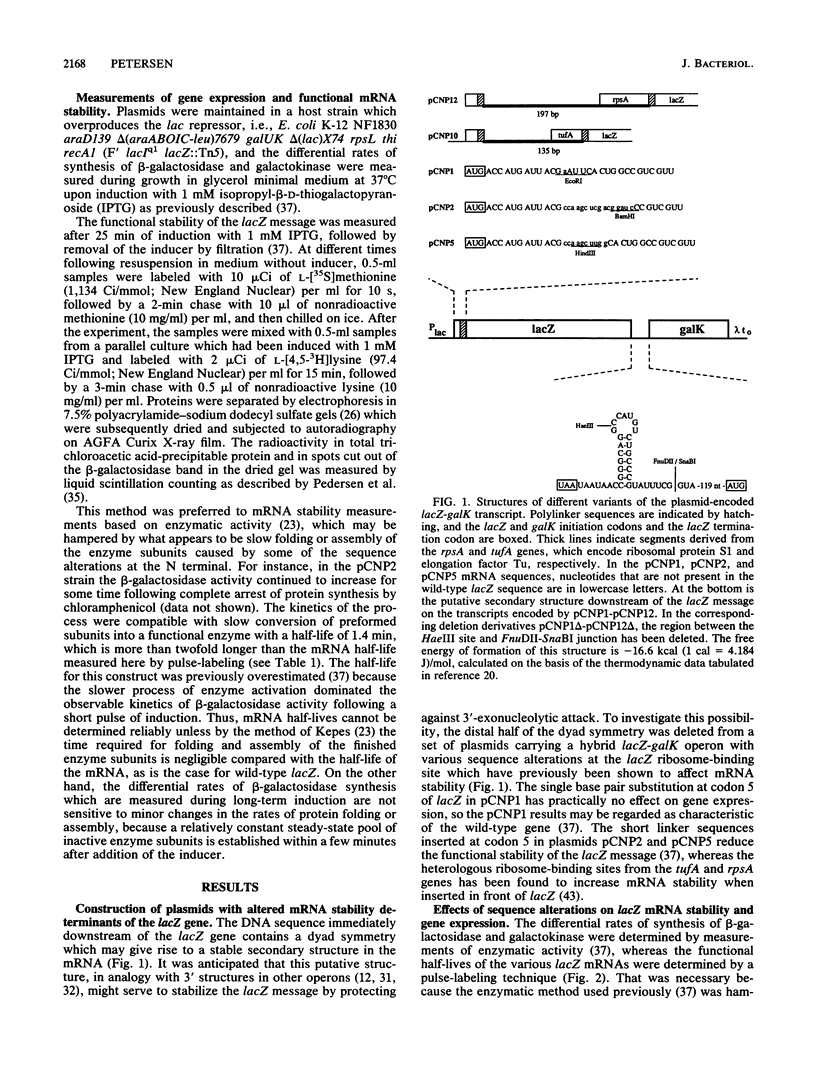
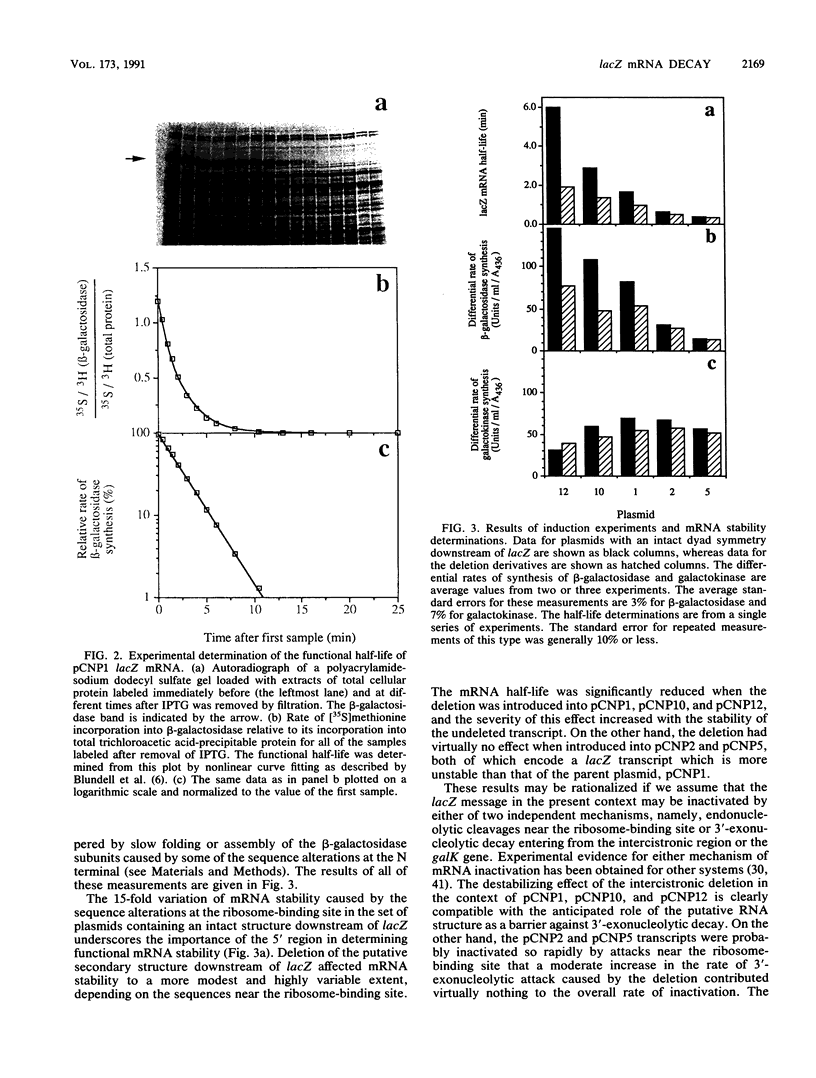
Abstract
The Escherichia coli lacZ gene was used as a model system to identify specific sequence elements affecting mRNA stability. Various insertions and substitutions at the ribosome-binding site increased or decreased the rate of mRNA inactivation by up to fourfold. Deletion of a dyad symmetry, which may give rise to a very stable secondary structure in the mRNA immediately downstream of the gene, decreased the functional stability of the lacZ message. The magnitude of the latter effect was strongly dependent on the sequences at the ribosome-binding site, ranging from practically no effect for the most labile transcripts to a threefold decrease in stability for the most stable one. The results suggest that the wild-type lacZ message is inactivated predominantly by attacks near the ribosome-binding site, presumably in part because the putative secondary structure downstream of the gene protects against 3'-exonucleolytic attack. Taken together, the data for all of the modified variants of lacZ were shown to be quantitatively compatible with a general model of mRNA inactivation involving multiple independent target sites.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambulos N. P., Jr, Duvall E. J., Lovett P. S. The mRNA for an inducible chloramphenicol acetyltransferase gene is cleaved into discrete fragments in Bacillus subtilis. J Bacteriol. 1987 Mar;169(3):967–972. doi: 10.1128/jb.169.3.967-972.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apirion D. Degradation of RNA in Escherichia coli. A hypothesis. Mol Gen Genet. 1973 May 28;122(4):313–322. doi: 10.1007/BF00269431. [DOI] [PubMed] [Google Scholar]
- Apirion D., Gitelman D. R. Decay of RNA in RNA processing mutants of Escherichia coli. Mol Gen Genet. 1980 Jan;177(2):339–343. doi: 10.1007/BF00267448. [DOI] [PubMed] [Google Scholar]
- Bartlett B. O., Scott Russell R., Jenkins W. Improved relationship between the deposition of strontium-90 and the contamination of milk in the United Kingdom. Nature. 1972 Jul 7;238(5358):46–48. doi: 10.1038/238046a0. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Higgins C. F. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988 Dec 10;72(1-2):15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Brawerman G. mRNA decay: finding the right targets. Cell. 1989 Apr 7;57(1):9–10. doi: 10.1016/0092-8674(89)90166-9. [DOI] [PubMed] [Google Scholar]
- Büchel D. E., Gronenborn B., Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980 Feb 7;283(5747):541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Kennell D. Evidence that the 5' end of lac mRNA starts to decay as soon as it is synthesized. J Bacteriol. 1985 Feb;161(2):820–822. doi: 10.1128/jb.161.2.820-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Castles J. J., Singer M. F. Degradation of polyuridylic acid by ribonuclease II: protection by ribosomes. J Mol Biol. 1969 Feb 28;40(1):1–17. doi: 10.1016/0022-2836(69)90292-7. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Beatty J. T., Cohen S. N., Belasco J. G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988 Feb 26;52(4):609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- Coffman R. L., Norris T. E., Koch A. L. Chain elongation rate of messenger and polypeptides in slowly growing Escherichia coli. J Mol Biol. 1971 Aug 28;60(1):1–19. doi: 10.1016/0022-2836(71)90442-6. [DOI] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Belasco J. G. The ompA 5' untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J Bacteriol. 1990 Aug;172(8):4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Helin K., Christensen O. W., Løbner-Olesen A. Translational control and differential RNA decay are key elements regulating postsegregational expression of the killer protein encoded by the parB locus of plasmid R1. J Mol Biol. 1988 Sep 5;203(1):119–129. doi: 10.1016/0022-2836(88)90096-4. [DOI] [PubMed] [Google Scholar]
- Gorski K., Roch J. M., Prentki P., Krisch H. M. The stability of bacteriophage T4 gene 32 mRNA: a 5' leader sequence that can stabilize mRNA transcripts. Cell. 1985 Dec;43(2 Pt 1):461–469. doi: 10.1016/0092-8674(85)90176-x. [DOI] [PubMed] [Google Scholar]
- Gross G., Hollatz I. Coliphage lambda to terminator lowers the stability of messenger RNA in Escherichia coli hosts. Gene. 1988 Dec 10;72(1-2):119–128. doi: 10.1016/0378-1119(88)90133-3. [DOI] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Cloned DNA sequences that determine mRNA stability of bacteriophage phi X174 in vivo are functional. Nucleic Acids Res. 1985 Aug 26;13(16):5937–5948. doi: 10.1093/nar/13.16.5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A. B., Good L., Simonetti J., Zuker M. Some simple computational methods to improve the folding of large RNAs. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):45–52. doi: 10.1093/nar/12.1part1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M., Kepes A. Initiation, elongation and inactivation of lac messenger RNA in Escherichia coli studied studied by measurement of its beta-galactosidase synthesizing capacity in vivo. J Mol Biol. 1971 Sep 28;60(3):453–472. doi: 10.1016/0022-2836(71)90181-1. [DOI] [PubMed] [Google Scholar]
- Klug G., Cohen S. N. Combined actions of multiple hairpin loop structures and sites of rate-limiting endonucleolytic cleavage determine differential degradation rates of individual segments within polycistronic puf operon mRNA. J Bacteriol. 1990 Sep;172(9):5140–5146. doi: 10.1128/jb.172.9.5140-5146.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lim L. W., Kennell D. Models for decay of Escherichia coli lac messenger RNA and evidence for inactivating cleavages between its messages. J Mol Biol. 1979 Dec 5;135(2):369–390. doi: 10.1016/0022-2836(79)90442-x. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Mott J. E., Galloway J. L., Platt T. Maturation of Escherichia coli tryptophan operon mRNA: evidence for 3' exonucleolytic processing after rho-dependent termination. EMBO J. 1985 Jul;4(7):1887–1891. doi: 10.1002/j.1460-2075.1985.tb03865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Robinson E. C., Hiles I. D., Higgins C. F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987 Jan 30;48(2):297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. V. Analysis of the proteins synthesized in ultraviolet light-irradiated Escherichia coli following infection with the bacteriophages lambdadrifd 18 and lambdadfus-3. Mol Gen Genet. 1976 Mar 30;144(3):339–343. doi: 10.1007/BF00341733. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Reeh S. Functional mRNA half lives in E. coli. Mol Gen Genet. 1978 Nov 9;166(3):329–336. doi: 10.1007/BF00267626. [DOI] [PubMed] [Google Scholar]
- Pedersen S., Skouv J., Kajitani M., Ishihama A. Transcriptional organization of the rpsA operon of Escherichia coli. Mol Gen Genet. 1984;196(1):135–140. doi: 10.1007/BF00334105. [DOI] [PubMed] [Google Scholar]
- Petersen C. The functional stability of the lacZ transcript is sensitive towards sequence alterations immediately downstream of the ribosome binding site. Mol Gen Genet. 1987 Aug;209(1):179–187. doi: 10.1007/BF00329856. [DOI] [PubMed] [Google Scholar]
- Plunkett G., 3rd, Echols H. Retroregulation of the bacteriophage lambda int gene: limited secondary degradation of the RNase III-processed transcript. J Bacteriol. 1989 Jan;171(1):588–592. doi: 10.1128/jb.171.1.588-592.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Régnier P., Grunberg-Manago M. Cleavage by RNase III in the transcripts of the met Y-nus-A-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J Mol Biol. 1989 Nov 20;210(2):293–302. doi: 10.1016/0022-2836(89)90331-8. [DOI] [PubMed] [Google Scholar]
- SINGER M. F., TOLBERT G. SPECIFICITY OF POTASSIUM-ACTIVATED PHOSPHODIESTERASE OF ESCHERICHIA COLI. Science. 1964 Aug 7;145(3632):593–595. doi: 10.1126/science.145.3632.593. [DOI] [PubMed] [Google Scholar]
- Schmeissner U., McKenney K., Rosenberg M., Court D. Removal of a terminator structure by RNA processing regulates int gene expression. J Mol Biol. 1984 Jun 15;176(1):39–53. doi: 10.1016/0022-2836(84)90381-4. [DOI] [PubMed] [Google Scholar]
- Wong H. C., Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci U S A. 1986 May;83(10):3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T., Sugisaki H., Takanami M., Kaziro Y. The nucleotide sequence of the cloned tufA gene of Escherichia coli. Gene. 1980 Dec;12(1-2):25–31. doi: 10.1016/0378-1119(80)90012-8. [DOI] [PubMed] [Google Scholar]