Abstract
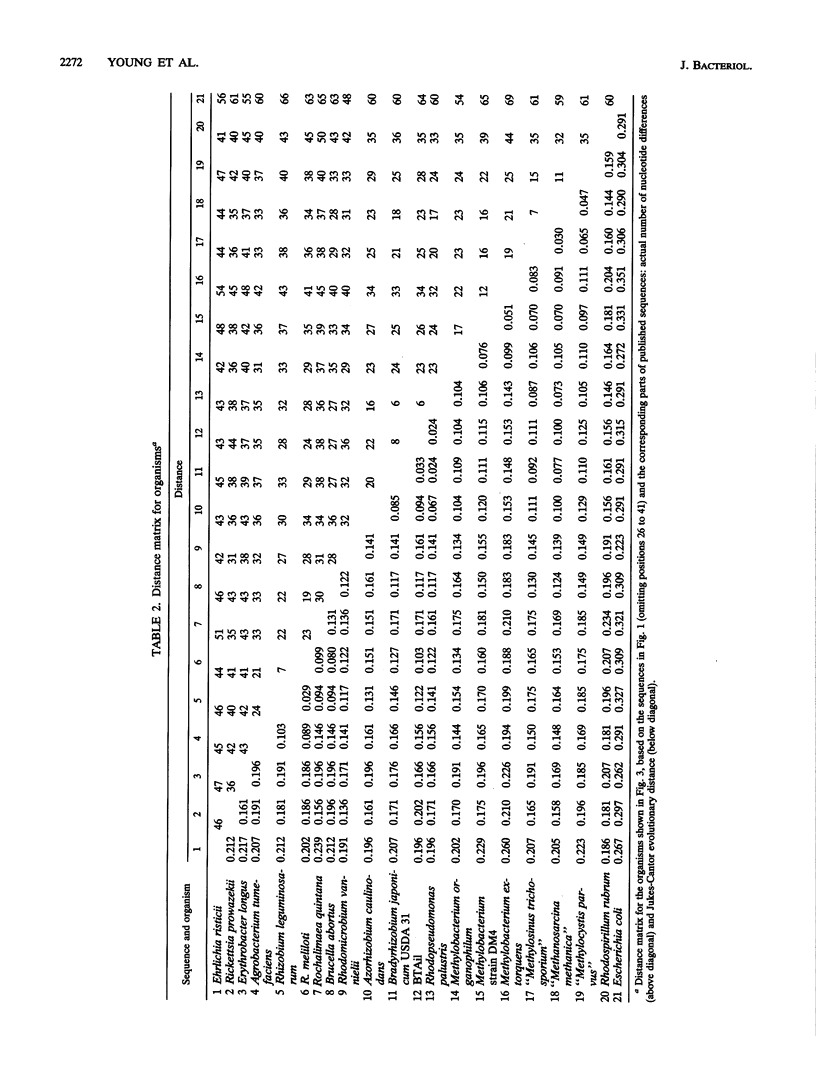
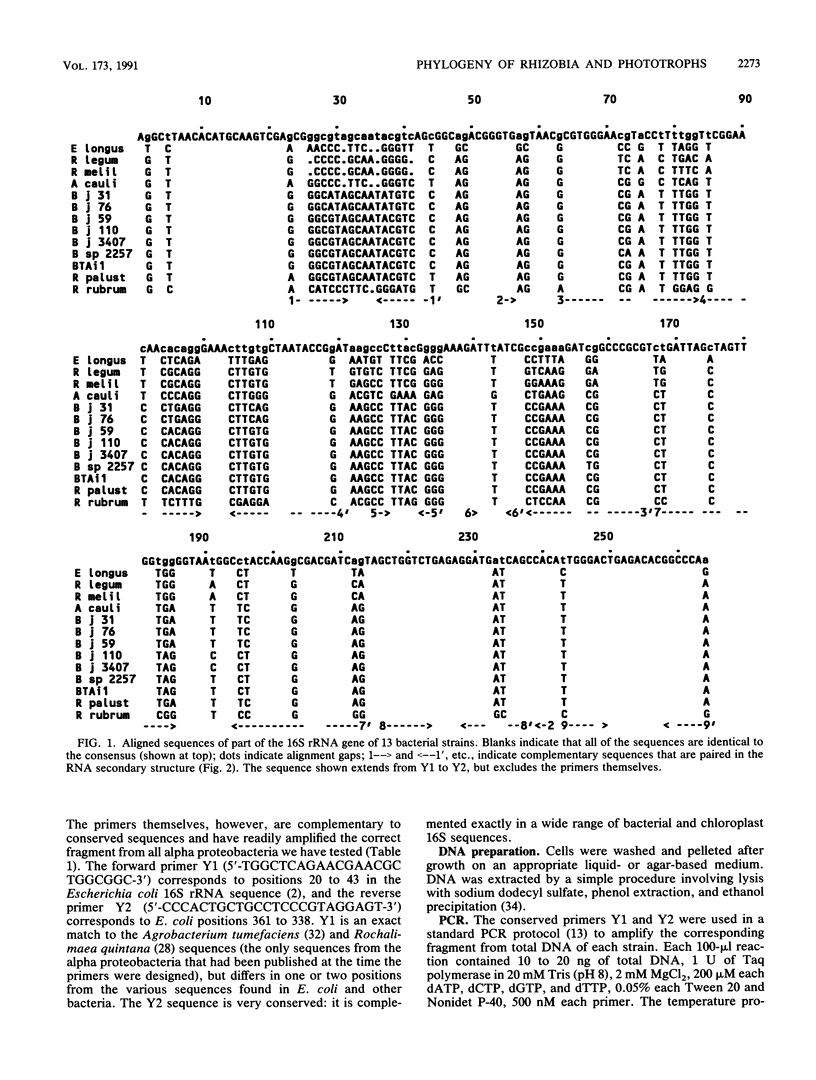
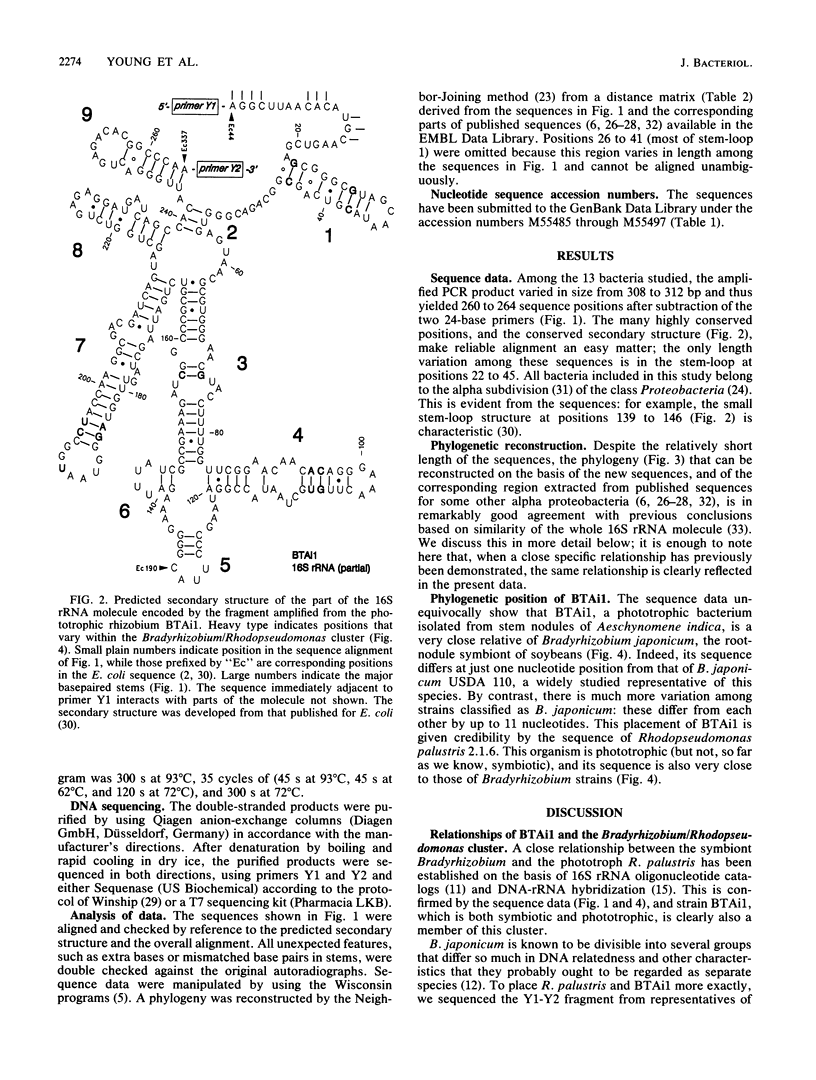
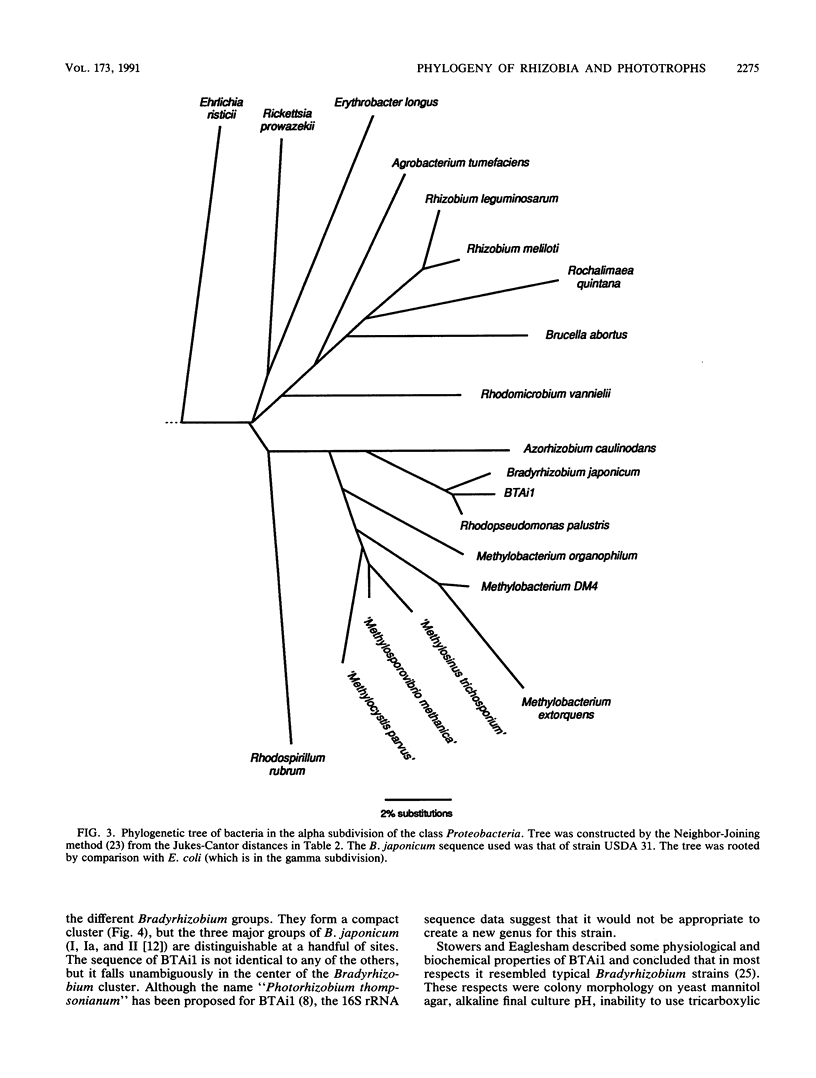
A 260-bp segment of the DNA that encodes 16S rRNA, corresponding to positions 44 to 337 in the Escherichia coli sequence, was amplified by the polymerase chain reaction and sequenced from each of 13 bacteria (rhizobia and purple phototrophs) in the alpha subdivision of the class Proteobacteria. The phylogenetic tree calculated from differences in the sequenced segment conforms well to our expectations based on other previously published data. The sequence from BTAi1 (a recently described phototrophic symbiont of the legume Aeschynomene) and that from the free-living phototroph Rhodopseudomonas palustris both fall within the range of variation found among strains of the soybean symbiont Bradyrhizobium japonicum. This suggests that it would be appropriate to include all of these organisms in a single genus.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Böttger E. C. Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol Lett. 1989 Nov;53(1-2):171–176. doi: 10.1016/0378-1097(89)90386-8. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsch M., Moreno E., Stackebrandt E. Nucleotide sequence of the 16S rRNA from Brucella abortus. Nucleic Acids Res. 1989 Feb 25;17(4):1765–1765. doi: 10.1093/nar/17.4.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans W. R., Fleischman D. E., Calvert H. E., Pyati P. V., Alter G. M., Rao N. S. Bacteriochlorophyll and Photosynthetic Reaction Centers in Rhizobium Strain BTAi 1. Appl Environ Microbiol. 1990 Nov;56(11):3445–3449. doi: 10.1128/aem.56.11.3445-3449.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis M. A., Myambo K. B., Gelfand D. H., Brow M. A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci U S A. 1988 Dec;85(24):9436–9440. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T. Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9355–9359. doi: 10.1073/pnas.86.23.9355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D. J., Pace B., Olsen G. J., Stahl D. A., Sogin M. L., Pace N. R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legocki R. P., Yun A. C., Szalay A. A. Expression of beta-galactosidase controlled by a nitrogenase promoter in stem nodules of Aeschynomene scabra. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5806–5810. doi: 10.1073/pnas.81.18.5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E., Stackebrandt E., Dorsch M., Wolters J., Busch M., Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J Bacteriol. 1990 Jul;172(7):3569–3576. doi: 10.1128/jb.172.7.3569-3576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Tsuji K., Tsien H. C., Hanson R. S., DePalma S. R., Scholtz R., LaRoche S. 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J Gen Microbiol. 1990 Jan;136(1):1–10. doi: 10.1099/00221287-136-1-1. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Dobson M. E., Samuel J. E., Dasch G. A., Mallavia L. P., Baca O., Mandelco L., Sechrest J. E., Weiss E., Woese C. R. Phylogenetic diversity of the Rickettsiae. J Bacteriol. 1989 Aug;171(8):4202–4206. doi: 10.1128/jb.171.8.4202-4206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Woese C. R., Dobson M. E., Weiss E. A common origin of rickettsiae and certain plant pathogens. Science. 1985 Nov 1;230(4725):556–558. doi: 10.1126/science.3931222. [DOI] [PubMed] [Google Scholar]
- Winship P. R. An improved method for directly sequencing PCR amplified material using dimethyl sulphoxide. Nucleic Acids Res. 1989 Feb 11;17(3):1266–1266. doi: 10.1093/nar/17.3.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Weisburg W. G., Paster B. J., Madigan M. T., Fowler V. J., Hahn C. M., Blanz P., Gupta R., Nealson K. H. The phylogeny of purple bacteria: the alpha subdivision. Syst Appl Microbiol. 1984;5:315–326. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- Yang D., Oyaizu Y., Oyaizu H., Olsen G. J., Woese C. R. Mitochondrial origins. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]


