Abstract
Schistosomiasis remains a largely neglected, global health problem. The morbid pathology of the disease stems from the host's inflammatory response to parasite eggs trapped in host tissues. Long term host/parasite survival is dependent upon the successful modulation of the acute pathological response, which is induced by egg antigens. In this study, using Multidimensional Protein Identification Technology, we identified the Schistosoma mansoni egg secretome consisting of 188 proteins. Notably we identified proteins involved in redox balance, molecular chaperoning and protein folding, development and signaling, scavenging and metabolic pathways, immune response modulation, and 32 novel, previously uncharacterized schistosome proteins. We localized a subset of previously-characterized schistosome proteins identified in egg secretions in this study, to the surface of live S. mansoni eggs using the circumoval precipitin reaction. The identification of proteins actively secreted by live schistosome eggs provides important new information for understanding immune modulation and the pathology of schistosomiasis.
Keywords: Schistosoma mansoni, Egg secretome, egg secreted proteins, MudPIT proteomics
1. Introduction
Schistosomiasis remains a neglected tropical parasitic disease with more than two hundred million human infections, and an estimated 280,000 deaths, per year in more than 70 countries [1]. Reassessment of disability due to schistosomiasis along with recent estimates of the global incidence of schistosome infection indicates that the actual public health burden due to schistosomiasis has been greatly underestimated [2]. The pathology of infection with the helminth Schistosoma mansoni is surprisingly varied, ranging from a relatively mild intestinal presentation to severe hepato-splenic disease, and is dependent upon not only parasitic antigens, but also the host's genetic milieu and state of concomitant schistosome infection or concurrent infection with other pathogens [3]. Specifically, morbid pathology stems from a complex host MHC-class II-restricted, CD4+ T cell-mediated hepato-intestinal granulomatous and fibrosing inflammatory response to parasite egg antigens [4]. In contrast, after initial infection, larval and adult parasites produce minimal inflammatory pathology in the host. Successful immunomodulation to limit excessive inflammation is critical to enhance the long term survival of both host and parasite [5,6].
After infection and a five week maturation period, schistosomes migrate to the mesenteric venous plexus, mate, and begin oviposition. Initial egg granuloma formation amidst pro-inflammatory cytokines normally shifts to a Th2-type anti-inflammatory cytokine environment [7], directly in response to secreted egg antigens [7,8]. The cumulative effect of granulomatous inflammation and fibrosis can lead to hepatic scarring, portal hypertension, gastro-intestinal hemorrhage, and death [9]. However, granulomas likely protect the host liver from egg secreted hepatotoxins [10] and possibly serve as foci of neovascularization that preserve host liver function and viability [11].
While jeopardizing host health, granulomatous inflammation facilitates the passage of live eggs from the mesenteric vasculature across endothelial and mucosal barriers to the lumen of the intestine with subsequent excretion, thus, allowing the parasitic lifecycle to continue. It is not clear how this process occurs, although it is dependent upon the inflammatory response, because egg excretion is minimal in immunocompromised mice [12]. Moreover, in HIV+ patients co-infected with S. mansoni, decreased CD4+ T-cell counts correlated with diminished egg excretion [13]. It is unclear how eggs initially attach to the endothelium and initiate penetration during extravasation, but factors released by eggs may bind extracellular matrix proteins.
Because of the role of egg antigens in regulating the cytokine environment and the subsequent immunopathology of schistosomiasis, identifying the components of soluble egg antigens (SEA) and egg secretory proteins (ESP) is of considerable interest. To date, only a handful of ESP and SEA components have been characterized. One of the most abundant egg proteins comprising 10% of total SEA is Schistosoma mansoni 40 kDa protein (Sm-p40, [14]), a protein with homology to small heat shock proteins (HSPs) and α-crystallin [15]. By examining the ability of SEA fractions to stimulate proliferation of CD4+ Th cells isolated from S. mansoni infected mice, not only Sm-p40, but p150/166 (albumin), phosphoenolpyruvate carboxykinase, glutathione S-transferase (GST)-(28K), peroxiredoxin 1, and SmEP25 [16] proteins have been identified. SmEP25 is identical to IL-4-inducing principle of S. mansoni eggs (IPSE)/alpha-1 (the name used henceforth) [17]. IPSE/alpha-1 is an egg-specific 20 kDa glycoprotein that forms a 40 kDa homodimer possessing the ability to stimulate basophils to degranulate and release IL-4, thus, perhaps playing a role in initiating the Th-2 phenotype [18]. High mobility group box 1 protein, an abundant nuclear protein involved in DNA replication, transcription, and chromatin assembly, also is found in egg secretions [19]. Additionally, the egg-specific 100 kDa protein kappa-5 remains poorly characterized [20], the egg-specific 31 kDa glycoprotein omega-1 [17,20] has been identified as a hepatotoxic ribonuclease [21], and 12 kDa thioredoxin has been identified in egg secretions by indirect immunofluorescence [22].
We hypothesized that antigens actively secreted by live eggs profoundly modulate the host-parasite interaction. To more fully characterize the proteinaceous components of ESP, we took a high throughput proteomic approach to identify live-egg secreted proteins by the extremely sensitive and comprehensive Multidimensional Protein Identification Technique (MudPIT, [23]. We identified 188 proteins, 156 with known identities and 32 novel, previously uncharacterized proteins. Having a comprehensive list of egg secreted proteins will facilitate our understanding of this complex host/parasite relationship.
2. Materials and methods
2.1 Collection of ESP and egg culture
For ESP collection, eggs were isolated and cultured as previously described [22]. Hatching experiments after culture and collection of ESP showed an >80% hatching rate indicating that the eggs used to make ESP were viable. Unhatched eggs (<20%) were either dead or unembryonated. Hatching of eggs during the collection period was <1%. Thus, the ESP collected for MudPIT analysis was representative of proteins secreted by live and not dead or hatched eggs.
2.2 MudPIT analysis of ESP
To identify the full proteomic signature of egg secreted proteins, ESP was treated as previously published [23]. Briefly, trichloroacetic acid-precipitated proteins were denatured (in 8 M urea) and cysteine residues were reduced (with 5 mM TCEP) and alkylated (with 20 mM iodoacetamide), to disrupt and prevent the reformation of disulfide bonds, and proteolytically digested at pH 8.5 with endoproteinase LysC (Roche) overnight at 37 °C. The samples were then digested overnight with trypsin (Roche) in 2M urea and 2 mM CaCl2 at 37 °C. The digestion reaction was quenched and acidified with 5% formic acid to aid in the ionization of the peptides for mass spectrometry. Peptides were loaded onto a fused-silica microcapillary column (100 μm inner diameter, 365 μm outer diameter, Polymicro) pulled with a laser puller (Model P-2000, Sutter Instrument Co.) to create a 5 μm tip for electrospray ionization, and packed first with 8-10 cm of 5 μm C18 reverse phase material (Aqua, Phenomenex) and followed by 4-5 cm of 5 μm strong cation exchange material (Partishpere SCX, Whattman).
Loaded biphasic microcapillary columns were installed in-line with a Quaternary Agilent 1100 series HPLC pump, such as to spray directly into an LCQ-Deca ion trap mass spectrometer (ThermoFinnigan, Palo Alto, CA) equipped with a nano-LC electrospray ionization source. Overflow tubing was used to decrease the flow rate to about 200-300 nl/min. Three different elution buffers were used: Buffer A (5% acetonitrile), Buffer B (80% acetonitrile, 0.1% formic acid), and Buffer C (500mM ammonium acetate, 5% acetonitrile, 0.1% formic acid). A fully automated, 12-step chromatography run was carried out whereby the peptides were sequentially eluted, first from the SCX resin to the RP resin, then from the RP resin directly into the mass spectrometer. The full MS spectra of eluted peptides were recorded over the 400 to 1600m/z range. Tandem MS (MS/MS) spectra were acquired in a data-dependent manner, sequentially on the first, second and third most intense ions selected from the full MS scan. SEQUEST [24] correlated MS/MS spectra to S. mansoni cDNA sequences in the S. mansoni Gene Index Release 6.0 (http://compbio.dfci.harvard.edu/tgi/). The SMGI database was six-frame translated and all open reading-frames larger than 50 amino acids were included in the database used for searching. In addition, the protein sequence database was concatenated to a reversed version of the database in order to define Xcorr and DeltaCN values for an appropriate false positive rate of protein identification. DTASelect [25] was used to apply these identification criteria in a manner similar to that employed by Peng et al. [26]. No cleavage bias was assumed when searching MS/MS spectra against the S. mansoni sequence database. It is widely accepted that endogenous proteases released upon cell lysis will contribute to proteolytic cleavage of proteins to be analyzed, thus it is not necessarily expected that all peptides be fully digested only at tryptic residues. The majority of peptides resulting in identification tended to contain at least one tryptic end. Proteins identified were required to have a minimum of two peptides per locus. For proteins whose corresponding EST sequences are incomplete or missing entirely, they will have underestimated sequence and spectral count values, but their identification would not be entirely hindered, as even partial sequences may be sufficient to identify peptides mapping to the regions of the gene that are represented in the EST database. Naturally, a complete genome sequence would be optimal to gain a more impartial view of all proteins, but unfortunately the S. mansoni genome sequence is not available at this time.
2.3 Bioinformatic analysis of S. mansoni ESP
Mass spectra, generated by LC/LC-MS/MS analysis of ESP, were assigned peptide identities based upon similarity to predicted fragmentation patterns of protein sequences by the SEQUEST algorithm [27]. Peptide sequences were then assigned to the S. mansoni Gene Index database (http://compbio.dfci.harvard.edu/tgi/) by sequence homology, BLASTed against the non-redundant database at NCBI (http://www.ncbi.nlm.nih.gov/) and annotated using Swiss-Prot/TrEMBL/KB primary accession numbers when possible. To quantify the relative abundance of any one protein in ESP, spectrum counts for the corresponding peptides belonging to each protein were combined.
Identified proteins were assigned annotated gene ontology using the Goblet server (http://goblet.molgen.mpg.de) and the human Swiss-Prot database [28,29]. Because the proteins were identified as secreted proteins, the presence or absence of N-terminal signal/targeting sequences was determined using SecretomeP, an algorithm capable of predicting the presence or absence of both classical (signal peptide, P) and non-classical (alternative sequence, +) secretion sequences [30].
2.4 RNA expression analysis
Controlled, semi-quantitative reverse transcription (RT)-PCR was performed using cDNA generated from 1 μg total RNA from each parasite lifecycle developmental stage (cercariae, skin (4 hour, 24 hour, 5 day) and lung (10 day) schistosomulae, liver (23 day), adult (male and female), and egg) as described [31]. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, M92359) was used as an internal loading control. PCR primers, reaction conditions, and cycle numbers are shown in Supplemental Table 1.
2.5 Circumoval precipitin reactions
Secretion by live eggs of selected proteins was confirmed by the circumoval precipitin reaction (COP) as described [22] using antibodies listed in Supplemental Table 2. For COP reactions using anti-Sm-p40 mouse monoclonal antibodies, consistently high background, suggestive of the presence of mouse IgG on the surface of the egg, was circumvented by using eggs isolated from hamster liver. Images were captured digitally as 1028 × 712 .tif files and compiled unaltered in Adobe Photoshop and Illustrator.
3. Results and discussion
3.1 Egg secretome analysis
Successful parasitism is the equilibrium between the necessity of survival of both host and parasite and the continuation of the parasite species. Adult schistosomes may survive within their vascular environment for up to 30 years [32]. The morbid pathology of schistosomiasis stems not from the resident adult parasites, but from a complex host immune response to parasite egg antigens [5]. This same granulomatous inflammatory reaction, which encapsulates the egg within an oxidatively stressful microenvironment, simultaneously mediates the passage of live parasite eggs across host intestinal tissues, and thus, is fundamental for the continuation of the parasite life cycle [12,13]. Because of the critical role of egg antigens during the schistosome infection, identifying the egg secretome is of interest.
The present study took a high-throughput, proteomic approach to comprehensively identify the secretome of the S. mansoni egg. The power of MudPIT analysis is precisely that proteins in a heterogeneous protein sample can be positively identified by this extremely sensitive method. The S. mansoni egg secretome was found to be surprisingly complex. Of the 188 proteins identified by MudPIT analysis (Supplemental Table 3), 156 proteins had similarity to previously annotated proteins and 32 proteins were uncharacterized, novel proteins. There is precedent for using spectrum count to reflect the relative abundance of individual proteins [33-35]. Original data correlating identified peptides to TC sequences are shown in Supplemental Table 4. It should be noted that this approach is not exactly quantitative but rather provides a semi-quantitative view of the components represented in a protein mixture. The advantage to other approaches is that the identification of components is much more comprehensive, but the relative abundance of those components can never be exactly quantified without absolute quantitative measures that are quite low-throughput and costly [36]. The proteins identified as components of ESP were ranked by descending total spectrum count (Spec ct, Table 1, Supplemental Table 3). This ranking does not represent a true molar concentration of proteins in ESP, but rather serves to delineate highly abundant proteins from those of lesser abundance. The number of unique peptides for each protein (Seq ct) and the minimum peptide coverage (% cov) were also listed (Table 1 and Supplemental Tables 3 and 4).
Table 1.
Abundant S. mansoni egg secreted proteins identified by MudPIT. Sequence count (Seq ct) is the number of unique peptides identified for each protein; % Cov: the percent of protein covered by the unique peptides. The total number of times all of the unique peptides were identified for the protein (Spec ct) is a quantitative measure of the protein's abundance in the sample. Relative abundance indicates high, medium, or low abundance in ESP. The prediction by secretome P (SecP) on the likelihood of the protein being secreted via the classical (P) or alternative (+), or not predicted to be secreted (−).
| Protein | Accession |
Seq ct |
% Cov |
Spec ct |
Relative Abundance |
SecP |
|---|---|---|---|---|---|---|
| IPSE/alpha-1 | Q869D4 | 36 | 61% | 301 | P | |
| Omega-1 | Q2Y2H5 | 29 | 60% | 172 | P | |
| Sm-p40 (major egg antigen) | P12812 | 58 | 70% | 150 | + | |
| HSP70 (major surface antigen) | P08418 | 41 | 59% | 108 | − | |
| Niemann-Pick C2 (NPC2) | TC20385 | 11 | 49% | 104 | P | |
| Fructose-bisphosphate aldolase | P53442 | 28 | 67% | 75 | High | − |
| Peptidylglycine alpha hydroxylating mono-oxygenase (PHM) |
Q86QQ6 | 21 | 45% | 74 | P | |
| Glutathione S-transferase 26 kDa | P35661 | 21 | 54% | 74 | − | |
| Phosphoenolpyruvate carboxykinase | Q9XYR4 | 35 | 56% | 73 | − | |
| Peroxiredoxin 1 | O97161 | 29 | 71% | 71 | + | |
| Actin-2 | P53471 | 25 | 56% | 62 | + | |
| Enolase | Q27877 | 14 | 49% | 62 | − | |
| 14-3-3 protein 1 | Q26540 | 24 | 64% | 61 | − | |
| Secretory glycoprotein kappa-5 | Q2KMJ3 | 27 | 51% | 58 | P | |
| Protein disulfide-isomerase | P38658 | 27 | 52% | 54 | P | |
| Thioredoxin 1 | Q8T9N5 | 22 | 74% | 54 | + | |
| Glycogenin | TC25407 | 14 | 50% | 52 | + | |
| Glutathione S-transferase 28 kDa | P09792 | 17 | 53% | 48 | − | |
| Polyubiquitin C | TC35851 | 15 | 29% | 47 | − | |
| Actin-1 | P53470 | 25 | 52% | 46 | + | |
| Beta-tubulin | Q7Z1I4 | 20 | 60% | 44 | + | |
| Retinaldehyde dehydrogenase | TC22118 | 22 | 48% | 42 | + | |
| Venom allergen-like protein 3 | Q1WMM9 | 9 | 65% | 41 | P | |
| Malate dehydrogenase | TC23687 | 11 | 43% | 40 | + | |
| Histone H2Ab | TC32673 | 6 | 42% | 40 | + | |
| Histone H2A | TC31134 | 6 | 31% | 40 | Medium | + |
| Cytochrome oxidase c subunit 1 | TC31128 | 8 | 56% | 38 | − | |
| HSP86 | Q26582 | 16 | 39% | 38 | − | |
| Unknown ESP 5 | TC25980 | 8 | 59% | 37 | + | |
| Unknown ESP 2 | TC25599 | 8 | 24% | 34 | − | |
| Purine-nucleoside phosphorylase | Q9BMI9 | 14 | 36% | 34 | + | |
| Alpha tubulin | Q26595 | 18 | 34% | 34 | − | |
| Thioredoxin glutathione reductase | Q962Y6 | 15 | 34% | 32 | − | |
| 14-3-3 protein zeta | TC30162 | 11 | 39% | 30 | − | |
| GAPDH (P-37) | P20287 | 16 | 46% | 28 | − | |
| Elongation factor 1-alpha | Q94747 | 15 | 35% | 28 | − | |
| Calreticulin precursor (SM4) | Q06814 | 14 | 40% | 27 | P | |
| Unknown ESP 1 | AW497769 | 10 | 64% | 26 | short | |
| Unknown ESP 6 | TC22057 | 11 | 61% | 26 | P | |
| Egg antigen SME16 | Q07167 | 7 | 49% | 26 | − | |
| ATP synthase beta chain | TC30449 | 14 | 34% | 26 | + | |
| Dj-1 beta | TC20573 | 12 | 74% | 25 | Low | − |
| Histone H4 | Q9GSS8 | 10 | 43% | 25 | − | |
| Transketolase | TC25273 | 14 | 36% | 24 | + | |
| Unknown ESP 7 | TC26653 | 9 | 44% | 23 | − | |
| Phosphoglycerate mutase | Q8WT66 | 6 | 32% | 23 | − | |
| Phosphoglycerate kinase | P41759 | 11 | 29% | 23 | − | |
Many abundant egg proteins are known to be heavily glycosylated [17,18,21]. During MudPIT analysis, glycoproteins cleaved by a protease generate glycopeptides that are part of the heterogeneous peptide mix. However, because the mass of glycosylated peptides cannot be predicted, they are not identified by MudPIT analysis. However, proteins known to be heavily glycosylated, such as IPSE/alpha-1, omega-1, and kappa-5 [17,18,21], were identified successfully by the MudPIT analysis here, via their non-glycosylated peptides. Although it is possible that every tryptic peptide in a protein could be glycosylated, and therefore the protein would not be identified by MudPIT, our results indicate that even heavily glycosylated proteins are not missed by MudPIT analysis.
In the present study, we noted that a small fraction of the total proteins comprised the bulk of the egg secretions. While spectrum count is not an exact molar representation of true abundance, it can be interpreted to support the notion that abundant proteins will generate a higher percentage of total peptides analyzed by mass spectral analysis, while less-abundant proteins will contribute far fewer peptides to the analysis, and thus, will have fewer relative spectrum counts. For analysis, we divided the egg secretome into thirds (high, medium, and low) to represent highly abundant proteins versus those of lesser abundance. The first 10 most-abundant proteins comprised the top third of total protein in ESP (Table 1). Among the most highly-abundant proteins identified in egg secretions were IPSE/alpha-1, omega-1, Sm-p40, HSP70, Niemann-Pick C2 (NPC2) and peptidylglycine alpha hydroxylating monooxygenase (PHM) [37]. A mere 31 proteins comprised the middle third of total ESP and the following 147 proteins comprised the low-abundance tier (Table 1). Proteins previously identified in ESP, namely, IPSE/alpha-1, omega-1, thioredoxin, and peroxiredoxin 1, were all identified as components of ESP by the analysis here. In contrast, high mobility group box-1, a nuclear protein also previously found in ESP [19], was not identified in this study. The absence of high mobility group box-1 may be explained by the presence of 42 potential trypsin cleavage sites that would generate peptides shorter than the peptide length minimum used in this analysis (7 residues, see Supplemental Table 4).
In addition to the proteins already mentioned, many other S. mansoni proteins in several functional categories were found in egg secretions: antioxidant proteins (peroxiredoxin 1, thioredoxin, thioredoxin glutathione reductase, GST-26K, GST-28K, GST-omega, superoxide dismutase), heat shock, chaperone and protein folding proteins (HSP70, HSP86/HSP90, polyubiquitin C, protein disulfide isomerase, 14-3-3 proteins), calcium-binding proteins (translationally controlled tumor protein, calpain, 16 kDa calcium-binding protein), inflammation-inducing proteins (IPSE/alpha-1, venom allergen-like proteins 2, 3, 5 and 9), glycolytic or glycolytic feeder pathway enzymes (fructose bisphosphate aldolase, enolase, phosphoenolpyruvate carboxykinase, GAPDH), and scavenging pathway proteins (NPC2 (cholesterol trafficking)).
3.2 Secretome P predictions
Secreted proteins would be expected to have a secretory sequence (either N-terminal or leaderless) and the presence of such sequences would further validate the identification of individual proteins in ESP. Protein sequences identified by MudPIT were analyzed for the presence or absence of secretion signals, including both the classic N-terminal signaling peptide and the non-classical or alternative internal secretion sequences [38]. Non-classically secreted proteins may be secreted by lysosomal or exosomal vesicular fusion, by membrane transporters, or by membrane blebbing [38].
Using the most complete protein sequences available, the presence or absence of either a classic, N-terminal signal peptide sequence or a non-classical, internal protein secretion sequence was predicted by the SecretomeP v. 2.0 algorithm [30], which assigned a neural network value to the protein sequences. Any protein sequence receiving an neural network value ≥0.5 was predicted to be secreted by either a classic signal sequence (P), or via a non-classical pathway (+). Protein sequences with a neural network value ≤0.5 were predicted to be non-secreted (−).
Of the 188 identified proteins in ESP, 118 proteins (63% of total) had an neural network value ≥0.5, and thus, were predicted by SecretomeP to be secreted (SecP in Table 1 and Supplemental Table 3); the proteins contained either a classic signal peptide leader sequence (P, 32 proteins, 17%) or were predicted to be secreted through a non-classical pathway (+, 86 proteins, 46%). Concomitantly, 67 proteins (35% of total) were predicted by SecretomeP to be not secreted via either pathway, and 3 proteins (short, 2%) were too short for the SecretomeP algorithm to analyze.
Because Sm-p40 was the third most-abundant protein in egg secretions contributing 4% of total spectrum counts, it was surprising that SecretomeP predicted Sm-p40 to be non-secreted. However, examination of available transcript and genomic databases revealed that Sm-p40 is a gene family. Of note, tentative consensus sequence TC28308 encoded a Sm-p40 family member encoding peptides identified by MudPIT. When this TC was used for the SecretomeP prediction, the neural network value was 0.774 and was therefore reported as (+) for Sm-p40 (Table 1, Supplemental Table 3).
One explanation for the number of proteins identified in ESP that were predicted by SecretomeP to be not secreted by either the classical or alternative pathway is that the Secretome P algorithm was trained using mammalian, and not platyhelminth, protein sequences [30], and that the sequences available in the databases potentially encode truncated ORFs, as described above for Sm-p40.
3.3 Gene ontology
Gene ontology was assigned to the array of identified proteins to gain a basic understanding of protein function and the biological processes in which they are involved. The gene ontology of the most-abundant egg secreted proteins (22 proteins comprising 50% of total ESP) was compared to the ontology of both the entire complement of egg secreted proteins (188 proteins, 100% of ESP) and the ontology of annotated Schistosoma mansoni proteins in the NCBI non-redundant database, as analyzed by the GOblet server previously (650 proteins, [39]). Table 2 displays the results of GO annotation for the three major categories of molecular function, biological process, and cellular component.
TABLE 2.
Functional Gene Ontology annotations of the S. mansoni egg secretome. GO annotations for the most abundant egg secreted proteins (22 proteins, Top 50% ESP) are compared to the GO annotations for both the entire egg secretome identified in this study (188 proteins, Total ESP) and the approximately 650 non-fragmentary schistosome proteins of the NCBI non-redundant data bank (NCBI nr).
| Top 50% ESP | Total ESP | NCBI nr* | ||||
|---|---|---|---|---|---|---|
| GO categories and subcategories | Total (22) | % total | Total (188) | % total | Total (650) | % Total |
| GO:0003674 molecular function | 19 | 100% | 129 | 100% | 448 | 100% |
| GO:0003774 motor activity | 0 | 0% | 3 | 2% | 29 | 6% |
| GO:0003824 catalytic activity | 14 | 74% | 79 | 61% | 267 | 60% |
| GO:0016491 oxidoreductase activity | 5 | 26% | 24 | 19% | 91 | 20% |
| GO:0016740 transferase activity | 3 | 16% | 18 | 14% | 73 | 16% |
| GO:0016787 hydrolase activity | 4 | 21% | 29 | 22% | 90 | 20% |
| GO:0004871 signal transducer activity | 2 | 11% | 23 | 18% | 20 | 4% |
| GO:0005198 structural molecule activity | 3 | 16% | 8 | 6% | 20 | 4% |
| GO:0005215 transporter activity | 3 | 16% | 19 | 15% | 83 | 19% |
| GO:0005488 binding | 13 | 68% | 102 | 79% | 278 | 62% |
| GO:0000166 nucleotide binding | 5 | 26% | 32 | 25% | 77 | 17% |
| GO:0005496 steroid binding | 1 | 5% | 2 | 2% | - | - |
| GO:0005515 protein binding | 10 | 53% | 70 | 54% | 49 | 11% |
| GO:0043167 ion binding | 3 | 16% | 26 | 20% | 91 | 20% |
| GO:0016209 antioxidant activity | 3 | 16% | 6 | 5% | 26 | 6% |
| GO:0004601 peroxidase activity | 1 | 5% | 4 | 3% | 8 | 2% |
| GO:0004791 thioredoxin-disulfide reductase act. | 2 | 11% | 3 | 2% | - | - |
| GO:0030234 enzyme regulator activity | 2 | 11% | 13 | 10% | 10 | 2% |
| GO:0008150 biological_process | 17 | 100% | 122 | 100% | 422 | 100% |
| GO:0000003 reproduction | 1 | 6% | 3 | 2% | 126 | 30% |
| GO:0007275 development | 2 | 12% | 14 | 11% | 76 | 18% |
| GO:0007582 physiological process | 16 | 94% | 117 | 96% | 330 | 78% |
| GO:0008152 metabolism | 16 | 94% | 101 | 83% | 217 | 51% |
| GO:0016265 death | 6 | 35% | 22 | 18% | - | - |
| GO:0051179 localization | 4 | 24% | 37 | 30% | - | - |
| GO:0007154 cell communication | 6 | 35% | 30 | 25% | 73 | 17% |
| GO:0050789 regulation of biological process | 6 | 35% | 45 | 37% | 6 | 1% |
| GO:0050896 response to stimulus | 4 | 24% | 26 | 21% | 34 | 8% |
| GO:0006950 response to stress | 3 | 18% | 19 | 16% | - | - |
| GO:0005575 cellular_component | 15 | 100% | 108 | 100% | 335 | 100% |
| GO:0005576 extracellular region | 3 | 20% | 14 | 13% | 13 | 4% |
| GO:0005623 cell | 14 | 93% | 102 | 94% | 279 | 83% |
| GO:0043226 organelle | 7 | 47% | 56 | 52% | - | - |
| GO:0043234 protein complex | 4 | 27% | 31 | 29% | - | - |
| GO:0000015 phosphopyruvate hydratase complex | 1 | 7% | 8 | 7% | - | - |
| GO:0005667 transcription factor complex | 3 | 20% | 3 | 3% | - | - |
as shown in Knudsen et al. 200639
Molecular function was assigned to 129 proteins. Fully 79% (102 proteins) had binding capacity (GO:0005488) including ion binding (GO:0043167), nucleic acid binding (GO:0000166), and protein binding (GO:0005515). Catalytic activity (GO:0003824) was identified for 61% of the proteins with hydrolase (GO:0016787), oxidoreductase (GO:0016491), and transferase (GO:0016740) activities specifically enriched. Similarly, 122 proteins were involved in annotated biological processes. Of these, 96% were involved in physiological processes (GO:0007582) such as cell death (GO:0016265), homeostasis (GO:0019725), metabolism (GO:0044237), and cell communication (GO:0007154). Finally, 108 proteins assigned to cellular component categories (GO: 0005575) were frequently localized to cytoplasmic organelles (GO:0043226) and protein complexes (GO:0043234).
3.4 RT-PCR results
MudPIT identified 36 unknown, previously uncharacterized proteins in egg secretions (unknown ESP 1-36, Table 1 and Supplemental Table 3). The developmental expression pattern of a subset of some of the more abundant unknown ESP proteins (Unknown ESP 2-5) was determined by RT-PCR. The results in Figure 1 show that the expression of unknowns ESP 2-4 were specific to egg, while unknown ESP 5 was expressed not only in egg, but also in all stages tested except for adult females. The expression pattern of unknown ESP 5 was interesting; expression was seen to be highest during stages adapting to new environments (Figure 1). Although unknown ESP 5 was expressed at very low levels in cercariae, its expression was rapidly induced after transformation to schistosomula. However, within 24 hour (24 hr) the expression of unknown ESP 5 was greatly reduced and remained so until 10 days (10 d) after transformation, at which point its expression increased and remained so through juvenile liver stage (23 d) until again being reduced in adult worms. Therefore unknown ESP 5 expression reflects a pattern in which maximum expression occurs as parasites adapt to the skin, lungs and liver of the host.
Figure 1. RT-PCR analysis of unknown egg secreted protein mRNA expression across Schistosoma mansoni life cycle stages.
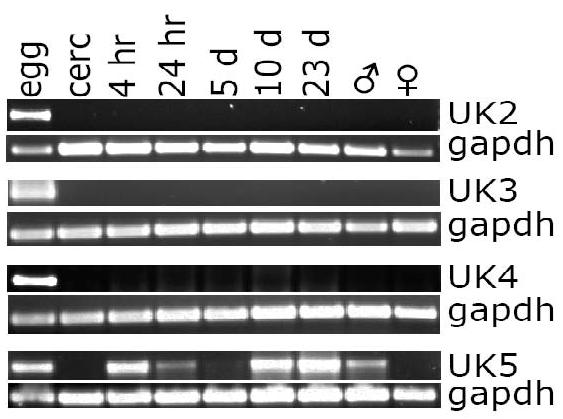
RT-PCR, using primer pairs specific for unknown ESP proteins 2-5 (UK 2-5), was used to visualize the expression of unknown egg protein expression throughout the S. mansoni life cycle. In each lane, 0.5 μl cDNA from one of nine developmental stages (egg (egg); cercariae (cerc); schistosomula cultured for either 4 hours (4 hr), 24 hours (24 hr), 5 days (5d), or 10 days (10d); 23 day larvae (23d); adult males (♂); adult females (♀)) was probed by RT-PCR using the primers and conditions listed in Supplemental Table 2.
3.5 Circumoval precipitin reaction results
The COP reaction was first described as a serodiagnostic tool for schistosome infection [40]. In the COP reaction, proteins exit the egg through pores in the eggshell, interact with host antibodies, and form a precipitate at the point of secretion [41]. We confirmed the MudPIT data by localizing some of the more abundant egg secreted proteins on the surface of live eggs by both direct and indirect immunofluorescent COP reactions (Figure 2 and Supplemental Figure 1). Figure 2 shows the COP reactions obtained using antibodies against HSP70 and Sm-p40, visualized by fluorochrome labeled secondary antibodies. Although it is not possible to use the COP reaction to confirm the secretion of every protein identified in this study, the localization to the surface of live eggs in a COP reaction of a subset of proteins identified in ESP supports the presence in ESP of other proteins identified in this study.
Figure 2. Egg secreted proteins can be visualized in circumoval precipitin reactions.
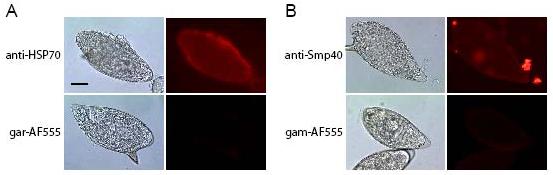
Panel (A) shows eggs cultured with rabbit polyclonal antibodies against HSP70 visualized with anti-rabbit IgG-Alexa Fluor 555-labeled secondary antibody, or with secondary antibody alone (goat anti rabbit-AF555). Panel (B) shows eggs cultured with either a mixture of equal amounts of several purified monoclonal antibodies against Sm-p40 visualized by anti-mouse IgG-Alexa Fluor 555-labeled secondary antibody, and the secondary antibody alone (goat anti mouse-AF555). Exposure times are: bright field (1/30 sec), immunofluorescence (1 sec). Bar equals 30 μm.
Additional proteins (thioredoxin, ubiquitin, PHM, tubulin, albumin, thioredoxin glutathione reductase, and GST(26K), all identified by MudPIT to be present in egg secretions, also were localized to the surface of live eggs via the COP reaction and are shown in Supplemental Figure 1. As an additional control, ribosomal subunit S6, which was not identified in ESP, also was not co-localized in a COP reaction indicating that the presence of antibodies alone during the in vitro culture of eggs does not produce a COP reaction (Supplemental Figure 1).
3.6 Functions of proteins found in ESP
The 188 egg secreted proteins fall into some general categories including energy production, cell signaling, and maintenance of redox balance (thioredoxin, peroxiredoxins, albumin, GST-26K, GST-28K, GST-omega). Previously identified egg-specific glycoproteins (IPSE/alpha-1, omega-1, kappa-5) were positively identified as components of ESP, and yet some of the most abundant proteins are still relatively uncharacterized. A comparison of our proteomic identification of egg secreted proteins and the results of both peptide mass fingerprinting of SEA [42] and cercarial secretions [39] revealed that many proteins were identified in all three studies: 14-3-3 proteins, actin, enolase, GAPDH, HSP70, thioredoxin, and GST −26K and −28K. However, in this study we also identified protein components not previously identified in parasitic secretions including NPC2 and venom allergen-like proteins.
One of the most abundant protein in ESP was Sm-p40. Sm-p40 shows similarity to α-crystallin, which can function as a molecular chaperone, prevent protein aggregation, and facilitates protein refolding [43]. Based upon Sm-p40's similarity to small heat shock proteins and α-crystallin, it is intriguing to speculate that this extremely abundant secreted egg protein may function as a molecular chaperone during protein folding or secretion, or may protect the egg in some capacity within the oxidatively stressful granuloma environment.
Similarly, heat shock proteins, including HSP86/90, -70, -60, -40, and -10, were found to be present in ESP. HSPs are typically immunodominant antigens and major targets of host immune responses, including autoimmunity [44]. As a group, heat shock proteins perform many different tasks within the same cellular compartments, thus, regulation of access to substrates is essential and relies upon the presence of co-chaperones to facilitate targeting to specific substrates and functions. Co-chaperones include J-domain proteins, such as Dj-1 beta (TC20573), also identified as a component of ESP in this study.
NPC2, a ubiquitous, essential protein involved in cholesterol trafficking, was also identified as one of the most-abundant protein in ESP; this is the first time NPC2 has been identified in Schistosoma. NPC2 is a small intralysosomal cholesterol-binding protein involved in the egress of cholesterol out of endosomal/lysosomal compartments [45,46]. Although NPC2, along with NPC1, is essential for proper cellular cholesterol trafficking, its precise function has remained elusive [45]. The S. mansoni and human NPC2 proteins shared 37% identity and 55% similarity, and Phe-66, Val-96, and Tyr-100, which have been found to be essential for cholesterol binding [47], were conserved in the S. mansoni protein, as was the predicted secondary structures of the proteins (Figure 3). Glycosylation has been shown to be essential for human NPC2 function [48] and a conserved N-glycosylation site was predicted in the S. mansoni NPC2 (Figure 3). Furthermore, molecular modeling of S. mansoni NPC2 generated a predicted structure similar to human NPC2 [49] that included conservation of the order of β-sheets, Cys-Cys bonds, and localization of the residues implicated in cholesterol binding to a hydrophobic pocket in the protein (Figure 3) suggesting a similar cholesterol-binding function for the S. mansoni NPC2 protein. One possible explanation for the presence of NPC2 in ESP is to facilitate the movement of cholesterol (which eggs must scavenge). However, NPC2 may also facilitate the egress of other glycolipids [50]. Moreover, parasite glycoproteins circulating on host low-density lipoprotein particles have been shown to disrupt lipid homeostasis in host immune cells and induce cellular apoptosis [51] and parasite glycolipids may influence pathogenesis and immunity to infection [52]. NPC2 may participate in this exchange of parasite-derived immunomodulatory molecules.
Figure 3.
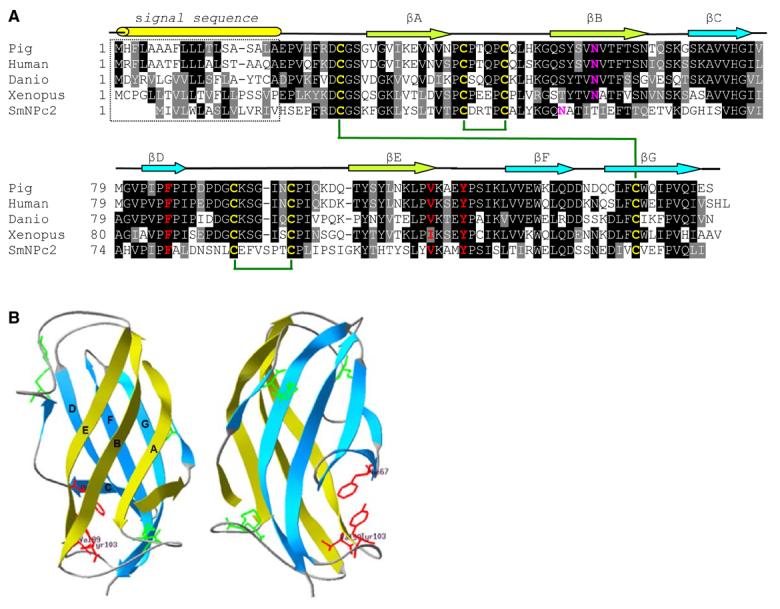
Panel A. Primary sequence alignment of S. mansoni and vertebrate NPC2 proteins. NPC2 proteins from pig (O97763), human (P61916), Danio rerio (Q9DGJ3), and Xenopus tropicalis (CAJ83028) were aligned with the S. mansoni NPC2 protein using ClustalW [57]. Identical residues are shown in reverse font and functionally conserved residues are shown with gray background. The predicted secondary structure elements [58] of S. mansoni NPC2 are shown above the sequence alignment. Predicted β-sheets (arrows) and leader peptide (tube) are indicated. Residues determined to be essential for cholesterol binding in human NPC2 are shown in red, conserved Cys residues are shown in yellow and potential disulfides are indicated by green lines, and potential N-glycosylation sites are shown in pink. Panel B. Structural model of S. mansoni NPC2. Ribbon diagrams of S. mansoni NPC2 with views related by a 90° rotation. One β-sheet is shown in lime and the other in turquoise. The side chains of cysteine residues that form three disulfide bonds are shown in green. Residues that have been found to be essential for cholesterol binding, Phe-66, Val-96, and Tyr-100, are shown in red. This figure was generated using Swiss-Model and DeepView/Swiss-PDB Viewer [59].
Several glycolytic enzymes, including enolase, were identified in ESP. Enolase, plays a key role in glycolysis and gluconeogenesis; however, it is also a multifunctional, non-classically secreted protein found on the surface of hematopoetic, epithelial, and endothelial cells, where it serves as not only a plasminogen receptor (plasminogen possesses fibrinolytic characteristics) and is implicated in tissue invasion, but as a heat-shock protein, a hypoxic stress protein, and a cytoskeletal and chromatin binding protein [53]. Enolase has been implicated in a wide variety of autoimmune diseases, including lupus erythematosus, ulcerative colitis, Crohn's disease, autoimmune hepatitis, apoptosis, and endometriosis [53]. The intriguing similarity between enolase, T-crystallins (monomers), and Sm-p40, which is similar to alpha crystallin, remains to be elucidated. Another glycolytic enzyme identified in ESP was GAPDH. GAPDH has been shown to bind fibronectin and laminin [54] and may be involved in mediating the attachment of the egg to host tissues, or somehow aid in the passage of live eggs across host tissues into the external environment.
Some of the proteins secreted by eggs may have pleiotropic effects upon the surrounding granuloma cells or the host's immunoflammatory response. For example, many neuropeptide transmitters require amidation for biological activity and PHM was identified in egg secretions [37]. Several 14-3-3 proteins, which act as molecular scaffolds or chaperones and may have isoform specific functions during development [55], were present in egg secretions. And finally, the presence of polyubiquitin C in egg secretions is intriguing in that ubiquination of proteins has diverse functions in a myriad of cellular processes such as protein stability and intracellular localization. Several dynamic classes of proteins involved in cell communication (connexins) and adhesion (cadherin/catenin complexes) are regulated by ubiquination [56]. What effects these egg secreted proteins have upon the host remains to be determined.
In conclusion, we used MudPIT to identify the S. mansoni egg secretome, a complex array of 188 proteins with an intriguing array of molecular functions. Additionally, 32 novel ESP proteins were identified. Since egg secreted proteins profoundly influence the Th1/Th2-cytokine environment and serve as the focus of the host immunoinflammatory response, the identification of proteins secreted by live eggs is of fundamental interest. Now that a comprehensive list of egg secreted proteins is available, future research may elucidate their specific role in the immunobiology and pathogenesis of schistosomiasis.
Supplementary Material
Acknowledgements
The authors thank Dr. Tim Day for the gift of anti-PHM antiserum and Dr. Philip LoVerde for the infected hamster liver. This work was funded by NIH/NIAID grant R03AI059037. Schistosome materials for this work were supplied in part through NIH-NIAID contract N01-AI-55270.
Abbreviations
- COP
circumoval precipitin
- ESP
egg secreted proteins
- GST
glutathione-S-transferase
- HSP
heat shock protein
- IPSE
IL-4-inducing principle of S. mansoni eggs
- MudPIT
multidimensional protein identification technique
- NPC2
Niemann-Pick C2
- PHM
peptidylglycine alpha-hydroxylating monooxygenase
- SEA
soluble egg antigens
- Sm-p40
S. mansoni egg protein 40
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hotez PJ, Molyneux DH, Fenwick A, Ottesen E, Ehrlich Sachs S, et al. Incorporating a rapid-impact package for neglected tropical diseases with programs for HIV/AIDS, tuberculosis, and malaria. PLoS Med. 2006;3:e102. doi: 10.1371/journal.pmed.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King CH, Dickman K, Tisch DJ. Reassessment of the cost of chronic helminthic infection: a meta-analysis of disability-related outcomes in endemic schistosomiasis. Lancet. 2005;365:1561–9. doi: 10.1016/S0140-6736(05)66457-4. [DOI] [PubMed] [Google Scholar]
- 3.Abath FGC, Morais CNL, Montenegro CEL, Wynn TA, Montenegro SML. Immunopathogenic mechanisms in schistosomiasis: what can be learnt from human studies? Trends Parasitol. 2006;22:86–91. doi: 10.1016/j.pt.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Hernandez HJ, Wang Y, Tzellas N, Stadecker MJ. Expression of class II, but not class I, major histocompatibility complex molecules is required for granuloma formation in infection with Schistosoma mansoni. Eur J Immunol. 1997;27:1170–6. doi: 10.1002/eji.1830270518. [DOI] [PubMed] [Google Scholar]
- 5.Pearce EJ, MacDonald AS. The immunobiology of schistosomiasis. Nat Rev Immunol. 2002;2:499–511. doi: 10.1038/nri843. [DOI] [PubMed] [Google Scholar]
- 6.Gause WC, Urban JF, Jr, Stadecker MJ. The immune response to parasitic helminths: insights from murine models. Trends Immunol. 2003;24:269–77. doi: 10.1016/s1471-4906(03)00101-7. [DOI] [PubMed] [Google Scholar]
- 7.Pearce E, Caspar P, Grzych J, Lewis F, Sher A. Downregulation of TH1 cytokine production accompanies induction of TH2 responses by a parasitic helminth, Schistosoma mansoni. J Exp Med. 1991;173:159–66. doi: 10.1084/jem.173.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grzych JM, Pearce E, Cheever A, Caulada ZA, Caspar P, et al. Egg deposition is the major stimulus for the production of Th2 cytokines in murine schistosomiasis mansoni. J Immunol. 1991;146:1322–7. [PubMed] [Google Scholar]
- 9.Boros DL. Immunopathology of Schistosoma mansoni infection. Clin Microbiol Rev. 1989;2:250–69. doi: 10.1128/cmr.2.3.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunne DW, Doenhoff MJ. Schistosoma mansoni egg antigens and hepatocyte damage in infected T-cell-deprived mice. Contrib Microbiol Immunol. 1983;7:22–9. [PubMed] [Google Scholar]
- 11.Loeffler DA, Lundy SK, Singh KP, Gerard HC, Hudson AP, Boros DL. Soluble egg antigens from Schistosoma mansoni induce angiogenesis-related processes by up-regulating vascular endothelial growth factor in human endothelial cells. J Inf Dis. 2002;185:1650–6. doi: 10.1086/340416. [DOI] [PubMed] [Google Scholar]
- 12.Doenhoff MJ. A role for granulomatous inflammation in the transmission of infections disease: schistosomiasis and tuberculosis. Parasitol. 1997;115:113–25. doi: 10.1017/s0031182097001972. [DOI] [PubMed] [Google Scholar]
- 13.Karanja DM, Colley DG, Nahlan BL, Ouma JH, Secor WE. Studies of schistosomiasis in western Kenya. I. Evidence for immune-facilitated excretion of schistosome eggs from patients with Schistosoma mansoni and human immunodeficiency virus coinfections. Am J Trop Med Hyg. 1997;56:515–21. doi: 10.4269/ajtmh.1997.56.515. [DOI] [PubMed] [Google Scholar]
- 14.Stadecker MJ, Hernandez HJ, Asahi H. The identification and characterization of new immunogenic egg components: implications for evaluation and control of the immunopathogenic T cell response in Schistosomiasis. Mem Inst Osw Cruz. 2001;96:29–34. doi: 10.1590/s0074-02762001000900004. [DOI] [PubMed] [Google Scholar]
- 15.Nene V, Dunne DW, Johnson KS, Taylor DW, Cordingley JS. Sequence and expression of a major egg antigen from Schistosoma mansoni. Homologies to heat shock proteins and alpha-crystallins. Mol Biochem Parasitol. 1986;21:179–88. doi: 10.1016/0166-6851(86)90021-6. [DOI] [PubMed] [Google Scholar]
- 16.Asahi H, Stadecker MJ. Analysis of egg antigens inducing hepatic lesions in schistosome infection. Parasitol Int. 2003;52:361–7. doi: 10.1016/s1383-5769(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 17.Dunne DW, Jones FM, Doenhoff MJ. The purification, characterization serological activity and hepatotoxic properties of two cationic glycoproteins (α1 and ω1) from Schistosoma mansoni eggs. Parasitol. 1991;103:225–36. doi: 10.1017/s0031182000059503. [DOI] [PubMed] [Google Scholar]
- 18.Schramm G, Gronow A, Knobloch J, Wippersteg V, Grevelding CG, et al. IPSE/alpha-1: a major immunogenic component secreted from Schistosoma mansoni eggs. Mol Biochem Parasitol. 2006;147:9–19. doi: 10.1016/j.molbiopara.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Gnanasekar M, Velusamy R, He Y-X, Ramaswamy K. Cloning and characterization of a high mobility group box 1 (HMGB1) homologue protein from Schistosoma mansoni. Mol Biochem Parasitol. 2006;145:137–46. doi: 10.1016/j.molbiopara.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Dunne DW, Agnew AM, Modha J, Doenhoff MJ. Schistosoma mansoni egg antigens: preparation of rabbit antisera with monospecific immunoprecipitating activity, and their use in antigen characterization. Parasite Immunol. 1986;8:575–86. doi: 10.1111/j.1365-3024.1986.tb00871.x. [DOI] [PubMed] [Google Scholar]
- 21.Fitzsimmons CM, Schramm G, Jones FM, Chalmers IW, Hoffman KF, et al. Molecular characterization of omega-1: a hepatotoxic ribonuclease from Schistosoma mansoni eggs. Mol Biochem Parasitol. 2005;144:123–27. doi: 10.1016/j.molbiopara.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 22.Alger HM, Sayed AA, Stadecker MJ, Williams DL. Molecular and enzymatic characterization of Schistosoma mansoni thioredoxin. Int J Parasitol. 2002;32:1285–92. doi: 10.1016/s0020-7519(02)00108-x. [DOI] [PubMed] [Google Scholar]
- 23.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 24.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–89. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 25.Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–6. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 27.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, et al. Direct analysis of protein complexes using mass spectrometry. Nature Biotechnol. 1999;17:676–82. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 28.Groth D, Lehrach H, Hennig S. GOblet: a platform for Gene Ontology annotation of anonymous sequence data. Nuc Acids Res. 2004;32:313–17. doi: 10.1093/nar/gkh406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hennig S, Groth D, Lehrach H. Automated Gene Ontology annotation for anonymous sequence data. Nuc Acids Res. 2003;31:3712–15. doi: 10.1093/nar/gkg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S. Feature-based prediction of non-classical and leaderless protein secretion. Protein Eng Design & Selection. 2004;17:49–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- 31.Sayed AA, Cook SK, Williams DL. Redox balance mechanisms in Schistosoma mansoni rely on peroxiredoxins and albumin and implicate peroxiredoxins as novel drug targets. J Biol Chem. 2006;281:17001–10. doi: 10.1074/jbc.M512601200. [DOI] [PubMed] [Google Scholar]
- 32.Vermund SH, Bradley DJ, Ruiz-Tiben E. Survival of Schistosoma mansoni in the human host: estimates from a community-based prospective study in Puerto Rico. Am J Trop Med Hyg. 1983;32:1040–8. doi: 10.4269/ajtmh.1983.32.1040. [DOI] [PubMed] [Google Scholar]
- 33.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 34.Gao J, Friedrichs MS, Dongre AR, Opiteck GJ. Guidelines for the routine application of the peptide hits technique. J Am Soc Mass Spectrom. 2005;16:1231–8. doi: 10.1016/j.jasms.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal Chem. 2005;77:6218–24. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 36.Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. PNAS. 2003;100:6940–5. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mair GR, Niciu MJ, Stewart MT, Brennan G, Omar H, et al. A functionally atypical amidating enzyme from the human parasite Schistosoma mansoni. FASEB J. 2004;1:114–21. doi: 10.1096/fj.03-0429com. [DOI] [PubMed] [Google Scholar]
- 38.Nickel W. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic. 2005;6:607–14. doi: 10.1111/j.1600-0854.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 39.Knudsen GM, Medzihradszky KF, Lim K-C, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol Cell Proteomics. 2005;4:1862–75. doi: 10.1074/mcp.M500097-MCP200. [DOI] [PubMed] [Google Scholar]
- 40.Oliver-González J. Anti-egg precipitins in the serum of humans infected with Schistosoma mansoni. J Inf Dis. 1954;95:86–91. doi: 10.1093/infdis/95.1.86. [DOI] [PubMed] [Google Scholar]
- 41.Demaree RS, Hillyer GV. Schistosoma species: transmission electron microscopy of the circumoval immune precipitin reaction on eggs. Exp Parasitol. 1981;52:77–85. doi: 10.1016/0014-4894(81)90063-1. [DOI] [PubMed] [Google Scholar]
- 42.Curwen RS, Ashton PD, Johnston DA, Wilson RA. The Schistosoma mansoni soluble proteome: a comparison across four life-cycle stages. Mol Biochem Parasitol. 2004;138:57–66. doi: 10.1016/j.molbiopara.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 43.Horwitz J. α-Crystallin can function as a molecular chaperone. Proc Natl Acad Sci USA. 1992;89:10449–53. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell & Mol Life Sci. 2005;62:670–84. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanier MT, Millat G. Structure and function of the NPC2 protein. Biochim Biophys Acta. 2004;1685:14–21. doi: 10.1016/j.bbalip.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membrane supports a role in lysosomal cholesterol transport. J Biol Chem. 2006;281:31594–604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- 47.Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc Natl Acad Sci USA. 2003;100:2518–25. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chikh K, Vey S, Simonot C, Vanier MT, Millat G. Niemann-Pick type C disease:importance of N-glycosylation sites for function and cellular location of the NPC2 protein. Mol Genet Metab. 2004;83:220–30. doi: 10.1016/j.ymgme.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 49.Friedland N, Liou H-L, Lobel P, Stock AM. Structure of a cholesterol-binding protein deficient in Niemann-Pick type C2 disease. Proc Nat Acad Sci USA. 2003;100:2512–7. doi: 10.1073/pnas.0437840100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang M, Sun M, Dwyer NK, Comly ME, Patel SC, et al. Differential trafficking of the Niemann-Pick C1 and 2 proteins highlights distinct roles in late endocytic lipid trafficking. Acta Paediatr Suppl. 2003;92:63–73. doi: 10.1111/j.1651-2227.2003.tb00224.x. [DOI] [PubMed] [Google Scholar]
- 51.Sprong H, Suchanek M, van Dijk SM, van Remoortere A, Klumperman J, et al. Aberrant receptor-mediated endocytosis of Schistosoma mansoni glycoproteins on host lipoproteins. PLoS Med. 2006;3:1360–70. doi: 10.1371/journal.pmed.0030253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Kleij D, van den Biggelsar AH, Kruize YC, Retra K, Fillie Y, et al. Responses to Toll-like receptor ligands in children living in areas where schistosome infections are endemic. J Infec Dis. 2004;15:1044–51. doi: 10.1086/382089. [DOI] [PubMed] [Google Scholar]
- 53.Pancholi V. Multifunctional α-enolase: its role in diseases. Cell Mol Life Sci. 2001;58:902–20. doi: 10.1007/PL00000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gozalbo D, Gil-Navarro I, Azorín I, Renau-Piqueras J, Martinez JP, et al. The cell wall-associated glyceraldehyde-3-phosphate dehydrogenase of Candida albicans is also a fibronectin and laminin binding protein. Infect Immun. 1998;66:2052–9. doi: 10.1128/iai.66.5.2052-2059.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGonigle S, Soschiavo M, Pearce E. 14-3-3 proteins in Schistosoma mansoni; identification of a second epsilon isoform. Int J Parasitol. 2002;32:685–93. doi: 10.1016/s0020-7519(01)00323-x. [DOI] [PubMed] [Google Scholar]
- 56.Berthoud VM, Minogue PJ, Laing JG, Beyer EC. Pathways for degradation of connexins and gap junctions. Cardiovas Res. 2004;62:256–67. doi: 10.1016/j.cardiores.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 57.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nuc Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–5. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 59.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 60.Alger HM, Williams DL. The disulfide redox system of Schistosoma mansoni and the importance of a multifunctional enzyme, thioredoxin glutathione reductase. Mol Biochem Parasitol. 2002;121:129–39. doi: 10.1016/s0166-6851(02)00031-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


