Summary of Recent Advances
Argonaute is at the heart of all effector complexes in RNA interference. In the classical RNAi pathway Argonaute functions as the Slicer enzyme that cleaves a mRNA target directed by a complementary siRNA. Two recently described Argonaute protein subfamilies mediate distinct functions in RNAi. The Piwi subfamily functions in the germline through a novel class of small RNAs that are longer than Argonaute specific si- and miRNAs. Piwi-interacting RNAs (piRNAs) carry a 2’ O-methylation on their 3’ end and appear to be synthesized by a Piwi Slicer dependent mechanism. Piwi/piRNA complexes in mammals and flies are directly linked to the control of transposable elements during germline development. Amplified RNAi in C. elegans is mediated by secondary siRNAs selectively bound to secondary Argonautes (SAGOs) that belong to a worm specific Argonaute subfamily (WAGO). Secondary siRNAs are 5’ triphosphorylated which may allow specific loading into SAGO complexes that are rate limiting for RNAi in C. elegans. Interestingly SAGOs lack conserved Slicer amino acid residues and probably act in a Slicer-independent fashion.
Introduction
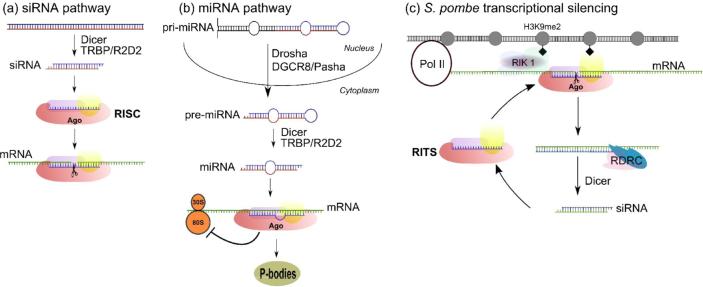
RNA interference (RNAi), the process by which genes are silenced by small RNAs, is a rapidly emerging field of research with contributions from molecular biology, genetics, biochemistry, bioinformatics and structural biology. Beyond the progress made in the basic research laboratory, RNAi technology is receiving deserved consideration as a clinical therapeutic strategy to specifically target genes associated with disease [1]. RNAi is accomplished through transcriptional (TGS) or post-transcriptional (PTGS) gene silencing (recently reviewed in [2]). One form of PTGS is induced by the introduction of long dsRNAs homologous to a target gene which produces small interfering RNAs (siRNA pathway). The RNA induced silencing complex (RISC) is guided by a siRNA complementary to mRNA targets and catalyzes a mRNA cleavage event (slicing) leading to message degradation (Figure 1a). PTGS also operates via translational repression induced by microRNAs (miRNA pathway) derived from hairpin precursors transcribed in the nucleus. Although controversial the miRNA silencing machinery directs targeted mRNAs into cytoplasmic foci called P-bodies, which are deprived of the translational machinery but retain proteins involved in mRNA degradation [3-8] (Figure 1b). TGS is best described in fission yeast where small RNAs and a specialized RISC complex (RITS) guide the formation of heterochromatin [9] (Figure 1c).
Figure 1.
RNA interference pathways involving the Argonaute protein subfamily. a) In “classical RNAi”, or the siRNA pathway, exogenous dsRNAs are processed into siRNAs by Dicer complexes. The siRNAs are loaded into Argonaute proteins which are at the center of the RNA-induced silencing complex (RISC). The siRNA charged RISC seeks out complementary mRNA and catalyzes endonucleolytic cleavage of the transcript, a process called slicing. Argonaute is the enzymatic component of RISC and is therefore called Slicer. b) MicroRNAs (miRNAs) are derived from endogenous hairpin precursors transcribed in the nucleus (pri-miRNA). Drosha containing complexes process the pri-miRNA into pre-miRNA which is transported to the cytoplasm where mature miRNAs are made by Dicer. Argonaute is loaded with miRNAs in miRNP complexes and are guided to the 3’ UTR of mature mRNA transcripts. miRNPs are believed to not slice, but instead inhibit translation or sequester transcripts to cytoplasmic foci called P-bodies. c) Transcriptional silencing and spreading in S. pombe is dependent on the RITS complex which contains Argonaute, Tas3 and Chp1 proteins. Silencing is achieved through the formation of heterochromatin induced by H3 Lys9 dimethylation (H3K9me2, black diamonds). An siRNA guided RITS complex slices nascent transcripts synthesized by Pol II. RDRC converts the cleaved transcript into a dsRNA substrate for Dicer which produces secondary siRNAs. Loading into new RITS complexes for additional targeting is coupled to the Rik1 complex, which contains a histone methyltransferase resulting in H3K9me2 and spreading of silencing.
This review will briefly summarize the structural and biochemical contributions made toward a mechanistic understanding of Argonaute, the Slicer enzyme component of RNAi effector complexes. The greater part of the review will highlight recent literature describing two new classes of small RNAs (Table 1) that interact specifically with the Piwi and worm-specific Argonaute (WAGO) protein subfamilies.
Table 1.
Distinct Classes of Small RNAs
Argonaute is Slicer
Argonaute proteins are present in all RISC complexes reported to date and are now the best defined protein component of the RNAi machinery. The critical step in “classic” RNAi is cleavage of mRNA transcripts through an endonucleolytic cleavage guided by a siRNA, termed Slicing (Figure 1). Argonaute was shown to be the enzyme within RISC that catalyzes this event and was thus identified as Slicer [10,11]. It is composed of four domains: an N-terminal, PAZ, Mid and a C-terminal PIWI domain (Figure 2a). The structure of a full length Argonaute protein from Pyrococcus furiosus (PfAgo) revealed marked similarity between the PIWI domain of Argonaute to the RNase H family of ribonucleases [10]. PIWI domains in subsequent structures of Aquifex aeolicus Argonaute [12] and a Mid-PIWI domain containing protein from Archaeoglobus fulgidus [13,14] also contain a RNase H-like fold. This structural fold along with two critical aspartates is shared by the catalytic domains of enzymes having nuclease or polynucleotidyl transferase activities such as RNase H, integrase and transposase [10] (Figure 2b). The active site of Argonaute is composed of a DDH metal coordinating triad [15] (Figure 2d). Argonaute is the Slicer enzyme as mutation of the aspartates and histidine residues in human Argonaute 2 abolishes activity [11,15]. The mechanism of Argonaute likely involves a Mg2+ activated water molecule which performs an SN2 like nucleophilic attack on the scissile phosphate which proceeds through a pentacovalent intermediate. Following collapse of this high energy intermediate a 5’ product RNA with a 3’ hydroxyl and a 3’ product carrying a 5’ phosphate are released. RNase H operates via a two metal ion mechanism; metal ion A is analogous to the metal ion binding site identified in Argonaute [15], while the role of metal ion B is to be in position to stabilize the transition state [16]. To date a second metal ion binding site has not been identified in Argonaute. It is possible that a second metal ion binding site may require the presence of a guide strand or target RNA for which a structure has not been reported. Despite a partial appreciation of the Slicer mechanism, insight into binding recognition of guide strand RNAs has made considerable progress in recent years. A combination of biochemistry and structures of the PAZ domain alone and in complex with RNA [17-20] illustrates its role in binding to the single stranded 3’ end of siRNAs in a sequence independent manner. The fidelity of Slicer activity depends on a 5’ monophosphate on the guide strand as determined for recombinant human Argonaute 2 [15]. Although there are no reported structures of a full-length eukaryotic Argonaute protein in complex with guide or target strand RNA the structure of a PIWI domain containing protein from Archaeoglobus fulgidus (AfPIWI) with an siRNA like duplex provides the first structural explanation for 5’ end recognition of the siRNA [13,14]. The 5’ phosphate of the guide strand is found in a conserved binding pocket contained within a cleft bridging the Mid and PIWI domains (Figure 2c). The 5’-monophosphate forms several hydrogen bonds with highly conserved residues including coordination to a metal ion bound to the C-terminal carboxylate of the protein. Unfortunately AfPIWI lacks the N-terminal and PAZ domains characteristic of eukaryotic Argonautes, as well as the metal binding residues essential for Slicer activity. Several models of full-length Argonaute proteins bound to guide and target strand RNAs have been proposed [10,12-14], although new structures of eukaryotic Argonaute proteins with RNA substrates will be essential to gain a true understanding of their mechanism. Several recent reviews elaborate in more detail on Argonaute structure and function [21-24].
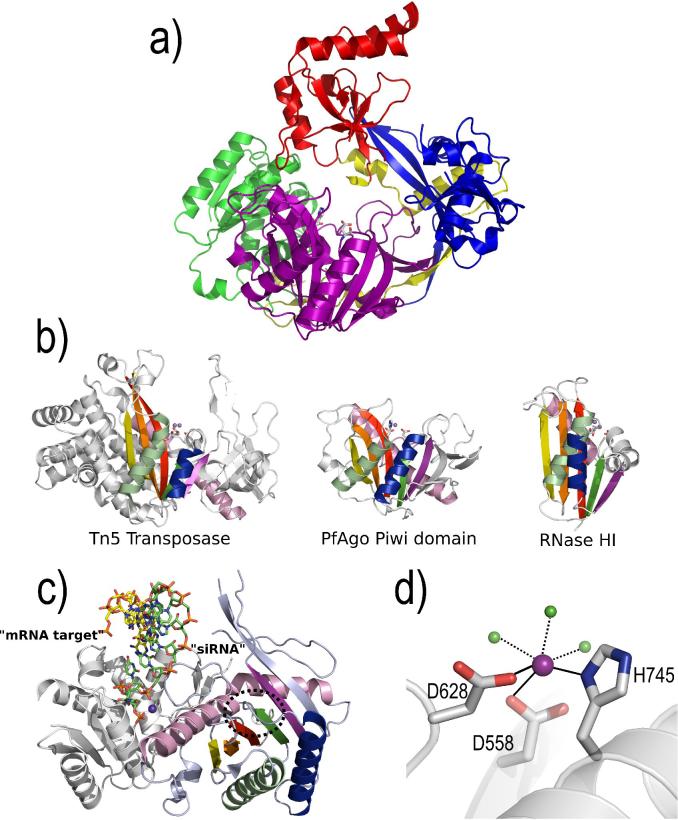
Figure 2.
Structure of Argonaute. a) The overall structure of a full-length Argonaute protein from Pyrococcus furiosus (PfAgo) [10]. PfAgo is composed of N-terminal (blue), PAZ (red), Mid (green), and PIWI domains (purple). The PIWI domain shares an RNase H-like fold. The active site residues (sticks) are located within the PIWI domain. b) A comparison of the RNase H-like fold for Tn5 Transposase [58], PfAgo PIWI domain [10], and E. coli RNase HI [59]. The equivalent secondary structural elements are presented in identical colors for each structure. The metal binding active site residues (sticks) and catalytic metal ions (spheres) are shown for each structure. Notice that the PfAgo PIWI domain shares more similar features with Tn5 transposase than to RNase HI. c) Structure of a PIWI domain protein from Archaeoglobus fulgidus complexed with siRNA duplex [13,14]. The strand representing the siRNA or guide strand (green sticks) is associated through its 5’-monophosphate to a conserved pocket between the Mid (white cartoon) and PIWI (colored according to secondary structure as in panel b) domains. Several highly conserved residues recognized the 5’-monophosphate, including a metal ion (purple sphere) distant from the active site (dashed circle). The mRNA target strand (yellow sticks) is thought to extend into the RNase H-like active site (dashed circle), but experimental data is not available to confirm this model. d) A close-up view of the PfAgo active site contained within the RNase H-like PIWI domain. The DDH motif in the PfAgo active site coordinates a metal ion [15] (purple sphere) that is further coordinated by three water molecules (green spheres).
Diversity of the Argonaute Family
At least three Argonaute protein subfamilies have been identified in eukaryotes [25] (see also [23,24]). The AGO subfamily is present in animals, plants, and fission yeast and is involved in both transcriptional and post-transcriptional silencing mechanisms (Figure 1). Piwi subfamily proteins have only been identified in animals and interact with a subset of small RNAs called Piwi-interacting RNAs (piRNAs) in the germline [26-30]. The role of Piwi proteins in silencing is now beginning to emerge with suggested functions in transcriptional silencing [31,32], translational repression [33], and silencing of transposons [34]. The Piwi protein size and domain structure is similar to the AGO subfamily with an N-terminal, PAZ, Mid and C-terminal RNase H-like PIWI domain. Most Piwi proteins have an intact DDH metal binding signature and may function in a similar capacity to Argonautes within germline cells. The WAGO subfamily (also called group III Argonautes) is exclusive to worms and is unusually high in number (∼18 out of 27 Argonautes in C. elegans) [25]. Unlike the AGO and Piwi subfamilies the majority of WAGOs lack conservation of the DDH metal binding residues, suggesting a Slicer-independent function.
Piwis, piRNAs and Silencing of Transposons
Mammalian Piwi proteins are important for male germline development particularly spermatogenesis [35-37]. In Drosophila, mutation of Piwi results in sterility and loss of germline cells in males and females [38]. In vertebrates and invertebrates, expression of Piwi-family proteins is restricted to the germline unlike the ubiquitous expression patterns observed for Argonaute. Until recently the RNAi function of Piwi proteins and the population of small RNAs that they bind to has remained obscure despite the implication that Piwi proteins are functionally similar to Argonautes.
Following immunoprecipitation of Piwi proteins from mammalian testes an abundant population of Piwi-interacting RNAs (piRNAs) were discovered [26-30]. Testes-specific mammalian piRNAs are distinctively longer than previously characterized Argonaute associated si- and miRNAs (26−31 nt vs. 21−24 nt). A unique subset of piRNAs in mice distribute between two Piwi proteins, MIWI [26,30] and MILI [27]. MIWI bound piRNAs are slightly longer (29−31 nt) than the MILI bound piRNAs (26−28 nt) but the biological significance of this difference remains unknown. RIWI, the rat homolog of MIWI also binds to 29−30 nt piRNAs in the testes [28]. Rat and mouse piRNA sequences are extremely diverse and map to their respective genomes in discrete genomic loci. Many piRNAs within a given loci are identified only once and have an extreme strand bias matching either the sense or antisense strand in a non-overlapping manner. In several instances adjacent piRNA loci abruptly switch to the opposing strand [26,28]. Sequenced piRNAs have a strong bias for a 5’ uridine similar to repeat-associated siRNAs (rasiRNAs) and miRNAs [39], although the significance of this bias is unknown.
In Drosophila the previously identified rasiRNAs [39] have been shown to bind the Piwi family members Piwi and Aubergine (Aub) [40,41] and thus represent a subset of Drosophila piRNAs. Piwi-associated rasiRNAs (now referred to as piRNAs) are longer (24−29 nt) than Argonaute bound siRNAs/miRNAs and are derived predominately from repetitive genomic loci like transposons or Satellite repeats [39]. The majority of piRNAs matching transposons, was shown to be antisense to the active transposon element [41]. In flies, loss of Dicer in the miRNA pathway and siRNA pathway has no effect on piRNA levels and/or transposon repression suggesting that Piwi associated small RNAs in Drosophila have an alternative biogenesis pathway [41]. The mystery of piRNA biogenesis is becoming clearer with recent sequence data reported for piRNAs interacting with all three Piwi family proteins (Piwi, Aubergine, Ago3) in Drosophila [42,43]. Sequences of piRNAs bound to Piwi and Aubergine are predominately antisense to regions of repeats and transposons in the fly genome similar to previous observations [34]. Analogous to mammalian piRNAs the Piwi/Aubergine piRNAs also have a strong preference for a 5’ uridine. The surprise came when piRNAs bound to the third Drosophila Piwi protein (Ago3) were examined. Ago3 piRNAs strongly map to the sense strand orientation and do not exhibit a 5’ uridine bias. Intriguingly the Ago3 associated 5’ end of the sense piRNAs frequently share 10 base pair complementarity with the 5’ end of Aubergine and to a lesser extent Piwi bound antisense piRNAs. Consequently the Ago3 bound piRNAs complementary relationship ensures a strong bias for an adenine at position 10 within the majority of Ago3 piRNA sequences. This observation led Brennecke et al. [42] and Gunawardane et al. [43] to propose a Slicer mediated mechanism for piRNA biogenesis (Figure 3). The proposed mechanism requires a pool of primary antisense piRNAs derived potentially from piRNA clusters by an unknown initiation step. Primary antisense piRNAs are loaded into Piwi or Aubergine, which guide targeting of active transposon transcripts. Slicer mediated cleavage between nucleotides 10 and 11 of a target RNA is proposed to yield the 5’ end of sense piRNAs. An unknown endonuclease must then process the 3’ end for loading into Ago3. A sense piRNA/Ago3 complex is poised for targeting complementary piRNA cluster transcripts, where Slicer cleavage by Ago3 builds the 5’ end of antisense piRNA substrate for Piwi/Aubergine complexes. Again 3’ end processing by an endonuclease must assist Ago3, leading to completion of the catalytic cycle. Piwi, Aubergine, and Ago3 each have in vitro Slicer activity [43] and are thus capable of catalyzing the reactions in the proposed model. It remains unclear how primary Piwi/Aubergine antisense piRNAs are produced to initiate the amplification loop. Brennecke et al. suggest that processing of a single stranded piRNA cluster transcript is likely, although maternal supply of piRNA complexes could also serve to initiate the cycle [42].
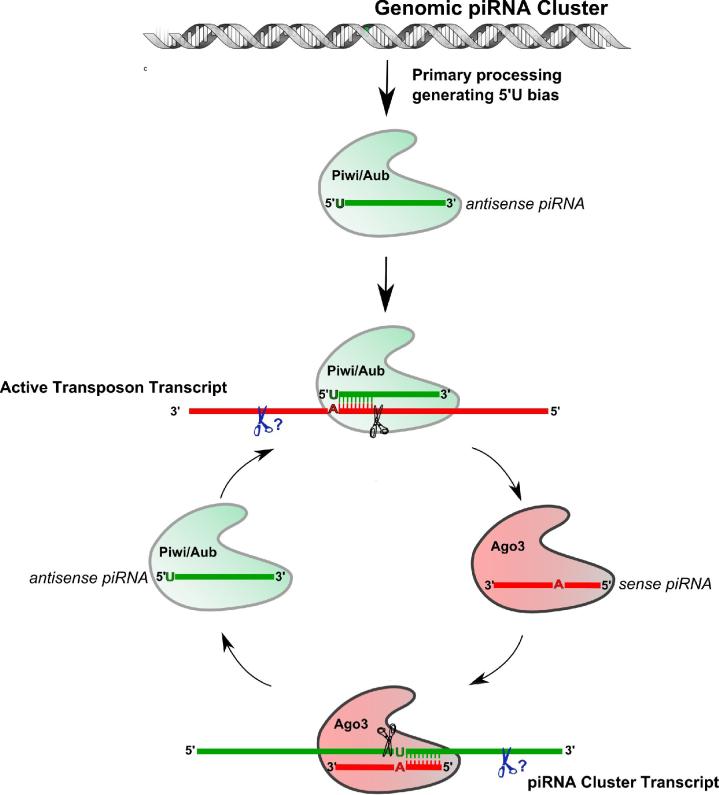
Figure 3.
Drosophila piRNA Biogenesis Mechanism. Antisense Piwi-interacting RNAs (piRNAs) in flies associate with Piwi and Aubergine (Aub) proteins. A 10 bp complementary relationship exists for antisense Piwi/Aub piRNAs with sense oriented Ago3 piRNAs, therefore Slicer cleavage of active transposon transcripts by Piwi/Aub is proposed to generate the 5’ end of sense piRNAs bound to Ago3. This process is coupled with an unknown endonuclease to generate the 3’ end of mature sense piRNA. Sense piRNAs (10 bp complementary to antisense piRNAs) bound to Ago3 target piRNA cluster transcripts that would initiate the biogenesis of antisense Piwi/Aub piRNAs.
The 3’ processing activity required for the piRNA ping-pong model was recently attributed to two putative nucleases encoded by the zucchini (zuc) and squash (squ) genes in flies [44]. Defects in piRNA biogenesis and the upregulation of transposons are evident in zuc and squ mutant flies. Additionally Zucchini and Squash proteins colocalize and interact directly with Aubergine [44]. These results strongly implicate these two putative nucleases as the 3’ processing activity in piRNA biogenesis. The current piRNA biogenesis model is based primarily on piRNA sequence data. Further biochemical support for the 5’ and 3’ processing activities responsible for piRNA biogenesis is essential.
Investigation of the relationship of piRNAs bound to the three mouse Piwi proteins (MIWI, MILI, MIWI2) should reveal insight into mammalian piRNA biogenesis. It should be noted that piRNAs are depleted in MIWI [30] and MILI [45] mutant mice providing further support for the direct involvement of Piwi proteins in mammalian piRNA biogenesis. As in Drosophila, evidence for a mammalian amplification loop for 5’ end generation of piRNAs is evident for MILI bound piRNAs [45] where a similar complimentary relationship between sense and antisense piRNAs are observed.
Transposon control in the germline by Piwi/piRNA complexes may be an evolutionarily conserved function. Characterization of MIWI2 indicates a role in transposon control in the mouse germline. Miwi2 deficient mice have increased transposon activity correlated with decreased DNA methylation [46]. Mili mutants also lose DNA methylation in transposons resulting in increased transposon activation [45].
In zebrafish, transposon derived piRNAs associate with the zebrafish Piwi protein Ziwi [47]. As in flies [41], zebrafish piRNAs are not depleted in Dicer deficient germ cells providing evidence that vertebrate piRNAs are also made by a distinct pathway from siRNA biogenesis. A strong 5’ uridine bias for antisense piRNAs is consistent with the sequenced piRNAs from mammals and flies. For sense oriented piRNAs derived from retroelements there is a tendency for loss of the 5’ uridine bias; however an increased incidence of adenine at position 10 mirrors the trend observed in Drosophila piRNAs, signifying a conserved biogenesis mechanism may exist for piRNA production in vertebrates. In contrast to genomic piRNA clusters in mammals, zebrafish piRNAs switch strand bias multiple times within a given cluster. This suggests that while a Piwi dependent mechanism of piRNA biogenesis may exist there are key differences between fish and mammals.
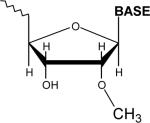
In addition to their longer length and unique biogenesis compared to siRNAs, piRNAs carry a 3’ end chemical modification, a feature initially observed in Drosophila [41]. Upon examination of the chemical nature of zebrafish piRNAs a similar modification was suggested. Using HPLC and mass spectrometry Houwing et al. determined that the more abundant rat piRNAs have a 2’-O-methyl modification on the 3’ end [47] (Table 1). Two additional studies on mouse piRNAs using alternative methods report an identical modification [48,49]. This modification is also seen in plant miRNAs [50]. It is mediated by Hen1 methyltransferase and lends stability to the small RNAs. Kirino and Mourelatos more recently describe data showing the mouse homolog of Hen1 contains piRNA methyltransferase activity [51]. Data from Saito et al. and Horwich et al. now show that the Drosophila homolog of Hen1 is the piRNA methyl transferase (Pimet) in flies [52,53]. It remains to be determined at which point in piRNA biogenesis methylation occurs. It is also not clear what role methylation has on piRNA function. One possibility is that methylation protects piRNAs from 3’−5’ exonucleases in the germline. Alternatively, methylated piRNAs carry a chemical signature that may be specifically recognized by Piwi proteins or other factors.
Silencing Amplification by Secondary Argonautes
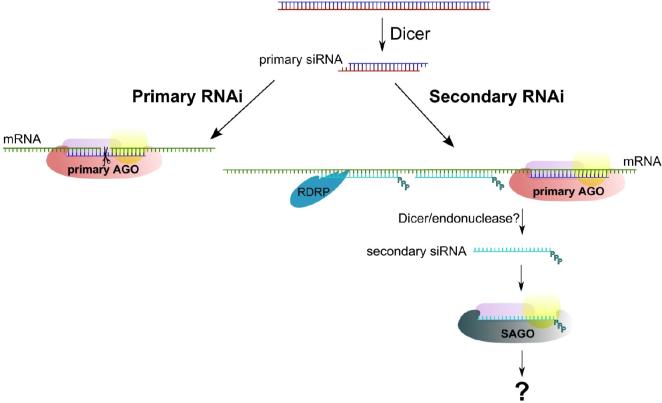
C. elegans has the largest number of Argonaute family members with 27 annotated Argonaute proteins. Both the Argonaute and Piwi protein subfamilies are represented in worms, but the majority (∼18) belong to a worm specific clade (WAGOs) [24]. Most WAGOs have an incomplete or missing DDH metal binding motif which is critical for enzymatic slicing by Argonautes that have been tested. Recently Yigit et al. identified a mutant worm (called MAGO, multiple-Ago mutant) deficient in RNAi resulting from knockdown of several previously uncharacterized WAGO subfamily proteins [25]. The RNAi deficiency in this mutant could not be rescued by RDE-1, the Argonaute protein involved in classical RNAi in worms. Furthermore, a RDE-1 mutant deficient in RNAi could not be rescued by WAGOs, suggesting that RNAi in worms occurs through at least two different pathways. Based on this hypothesis Yigit et al. propose that C. elegans Argonaute proteins perform RNAi in a sequential manner, beginning with a primary Argonaute (such as RDE-1) guided by primary siRNAs. Primary siRNAs are generated by Dicer processing of long dsRNA to initiate the RNAi trigger event. Amplification of the silencing signal in C. elegans is accomplished by the production of secondary siRNAs upstream of the primary siRNA guided trigger site which is dependent on RNA dependent RNA polymerase activity (RdRP) [54,55]. Secondary siRNAs are produced by RdRP using cleaved mRNA upstream of the initial trigger as a template [56,57]. Processing of the resulting dsRNA product by Dicer yields secondary siRNAs. Yigit et al. report that secondary siRNAs produced in this manner are discriminately bound to secondary Argonaute proteins (SAGOs) [25] (Figure 4). SAGOs are a subset of the WAGO subfamily and lack residues in the metal binding motif characteristic of Slicer active Argonautes. Over expression of SAGOs enhances RNAi and so likely represent the rate limiting step in the RNAi pathway in worms [25]. How do secondary Argonaute proteins specifically recognize and recruit secondary siRNAs? Sijen et al. and Pak et al. each developed alternative strategies to sequence secondary siRNAs [56,57]. Previous sequencing data failed to adequately represent secondary siRNAs due to absence of a 5’ monophosphate that is required in the sequencing protocol. Secondary siRNAs instead carry a 5’ triphosphate modification (Table 1) which is not the typical product of Dicer. Instead secondary siRNAs in C. elegans could be made by self-termination by RDRP, or an unknown endonuclease acts on double stranded substrates made by RdRP to produce mature secondary siRNA [56,57]. It is tempting to speculate that the 5’-triphosphate modification serves as a recognition element for the SAGOs to specifically bind and mediate silencing while excluding binding by primary Argonautes such as RDE-1. While all Argonautes examined have highly conserved amino acids that bind to the 5’ monophosphate of siRNAs, the WAGOs in C. elegans are slightly divergent in this region [23]. It is plausible that it is this divergence that lends specificity to SAGOs for binding to concentrated pools of secondary siRNAs.
Figure 4.
Amplified Silencing by Secondary Argonautes in C. elegans. Primary RNAi in C. elegans (“classical” RNAi) involves primary siRNAs bound to primary Argonaute containing complexes to mediate mRNA transcript cleavage. Secondary RNAi (silencing amplification) is first triggered by a primary Argonaute complex. In order for amplification to occur, secondary siRNAs are synthesized by RdRP upstream from the initial trigger site. Secondary siRNAs contain a 5’ triphosphate and are exclusively loaded into secondary Argonautes (SAGOs) which mediate downstream silencing. SAGOs lack critical active site amino acids and may function as non-Slicers.
New Classes of Small RNAs Distribute into Distinct Argonaute Proteins
The scope and diversity of RNAi-related processes continues to grow. The work described in this review shows that chemically modified piRNAs and secondary siRNAs in worms represent distinct classes of small RNAs which operate in discrete pathways. Not only are we getting a better mechanistic understanding of these pathways, but we are discovering that the Argonautes, central to all RNAi pathways are stretching their tentacles into diverse forms of gene silencing.
Acknowledgements
Thanks to Julius Brennecke and Niraj Tolia for helpful comments on the manuscript. This work was supported by a grant from the US National Institute of Health to L.J. (R01-GM072659)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
- 1.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat Rev Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 2.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 3.Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. Rna. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 7.Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs Silence Gene Expression by Repressing Protein Expression and/or by Promoting mRNA Decay. Cold Spring Harb Symp Quant Biol. 2006;71:523–530. doi: 10.1101/sqb.2006.71.013. [DOI] [PubMed] [Google Scholar]
- 9.Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 12.Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel DJ. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a Piwi domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat Struct Mol Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- 16.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. Embo J. 2006;25:1924–1933. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- 18.Ma JB, Ye KQ, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- 20.Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3 '-end recognition by the Argonaute2 PAZ domain. Nature Structural & Molecular Biology. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- 21.Song JJ, Joshua-Tor L. Argonaute and RNA - getting into the groove. Current Opinion in Structural Biology. 2006;16:5–11. doi: 10.1016/j.sbi.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 22.Hall TM. Structure and function of argonaute proteins. Structure. 2005;13:1403–1408. doi: 10.1016/j.str.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Parker JS, Barford D. Argonaute: A scaffold for the function of short regulatory RNAs. Trends Biochem Sci. 2006;31:622–630. doi: 10.1016/j.tibs.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- ••25.Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [The first study showing that RNAi in C. elegans utilizes at least two distinct pathways that operates in a sequential manner. Primary Argonaute proteins use primary siRNAs to initiate the RNAi response. Secondary siRNAs are synthesized by an RNA-dependent RNA polymerase and are loaded into secondary Argonaute proteins for downstream silencing. Secondary Argonautes lack conserved active site amino acid residues and may function in a Slicer independent manner.] [DOI] [PubMed] [Google Scholar]
- •26.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [Several groups (including references 26−30) report the discovery of Piwi-interacting RNAs (piRNAs) in the mammalian germline. piRNAs specifically interact with the Piwi protein subfamily and not members of the Argonaute subfamily.] [DOI] [PubMed] [Google Scholar]
- •27.Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [For annotation see reference 26.] [DOI] [PubMed] [Google Scholar]
- •28.Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363–367. doi: 10.1126/science.1130164. [For annotation see reference 26.] [DOI] [PubMed] [Google Scholar]
- •29.Watanabe T, Takeda A, Tsukiyama T, Mise K, Okuno T, Sasaki H, Minami N, Imai H. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20:1732–1743. doi: 10.1101/gad.1425706. [For annotation see reference 26.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- •30.Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20:1709–1714. doi: 10.1101/gad.1434406. [For annotation see reference 26.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- 32.Pal-Bhadra M, Leibovitch BA, Gandhi SG, Rao M, Bhadra U, Birchler JA, Elgin SC. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science. 2004;303:669–672. doi: 10.1126/science.1092653. [DOI] [PubMed] [Google Scholar]
- 33.Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and Piwi-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol. 2001;11:1017–1027. doi: 10.1016/s0960-9822(01)00299-8. [DOI] [PubMed] [Google Scholar]
- 35.Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mechanisms of Development. 2001;108:121–133. doi: 10.1016/s0925-4773(01)00499-3. [DOI] [PubMed] [Google Scholar]
- 36.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 37.Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 38.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- •40.Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes & Development. 2006;20:2214–2222. doi: 10.1101/gad.1454806. [This reference (and 41) report that repeat associated siRNAs interact with Piwi protein subfamily members in Drosophila.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- •41.Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–324. doi: 10.1126/science.1129333. [For annotation see reference 40.] [DOI] [PubMed] [Google Scholar]
- ••42.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. doi: 10.1016/j.cell.2007.01.043. [The first description of piRNA biogenesis (along with reference 43) involves a Piwi protein Slicer dependent mechanism in Drosophila. Piwi/piRNA complexes are shown to be involved in the suppression of retrotransposons.] [DOI] [PubMed] [Google Scholar]
- ••43.Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. A slicer-mediated mechanism for repeat-associated siRNA 5' end formation in Drosophila. Science. 2007;315:1587–1590. doi: 10.1126/science.1140494. [For annotation see reference 42.] [DOI] [PubMed] [Google Scholar]
- •44.Pane A, Wehr K, Schupbach T. zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell. 2007;12:851–862. doi: 10.1016/j.devcel.2007.03.022. [This study shows that piRNA abundance is dependent on two putative nucleases, Zucchini and Squash. It is proposed that these two nucleases are involved in 3'-end formation of mature piRNAs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- •45.Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [References 45 and 46 are the first to show that Piwi proteins control transposons in mammals. Evidence is also presented that suggest a similar piRNA biogenesis mechanism as observed in flies.] [DOI] [PubMed] [Google Scholar]
- •46.Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [For annotation see reference 45.] [DOI] [PubMed] [Google Scholar]
- •47.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. A Role for Piwi and piRNAs in Germ Cell Maintenance and Transposon Silencing in Zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [Piwi proteins in zebrafish are involved in transposons silencing. The authors (along with references 48 and 49) also report that mammalian piRNAs are 2'-O-methylated.] [DOI] [PubMed] [Google Scholar]
- •48.Kirino Y, Mourelatos Z. Mouse Piwi-interacting RNAs are 2'-O-methylated at their 3' termini. Nat Struct Mol Biol. 2007;14:347–348. doi: 10.1038/nsmb1218. [For annotation see reference 47.] [DOI] [PubMed] [Google Scholar]
- •49.Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T. The 3' termini of mouse Piwi-interacting RNAs are 2'-O-methylated. Nat Struct Mol Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [For annotation see reference 47.] [DOI] [PubMed] [Google Scholar]
- 50.Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •51.Kirino Y, Mourelatos Z. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. Rna. 2007 doi: 10.1261/rna.659307. [Mouse piRNAs are methylated by a homolog of HEN1 methyltransferase.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- •52.Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA Methyltransferase, DmHen1, Modifies Germline piRNAs and Single-Stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [References 52 and 53 show that Drosophila piRNAs are 2'-O-methylated by the Drosophila HEN1 homolog.] [DOI] [PubMed] [Google Scholar]
- •53.Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2'-O-methylation of Piwi- interacting RNAs at their 3' ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [For annotation see reference 52.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001;107:465–476. doi: 10.1016/s0092-8674(01)00576-1. [DOI] [PubMed] [Google Scholar]
- 55.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- ••56.Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [Two references (56 and 57) describe the sequencing of secondary siRNAs in C. elegans. Secondary siRNAs are likely processed independently of Dicer and carry a 5'-triphosphate.] [DOI] [PubMed] [Google Scholar]
- ••57.Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [For annotation see reference 56.] [DOI] [PubMed] [Google Scholar]
- 58.Steiniger-White M, Bhasin A, Lovell S, Rayment I, Reznikoff WS. Evidence for ”unseen” transposase--DNA contacts. J Mol Biol. 2002;322:971–982. doi: 10.1016/s0022-2836(02)00877-x. [DOI] [PubMed] [Google Scholar]
- 59.Katayanagi K, Okumura M, Morikawa K. Crystal structure of Escherichia coli RNase HI in complex with Mg2+ at 2.8 A resolution: proof for a single Mg(2+)-binding site. Proteins. 1993;17:337–346. doi: 10.1002/prot.340170402. [DOI] [PubMed] [Google Scholar]