Abstract
Heparanase is an endoglycosidase that specifically cleaves heparan sulfate (HS) side chains of heparan sulfate proteoglycans, the major proteoglycan in the extracellular matrix (ECM) and cell surfaces. Heparanase upregulation was documented in an increasing number of primary human tumors, correlating with reduced postoperative survival rate and enhanced tumor angiogenesis. The purpose of the current study was to determine heparanase levels in blood samples collected from pediatric cancer patients using an ELISA method. Heparanase levels were elevated four-fold in the plasma of cancer patients compared with healthy controls (664 ± 143 vs 163 ± 18 pg/ml, respectively). Evaluating plasma samples following anticancer therapy revealed reduced heparanase levels (664 ± 143 vs 429 ± 82 pg/ml), differences that are statistically highly significant (P = .0048). Of the 55 patients with complete remission (CR) or very good partial remission (VGPR) at restaging, 41 (74.5%) had lower heparanase amounts, whereas 14 patients (25.5%) had similar or higher amounts of plasma heparanase. All nine patients with stable or advancing disease had similar or elevated levels of heparanase on restaging. The results show that heparanase levels are elevated in the plasma of pediatric cancer patients and closely correlate with treatment responsiveness, indicating that heparanase levels can be used to diagnose and monitor patient's response to anticancer treatment.
Keywords: Heparanase, ELISA, marker, anticancer treatment
Introduction
Heparanase is an endo-β-glucuronidase that cleaves heparan sulfate (HS) side chains at a limited number of sites, yielding HS fragments of still appreciable size (∼5–7 kDa) [1–3]. Cleavage of HS, an important constituent of the extracellular matrix (ECM) and basement membranes, is considered critical for cell invasion associated with inflammation, angiogenesis, and tumor metastasis. This notion gained further support by employing antisense, small interfering RNA, and ribozyme technologies, clearly depicting heparanase-mediated HS cleavage and ECM remodeling as critical requisites for inflammation, angiogenesis, and metastatic spread [4–6]. Furthermore, heparanase upregulation was documented in an increasing number of primary human tumors, correlating with enhanced local and distant metastasis, increased microvessel density, and reduced postoperative survival of cancer patients [7–9]. Collectively, these studies provide compelling evidence for the clinical relevance of the enzyme, making it an attractive target for the development of anticancer drugs [10,11]. Heparanase induction in human malignancies, as well as in several other pathologies such as cirrhosis, nephrosis, and diabetes [12–18], implies that the enzyme may serve as a valuable marker for diagnosing the development of the diseases and, possibly, to follow treatment efficiency. Recently, we reported the development of an ELISA method capable of detection and quantification of heparanase in urine samples and demonstrated an elevation of heparanase levels in the urine of cancer and diabetes patients [19]. Here, we examined the ability of the ELISA method to detect and quantify heparanase levels in blood samples. We provide evidence that heparanase can be quantified in plasma but not in serum samples. Heparanase levels were significantly elevated in the plasma of pediatric cancer patients and, moreover, correlated with therapy effectiveness. These results indicate that heparanase levels can be used to diagnose and monitor patient's response to anticancer treatments.
Materials and Methods
Patients
The study group consisted of 64 pediatric patients [19 patients with Hodgkin's disease (HD), 11 with acute lymphoblastic leukemia, 7 with osteosarcoma, 7 with non-Hodgkin lymphoma, 4 each with brain glioma (2 with optic glioma and 2 with brain glioma), neuroblastoma, rhabdomyosarcoma, and Ewing's sarcoma (ES), and 1 each with Langerhan's cell histocytosis, germ cell tumor, Wilm's tumor, nasopharynx carcinoma, and glioblastoma multiform. Mean age was 10.3 years (0.5–21), with 28 girls and 36 boys. This study was carried out in accordance with Helsinki principles, with the approval of the local Ethics Committee, and informed parental consent. Children were diagnosed and treated in the Department of Pediatric Hematology-Oncology at the Meyer Children's Hospital, Rambam Health Care Campus, Haifa, Israel.
Sample Collection
Blood samples were collected at diagnosis and at restaging (i.e., after the first month of induction for leukemia patients; two to three courses of chemotherapy for the other malignancies) [20,21]. A total of 3 ml of peripheral blood was collected, applying EDTA as an anticoagulant; samples were cooled and plasma was obtained by centrifugation (1500g for 15 minutes at 4°C). All samples were frozen and thawed once.
Antibodies and Reagents
Antiheparanase 1E1 monoclonal antibody and polyclonal antibodies 1453 and 733 have been previously described [19,22]. HRP-conjugated goat anti-rabbit antibody was purchased from Jackson ImmunoResearch (West Grove, PA). Microtiter 96-well plates (Maxisorp) were from Nunc (Roskilde, Denmark). HRP colorimetric substrate 3,30,5,50-tetramethylbenzidine was purchased from Dako (Glostrup, Denmark). Bovine serum albumin (BSA) was from MP Biomedicals (Illkirch, France). Single-chain active heparanase (GS3) gene construct was kindly provided by Dr. Christian Steinkuhler (Instituto di Ricerche di Biologia Moleculare/Merck Research Laboratories, Pomezia, Italy) [23], and the protein was purified from the conditioned medium of baculovirus-infected insect cells, as described [23].
ELISA Method
The ELISA method was carried out as described [19]. Briefly, wells of microtiter plates were coated (for 18 hours at 4°C) with 1 µg/ml 1E1 monoclonal antiheparanase antibody in 50 µl of coating buffer (0.05 M Na2CO3 and 0.05 M NaHCO3, pH 9.6) and were then blocked with 1% BSA in PBS for 1 hour at 37°C. Samples were diluted with 0.5% BSA (1:1) and a total of 100 ml was loaded in duplicates and incubated for 2 hours at room temperature, followed by the addition of 100 µl antibody 1453 (1 µg/ml) for an additional 2 hours at room temperature. HRP-conjugated goat anti- rabbit IgG (1:20,000) in blocking buffer was added (for 1 hour at room temperature) and the reaction was visualized by the addition of 100 µl of the chromogenic substrate (3,30,5,50- tetramethylbenzidine) for 30 minutes. The reaction was stopped with 100 µl H2SO4 and absorbance at 450 nm was measured with a reduction at 630 nm using ELISA plate reader. Plates were washed five times with washing buffer (PBS, pH 7.4, containing 0.1% (v/v) Tween 20) after each step. As a reference for quantification, a standard curve was established by a serial dilution of recombinant single-chain (GS3) active heparanase, ranging from 180 pg/ml to 5 ng/ml.
Immunohistochemistry
Staining of formalin-fixed, paraffin-embedded, 5-µm tissue sections for heparanase was performed essentially as described [22,24,25]. Briefly, slides were deparaffinized and rehydrated, and endogenous peroxidase activity was quenched (for 30 minutes) by 3% hydrogen peroxide in methanol. Slides were then subjected to antigen retrieval by boiling (for 20 minutes) in 10 mM citrate buffer, pH 6. Slides were incubated with 10% normal goat serum in PBS for 60 minutes to block nonspecific binding and were incubated (for 20 hours at 4°C) with antiheparanase 733 antibody diluted 1:100 in blocking solution. Slides were extensively washed with PBS containing 0.01% Triton X-100 and incubated with a secondary reagent (Envision Kit; Dako) according to the manufacturer's instructions. Following additional washes, color was developed with 3-amino-9- ethylcarbazole reagent (Dako), and sections were counterstained with hematoxylin and mounted, as described [22,24,25].
Statistics
Data were analyzed using the Prism software (GraphPad, San Diego, CA). One-tailed paired t-test and the nonparametric Mann-Whitney test were employed. P < .05 was considered statistically significant.
Results
Detection and Quantification of Heparanase in Blood Samples
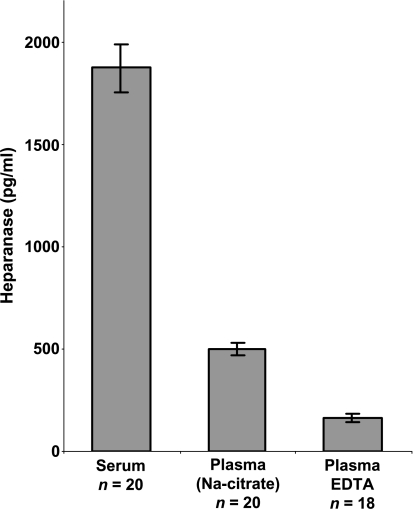
We have recently developed an ELISA method capable of detecting and quantifying heparanase in urine samples [19]. To expand the applications of our ELISA method, we evaluated its ability to quantify heparanase levels in blood samples. Heparanase levels in serum of control donors were found to be significantly higher than in plasma collected with sodium citrate (1871 ± 116 vs 500 ± 29 pg/ml, respectively), likely due to platelet activation and release of heparanaserich platelet dense granules [26,27] (Figure 1). Detection of heparanase in plasma was also noted to be affected by the anticoagulant employed, with lower levels quantified in plasma collected with EDTA compared with sodium citrate (163 ± 18 vs 500 ± 29 pg/ml, respectively; Figure 1). Because plasma collected with EDTA gave the lowest levels of heparanase in control healthy donors, this procedure was adopted for the rest of the study.
Figure 1.
Determination of heparanase levels in blood. Blood samples were obtained from control healthy volunteers and allowed to clot to generate serum (n = 20) or collected into tubes containing sodium citrate (n = 20) or EDTA (n = 18) as anticoagulants to produce plasma. Samples were diluted 1:2 with PBS and heparanase levels were quantified by a sandwich ELISA method, as described in the Materials and Methods section.
Detection and Quantification of Heparanase in Plasma of Pediatric Cancer Patients
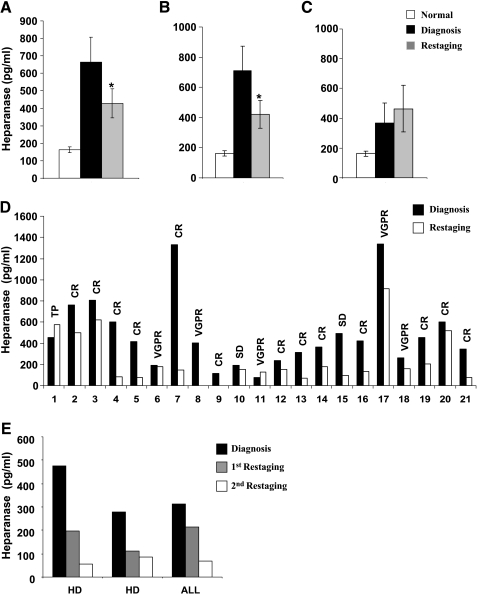
Encouraged by the ability of the ELISA method to quantify heparanase in plasma, we next evaluated the level of heparanase in samples collected from pediatric cancer patients. Heparanase levels were elevated four-fold in the plasma of cancer patients compared with healthy controls (664 ± 143 vs 163 ± 18 pg/ml; Figure 2A). Evaluating plasma samples following anticancer therapy revealed reduced heparanase levels (664 ± 143 vs 429 ± 82 pg/ml; Figure 2A), differences that are statistically highly significant (P = .0048). Plasma levels of heparanase as indicators of treatment effectiveness are further revealed by subgrouping the patients according to who responded to the treatment and exhibited complete remission (CR) or very good partial remission (VGPR) and who did not respond well and showed tumor progression (TP). Patients in CR or with VGPR had significantly lower levels of heparanase at restaging (710 ± 162 vs 421 ± 91 pg/ml, respectively; Figure 2B). Moreover, whereas the group that experienced disease remission exhibited reduced levels of heparanase (Figure 2B), patients with disease progression were diagnosed to bear a slight elevation of heparanase levels (Figure 2C). Even more impressive results were observed when heparanase levels were compared for each patient individually at diagnosis and on restaging (Figure 2D). Of the 64 patients included, 45 (70.3%) had lower amounts (at least 30% decrease) of heparanase levels at restaging. Of these, 43 (95.6%) underwent CR. Of the 64 patients, 19 (29.7%) had higher or equal amounts of heparanase, and of these 7 (37%) exhibited stable disease (SD) or TP, whereas the other 12 patients exhibited disease remission (Table 1). Of the 55 patients with CR or VGPR at restaging, 41 (74.5%) had lower heparanase amounts, whereas 14 patients (25.5%) had similar or higher amounts of plasma heparanase (Table 1). All 9 patients with stable or advancing disease had similar or elevated levels of heparanase at restaging (Table 1). Interestingly, in a few cases where an extended follow-up was available, heparanase levels were noted to be further decreased, reaching its level in control plasma (Figure 2E). These results confirm the correlation between treatment response and heparanase levels in the plasma of pediatric cancer patients.
Figure 2.
Elevation of heparanase levels in the plasma of pediatric cancer patients. (A) Plasma samples were collected from 64 pediatric patients with various malignancies at (■) diagnosis and ( ) restaging, following the recommended antitumor treatment, and heparanase levels were quantified by the ELISA method, as described above. Average heparanase levels are also shown for the subgroups of patients that were diagnosed (B) to undergo CR or (C) to exhibit TP. (D) Representative heparanase levels at (■) diagnosis and (□) restaging are shown individually for 21 patients. (E) For two HD and one acute lymphoblastic leukemia patients, plasma samples were available at (■) diagnosis, (
) restaging, following the recommended antitumor treatment, and heparanase levels were quantified by the ELISA method, as described above. Average heparanase levels are also shown for the subgroups of patients that were diagnosed (B) to undergo CR or (C) to exhibit TP. (D) Representative heparanase levels at (■) diagnosis and (□) restaging are shown individually for 21 patients. (E) For two HD and one acute lymphoblastic leukemia patients, plasma samples were available at (■) diagnosis, ( ) restaging, and (□) follow-up. CR, complete remission; TP, tumor progression; VGPR, very good partial remission; SD, stable disease.
) restaging, and (□) follow-up. CR, complete remission; TP, tumor progression; VGPR, very good partial remission; SD, stable disease.
Table 1.
Heparanase Levels in Pediatric Cancer Patients.
| All Patients (N = 64) | Heparanase | ||
| At Diagnosis (pg/ml) | At Restaging (pg/ml) | Normal (pg/ml) | |
| Overall average | 664 ± 143 | 429 ± 82 | 163 ± 18 |
| P (diagnosis vs restaging) | .0048 | < .0001 (diagnosis) | |
| .0638 (restaging) | |||
| Median | 331 | 195 | 155 |
| Average patient at CR (n = 55) | 710 ± 162 | 421 ± 91 | 0.0858 (restaging) |
| P | .0033 | ||
| Median patient at CR | 357 | 189 | |
| Correlation | 43 of 55 patients at CR had reduced heparanase levels (78.2%) | ||
| Average patient at SD or TP (n = 9) | 406 ± 55 | 508 ± 126 | |
| P | .422 | ||
| Median patient at SD or TP | 217 | 253 | |
| Correlation | 7 of 9 patients at SD or TP had elevated heparanase levels (77.8%) | ||
CR, complete remission; SD, stable disease; TP, tumor progression.
Heparanase Levels in HD
Because the study group available to us was heterogeneous and included a variety of hematological and solid malignancies, we decided to confirm our findings in a more homogenous group of patients. The largest group of patients tested in the current study was HD (n = 19) (Table 2). The average level of plasma heparanase in these patients was 1019 ± 334 pg/ml at diagnosis and 588 ± 173 pg/ml at restaging (Figure 3A), a decrease that is statistically significant (P = .035). Remarkably, all 13 (68.4%) patients with lower heparanase levels at restaging were at CR (Table 2). Of the six patients with higher heparanase levels, three exhibited stable disease or disease progression and showed modest elevation of heparanase levels (Figure 3C). Heparanase levels at diagnosis and restaging is shown for each individual HD patient in Figure 3D. These results further emphasize the correlation between response to treatment and heparanase levels in the plasma of pediatric cancer patients.
Table 2.
Heparanase Levels in the Plasma of Patients with Hodgkin's Disease.
| HD Patients (n = 19) | Heparanase | ||||
| At Diagnosis (pg/ml) | At Restaging (pg/ml) | Normal (pg/ml) | |||
| Overall average | 1019 ± 334 | 588 ± 173 | 163 ± 18 | ||
| P (diagnosis vs restaging) | .035 | < .0001 (diagnosis) | |||
| 0.0085 (restaging) | |||||
| Median | 433 | 259 | 155 | ||
| Average patient at CR (n = 16) | 1104 ± 390 | 586 ± 195 | 0.013 (restaging) | ||
| P (diagnosis vs restaging) | .033 | ||||
| Median patient at CR | 455 | 342 | |||
| Correlation | 13 of 16 patients at CR had reduced heparanase levels (81.3%) | ||||
CR, complete remission.
Figure 3.
Elevated levels of heparanase in HD. Plasma samples of pediatric patients with HD were obtained at (■) diagnosis and at ( ) restaging following chemotherapy. Heparanase levels were quantified for the (A) entire group (n = 19), (B) patients that were diagnosed to undergo CR (n = 16), and (C) patients that exhibit disease progression. (D) Heparanase levels at (■) diagnosis and (□) restaging are shown for each individual patient.
) restaging following chemotherapy. Heparanase levels were quantified for the (A) entire group (n = 19), (B) patients that were diagnosed to undergo CR (n = 16), and (C) patients that exhibit disease progression. (D) Heparanase levels at (■) diagnosis and (□) restaging are shown for each individual patient.
Heparanase Expression By Sarcomas
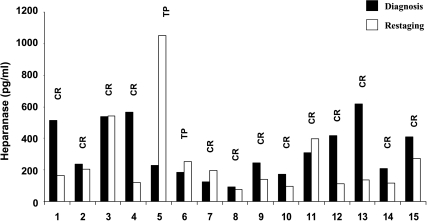
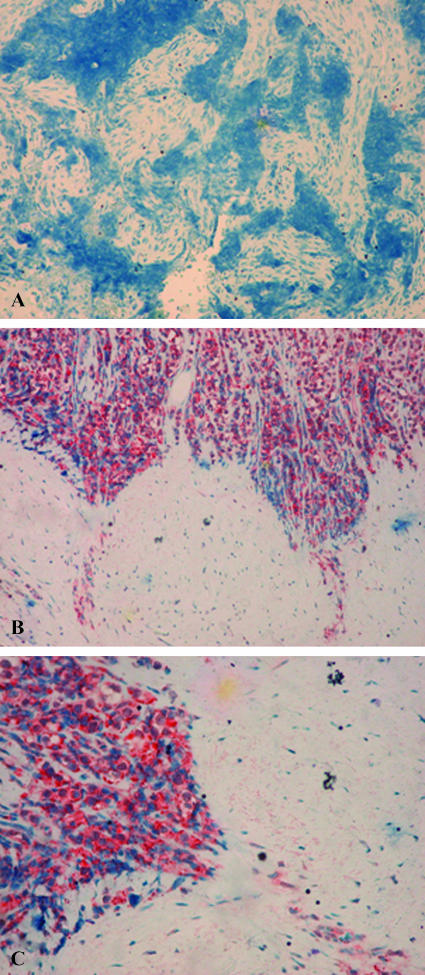
Whereas upregulation of heparanase is well documented in an increasing number of human carcinomas [8,9,11], heparanase expression by sarcomas has not been reported yet. Notably, heparanase was readily detected in the plasma of sarcoma patients included in our study group (n = 15), although heparanase elevation was modest (324 ± 44 pg/ml; Table 3) in comparison to HD and no significant average decrease was quantified following treatment (324 ± 44 vs 258 ± 65 pg/ml; Table 3). Plotting each patient individually indicated however that, of the 13 patients who experienced CR following treatment, 8 exhibited decreased levels of heparanase (Figure 4). In addition, one of two patients who underwent TP showed elevation of heparanase levels (Figure 4 and Table 3), suggesting that similar to HD, heparanase levels closely reflect treatment responsiveness also in sarcoma patients. To further confirm heparanase expression by sarcomas, paraffin-embedded sections of ES tumors were stained with antiheparanase antibody. Positive staining of heparanase was observed in five of eight ES biopsy specimens that were available to us, whereas three specimens were negatively stained for heparanase (Figure 5A). Heparanase staining was primarily evident in areas that appear to represent the invasive leading edge of the tumor (Figure 5, B and C), in agreement with a similar staining pattern observed in carcinomas [7]. Altogether our results suggest that heparanase is upregulated in hematological malignancies and solid tumors, including sarcomas, and that plasma levels of heparanase may assist in cancer diagnosis and assessment of treatment efficiency.
Table 3.
Heparanase Levels in the Plasma of Sarcoma Patients.
| Sarcoma Patients (n = 15) | Heparanase | ||
| At Diagnosis (pg/ml) | At Restaging (pg/ml) | Normal (pg/ml) | |
| Overall average | 324 ± 44 | 258 ± 65 | 163 ± 18 |
| P (diagnosis vs restaging) | .2107 | .0023 (diagnosis) | |
| .2293 (restaging) | |||
| Median | 246 | 163 | 155 |
| Average of patients at CR (n = 13) | 342 ± 49 | 198 ± 37 | 0.4285 (restaging) |
| P (diagnosis vs restaging) | .009 | ||
| Median of patients at CR | 308 | 140 | |
| Correlation | 10 of 13 patients at CR had reduced heparanase levels (77%) | ||
CR, complete remission.
Figure 4.
Elevation of heparanase levels in sarcomas. Heparanase levels in the plasma of pediatric sarcoma patients (n = 15) are shown for each individual patient at (■) diagnosis and (□) restaging. CR, complete remission; TP, tumor progression.
Figure 5.
Heparanase expression by ES. Formalin-fixed, paraffin-embedded 5-µm sections of ES tumor biopsies were subjected to immunostaining of heparanase, applying antiheparanase 733 antibody, as described in the Materials and Methods section. Shown are representative photomicrographs of (A) heparanase-negative and (B and C) heparanase-positive specimens at (A and B) low (x 20) and (C) high (x 40) magnification.
Discussion
Heparanase activity has been traditionally correlated with cell invasion associated with cancer metastasis, a consequence of structural modifications that loosen the ECM barrier [28–30]. More recently, heparanase upregulation was documented in an increasing number of human carcinomas and hematological malignancies [7,9,11,31–33]. In many cases, heparanase induction correlated with increased tumor metastasis, vascular density, and shorter postoperative survival rate, thus providing a strong clinical support for the prometastatic and proangiogenic function of the enzyme [7,9,10]. In addition to the well-studied catalytic feature of the enzyme, heparanase was noted to exert biological functions apparently independent of its enzymatic activity. This feature requires heparanase secretion and is thought to be mediated by as yet unidentified heparanase receptor. Thus, active and inactive heparanase secreted by tumor cells or the tumor microenvironment may exert local and systemic effects and is the subject of anticancer drug development programs [34,35]. We have recently reported the development of an ELISA method capable of quantifying heparanase levels in urine samples [19]. Here, we extended the utility of the ELISA method and demonstrate that it can also be successfully applied to quantify heparanase in plasma. Heparanase levels were significantly elevated in plasma obtained from pediatric cancer patients. Few cases exhibited exceptionally high levels of heparanase levels that decreased following treatment to levels still relatively high (i.e., Figure 2D, no. 17; Figure 3D, no. 2). Clinical aspects related to cases exhibiting high versus low levels of heparanase are currently under intense analysis. Importantly, heparanase levels in the plasma closely reflect the patients' status following anticancer treatment. Thus, heparanase levels were markedly decreased in most patients undergoing remission, but remained stable or slightly elevated in patients that responded poorly to treatment and exhibited TP (Table 1 and Figure 2), suggesting that plasma heparanase originates primarily from tumor cells, although minor contribution by immune cells and platelets cannot be excluded. The correlation between plasma heparanase levels and patient's outcome was evident for the entire study group (Figure 2) and for the more homogenous group of HD patients (Table 2 and Figure 3), revealing, for the first time, a possible involvement of heparanase in this malignancy. Heparanase enzymatic activity was noted in several types of hematopoietic cells, including neutrophils, monocytes, megakaryocytes, and activated lymphocytes [36–39], actively participating in their extravasation during inflammation [5], and modulating cellular adhesion [40,41]. Elevated levels of heparanase were found in leukemias, predominantly in cells of the myeloid lineage [31], possibly involving single nucleotide polymorphism [42]. Interestingly, elevation of heparanase levels correlated with enhanced tissue factor expression in blasts collected from acute leukemia patients [43], induction that appears to require heparanase secretion and to be mediated by the p38 signaling pathway [43]. This and other reports describing signal transduction initiation by extracellular heparanase [40,44–46] clearly depict the duality of secreted heparanase, functioning as an enzyme and as a signaling molecule, and the potential clinical significance of monitoring extracellular heparanase levels. Furthermore, heparanase elevation was also noted, for the first time, in the plasma of sarcoma patients (Table 3 and Figure 4), an observation that was further confirmed by immunostaining (Figure 5). In some cases, staining of heparanase was clearly evident in invasive cancer cells (Figure 5C), in agreement with a similar localization pattern noted in carcinomas [7], and the role of heparanase in cancer cell invasion and metastasis [8,9]. Thus, heparanase elevation appears to be common for hematological and solid malignancies, including sarcomas, making it a potential diagnostic and prognostic marker. The ELISA method and the ability to quantify heparanase in urine [19] and plasma are the first steps toward validating its significance as a tumor marker.
Abbreviations
- CR
complete remission
- ECM
extracellular matrix
- ES
Ewing's sarcoma
- HD
Hodgkin's disease
- HS
heparan sulfate
- TP
tumor progression
- VGPR
very good partial remission
Footnotes
This work was supported by grants from the Israel Science Foundation (grant 549/06); National Cancer Institute, National Institutes of Health (grant ROI-CA106456); the Israel Cancer Research Fund; and the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum and Israel's Ministry of Sciences and Technology.
References
- 1.Dempsey LA, Brunn GJ, Platt JL. Heparanase, a potential regulator of cell-matrix interactions. Trends Biochem Sci. 2000;25:349–351. doi: 10.1016/s0968-0004(00)01619-4. [DOI] [PubMed] [Google Scholar]
- 2.Parish CR, Freeman C, Hulett MD. Heparanase: a key enzyme involved in cell invasion. Biochim Biophys Acta. 2001;1471:M99–M108. doi: 10.1016/s0304-419x(01)00017-8. [DOI] [PubMed] [Google Scholar]
- 3.Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108:341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]
- 5.Edovitsky E, Lerner I, Zcharia E, Peretz T, Vlodavsky I, Elkin M. Role of endothelial heparanase in delayed-type hypersensitivity. Blood. 2006;107:3609–3616. doi: 10.1182/blood-2005-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy M, Reiland J, Murry BP, Chouljenko V, Kousoulas KG, Marchetti D. Antisense-mediated suppression of heparanase gene inhibits melanoma cell invasion. Neoplasia. 2005;7:253–262. doi: 10.1593/neo.04493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ilan N, Elkin M, Vlodavsky I. Regulation, function and clinical significance of heparanase in cancer metastasis and angiogenesis. Int J Biochem Cell Biol. 2006;38:2018–2039. doi: 10.1016/j.biocel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Vlodavsky I, Ilan N, Naggi A, Casu B. Heparanase: structure, biological functions, and inhibition by heparin-derived mimetics of heparan sulfate. Curr Pharm Des. 2007;13:2057–2073. doi: 10.2174/138161207781039742. [DOI] [PubMed] [Google Scholar]
- 9.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferro V, Hammond E, Fairweather JK. The development of inhibitors of heparanase, a key enzyme involved in tumour metastasis, angiogenesis and inflammation. Mini Rev Med Chem. 2004;4:693–702. doi: 10.2174/1389557043403729. [DOI] [PubMed] [Google Scholar]
- 11.McKenzie EA. Heparanase: a target for drug discovery in cancer and inflammation. Br J Pharmacol. 2007;151:1–14. doi: 10.1038/sj.bjp.0707182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldshmidt O, Yeikilis R, Mawasi N, Paizi M, Gan N, Ilan N, Pappo O, Vlodavsky I, Spira G. Heparanase expression during normal liver development and following partial hepatectomy. J Pathol. 2004;203:594–602. doi: 10.1002/path.1554. [DOI] [PubMed] [Google Scholar]
- 13.Katz A, Van-Dijk DJ, Aingorn H, Erman A, Davies M, Darmon D, Hurvitz H, Vlodavsky I. Involvement of human heparanase in the pathogenesis of diabetic nephropathy. Isr Med Assoc J. 2002;4:996–1002. [PubMed] [Google Scholar]
- 14.Levidiotis V, Freeman C, Punler M, Martinello P, Creese B, Ferro V, van der Vlag J, Berden JH, Parish CR, Power DA. A synthetic heparanase inhibitor reduces proteinuria in passive Heymann nephritis. J Am Soc Nephrol. 2004;15:2882–2892. doi: 10.1097/01.ASN.0000142426.55612.6D. [DOI] [PubMed] [Google Scholar]
- 15.Levidiotis V, Freeman C, Tikellis C, Cooper ME, Power DA. Heparanase is involved in the pathogenesis of proteinuria as a result of glomerulonephritis. J Am Soc Nephrol. 2004;15:68–78. doi: 10.1097/01.asn.0000103229.25389.40. [DOI] [PubMed] [Google Scholar]
- 16.Levidiotis V, Freeman C, Tikellis C, Cooper ME, Power DA. Heparanase inhibition reduces proteinuria in a model of accelerated anti-glomerular basement membrane antibody disease. Nephrology. 2005;10:167–173. doi: 10.1111/j.1440-1797.2005.00388.x. [DOI] [PubMed] [Google Scholar]
- 17.Levidiotis V, Kanellis J, Ierino FL, Power DA. Increased expression of heparanase in puromycin aminonucleoside nephrosis. Kidney Int. 2001;60:1287–1296. doi: 10.1046/j.1523-1755.2001.00934.x. [DOI] [PubMed] [Google Scholar]
- 18.Xiao Y, Kleeff J, Shi X, Buchler MW, Friess H. Heparanase expression in hepatocellular carcinoma and the cirrhotic liver. Hepatol Res. 2003;26:192–198. doi: 10.1016/s1386-6346(03)00107-4. [DOI] [PubMed] [Google Scholar]
- 19.Shafat I, Zcharia E, Nisman B, Nadir Y, Nakhoul F, Vlodavsky I, Ilan N. An ELISA method for the detection and quantification of human heparanase. Biochem Biophys Res Commun. 2006;341:958–963. doi: 10.1016/j.bbrc.2006.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben Arush MW, Barak AB, Maurice S, Livne E. Serum VEGF as a significant marker of treatment response in Hodgkin lymphoma. Pediatr Hematol Oncol. 2007;24:111–115. doi: 10.1080/08880010601052381. [DOI] [PubMed] [Google Scholar]
- 21.Ben Arush MW, Schenzer P, Maurice S, Elhasid R, Postovsky S, Ben Barak A, Haimi M, Zeidman I, Hayari L, Livne E. Serum vascular endothelial growth factor as a significant marker of treatment response in pediatric malignancies. Pediatr Hematol Oncol. 2005;22:513–524. doi: 10.1080/08880010591002387. [DOI] [PubMed] [Google Scholar]
- 22.Zetser A, Levy-Adam F, Kaplan V, Gingis-Velitski S, Bashenko Y, Schubert S, Flugelman MY, Vlodavsky I, Ilan N. Processing and activation of latent heparanase occurs in lysosomes. J Cell Sci. 2004;117:2249–2258. doi: 10.1242/jcs.01068. [DOI] [PubMed] [Google Scholar]
- 23.Nardella C, Lahm A, Pallaoro M, Brunetti M, Vannini A, Steinkuhler C. Mechanism of activation of human heparanase investigated by protein engineering. Biochemistry. 2004;43:1862–1873. doi: 10.1021/bi030203a. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Izhak O, Kaplan-Cohen V, Ilan N, Gan S, Vlodavsky I, Nagler R. Heparanase expression in malignant salivary gland tumors inversely correlates with long-term survival. Neoplasia. 2006;8:879–884. doi: 10.1593/neo.06382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doweck I, Kaplan-Cohen V, Naroditsky I, Sabo E, Ilan N, Vlodavsky I. Heparanase localization and expression by head and neck cancer: correlation with tumor progression and patient survival. Neoplasia. 2006;8:1055–1061. doi: 10.1593/neo.06577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eldor A, Bar-Ner M, Yahalom J, Fuks Z, Vlodavsky I. Role of heparanase in platelet and tumor cell interactions with the subendothelial extracellular matrix. Semin Thromb Hemost. 1987;13:475–488. doi: 10.1055/s-2007-1003524. [DOI] [PubMed] [Google Scholar]
- 27.Ishai-Michaeli R, Eldor A, Vlodavsky I. Heparanase activity expressed by platelets, neutrophils, and lymphoma cells releases active fibroblast growth factor from extracellular matrix. Cell Regul. 1990;1:833–842. doi: 10.1091/mbc.1.11.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bar-Ner M, Kramer MD, Schirrmacher V, Ishai-Michaeli R, Fuks Z, Vlodavsky I. Sequential degradation of heparan sulfate in the subendothelial extracellular matrix by highly metastatic lymphoma cells. Int J Cancer. 1985;35:483–491. doi: 10.1002/ijc.2910350411. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima M, Irimura T, DiFerrante D, DiFerrante N, Nicolson GL. Heparan sulfate degradation: relation to tumor invasion and metastatic properties of mouse B 16 melanoma sublines. Science. 1983;220:611–613. doi: 10.1126/science.6220468. [DOI] [PubMed] [Google Scholar]
- 30.Vlodavsky I, Fuks Z, Bar-Ner M, Ariav Y, Schirrmacher V. Lymphoma cells mediated degradation of sulfated proteoglycans in the subendothelial extracellular matrix: relation to tumor cell metastasis. Cancer Res. 1983;43:2704–2711. [PubMed] [Google Scholar]
- 31.Bitan M, Polliack A, Zecchina G, Nagler A, Friedmann Y, Nadav L, Deutsch V, Pecker I, Eldor A, Vlodavsky I, et al. Heparanase expression in human leukemias is restricted to acute myeloid leukemias. Exp Hematol. 2002;30:34–41. doi: 10.1016/s0301-472x(01)00766-4. [DOI] [PubMed] [Google Scholar]
- 32.Miao HQ, Liu H, Navarro E, Kussie P, Zhu Z. Development of heparanase inhibitors for anti-cancer therapy. Curr Med Chem. 2006;13:2101–2111. doi: 10.2174/092986706777935230. [DOI] [PubMed] [Google Scholar]
- 33.Sanderson RD, Yang Y, Suva LJ, Kelly T. Heparan sulfate proteoglycans and heparanase—partners in osteolytic tumor growth and metastasis. Matrix Biol. 2004;23:341–352. doi: 10.1016/j.matbio.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Ferro V, Dredge K, Liu L, Hammond E, Bytheway I, Li C, Johnstone K, Karoli T, Davis K, Copeman E, et al. PI-88 and novel heparan sulfate mimetics inhibit angiogenesis. Semin Thromb Hemost. 2007;33:557–568. doi: 10.1055/s-2007-982088. [DOI] [PubMed] [Google Scholar]
- 35.Vlodavsky I, Abboud-Jarrous G, Elkin M, Naggi A, Casu B, Sasisekharan R, Ilan N. The impact of heparanase and heparin on cancer metastasis and angiogenesis. Pathophysiol Haemost Thromb. 2006;35:116–127. doi: 10.1159/000093553. [DOI] [PubMed] [Google Scholar]
- 36.Matzner Y, Bar-Ner M, Yahalom J, Ishai-Michaeli R, Fuks Z, Vlodavsky I. Degradation of heparan sulfate in the subendothelial extracellular matrix by a readily released heparanase from human neutrophils. Possible role in invasion through basement membranes. J Clin Invest. 1985;76:1306–1313. doi: 10.1172/JCI112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naparstek Y, Cohen IR, Fuks Z, Vlodavsky I. Activated T lymphocytes produce a matrix-degrading heparan sulphate endoglycosidase. Nature. 1984;310:241–244. doi: 10.1038/310241a0. [DOI] [PubMed] [Google Scholar]
- 38.Savion N, Disatnik MH, Nevo Z. Murine macrophage heparanase: inhibition and comparison with metastatic tumor cells. J Cell Physiol. 1987;130:77–84. doi: 10.1002/jcp.1041300112. [DOI] [PubMed] [Google Scholar]
- 39.Vlodavsky I, Eldor A, Haimovitz-Friedman A, Matzner Y, Ishai-Michaeli R, Lider O, Naparstek Y, Cohen IR, Fuks Z. Expression of heparanase by platelets and circulating cells of the immune system: possible involvement in diapedesis and extravasation. Invasion Metastasis. 1992;12:112–127. [PubMed] [Google Scholar]
- 40.Sotnikov I, Hershkoviz R, Grabovsky V, Ilan N, Cahalon L, Vlodavsky I, Alon R, Lider O. Enzymatically quiescent heparanase augments T cell interactions with VCAM-1 and extracellular matrix components under versatile dynamic contexts. J Immunol. 2004;172:5185–5193. doi: 10.4049/jimmunol.172.9.5185. [DOI] [PubMed] [Google Scholar]
- 41.Vaday GG, Lider O. Extracellular matrix moieties, cytokines, and enzymes: dynamic effects on immune cell behavior and inflammation. J Leukoc Biol. 2000;67:149–159. doi: 10.1002/jlb.67.2.149. [DOI] [PubMed] [Google Scholar]
- 42.Ostrovsky O, Korostishevsky M, Levite I, Leiba M, Galski H, Vlodavsky I, Nagler A. Association of heparanase gene (HPSE) single nucleotide polymorphisms with hematological malignancies. Leukemia. 2007 doi: 10.1038/sj.leu.2404821. [Epub July 5] [DOI] [PubMed] [Google Scholar]
- 43.Nadir Y, Brenner B, Zetser A, Ilan N, Shafat I, Zcharia E, Goldshmidt O, Vlodavsky I. Heparanase induces tissue factor expression in vascular endothelial and cancer cells. J Thromb Haemost. 2006;4:2443–2451. doi: 10.1111/j.1538-7836.2006.02212.x. [DOI] [PubMed] [Google Scholar]
- 44.Gingis-Velitski S, Zetser A, Flugelman MY, Vlodavsky I, Ilan N. Heparanase induces endothelial cell migration via protein kinase B/Akt activation. J Biol Chem. 2004;279:23536–23541. doi: 10.1074/jbc.M400554200. [DOI] [PubMed] [Google Scholar]
- 45.Zetser A, Bashenko Y, Edovitsky E, Levy-Adam F, Vlodavsky I, Ilan N. Heparanase induces vascular endothelial growth factor expression: correlation with p38 phosphorylation levels and Src activation. Cancer Res. 2006;66:1455–1463. doi: 10.1158/0008-5472.CAN-05-1811. [DOI] [PubMed] [Google Scholar]
- 46.Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]