Abstract
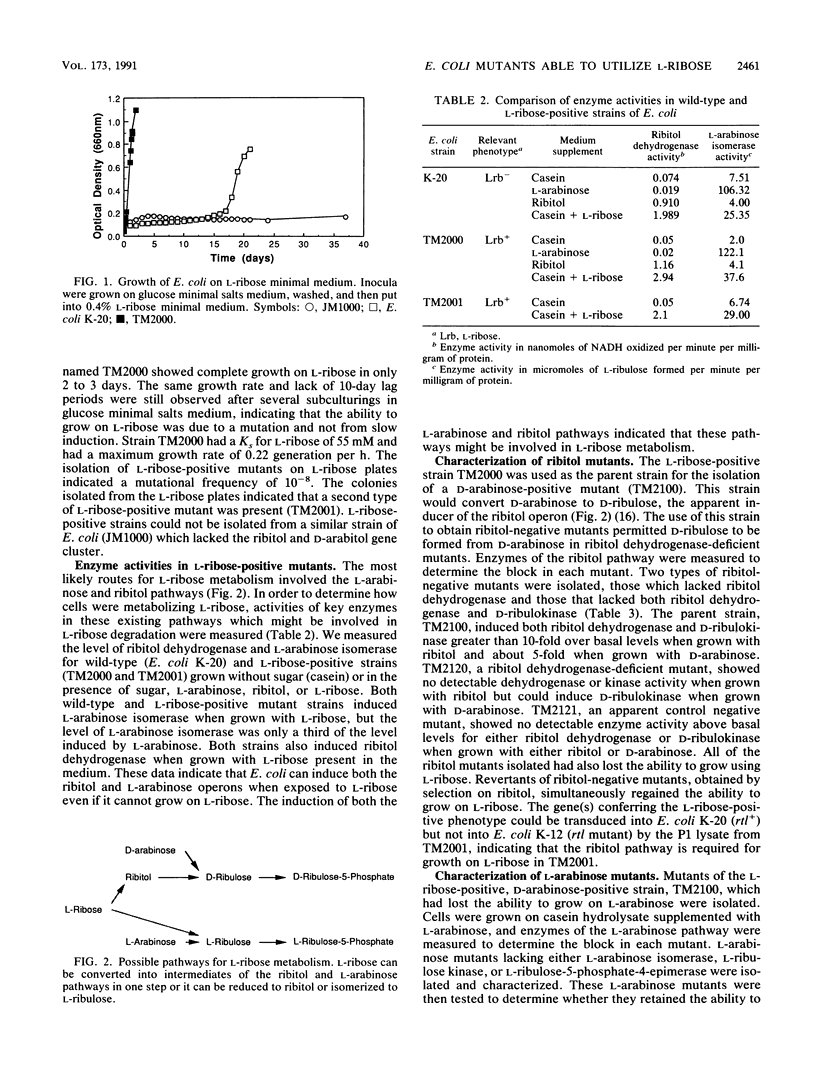
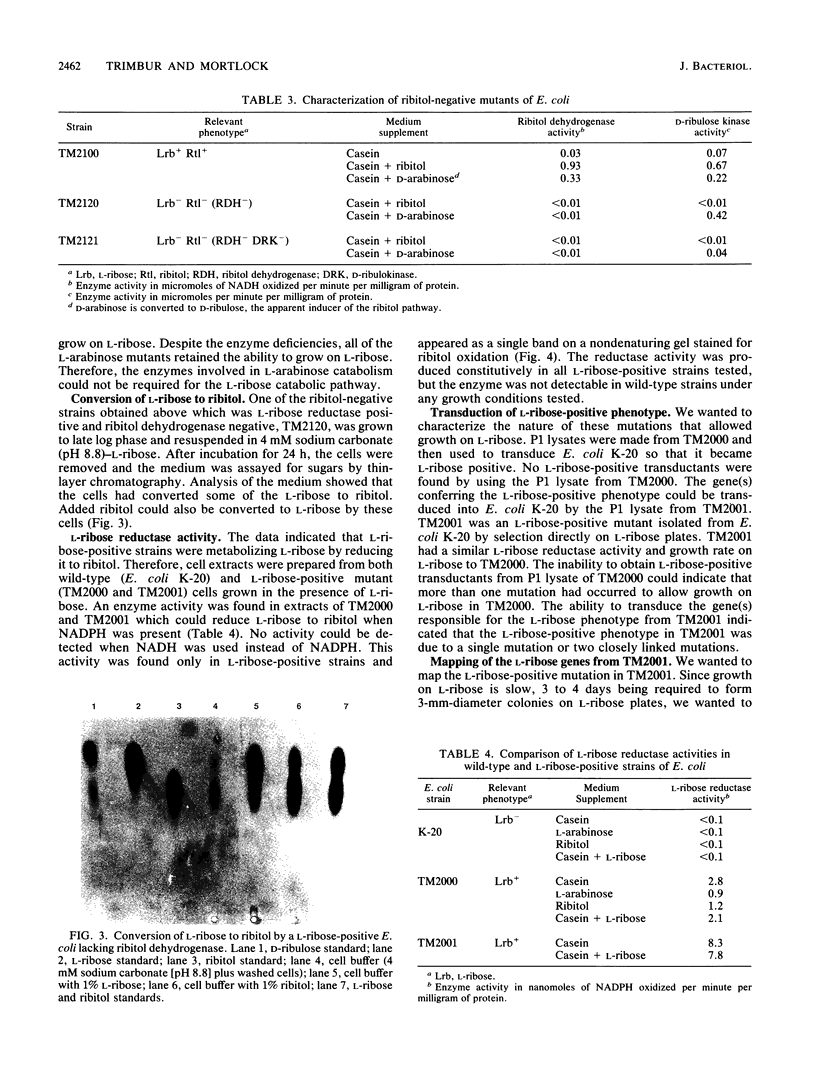
Wild-type strains of Escherichia coli were unable to utilize L-ribose for growth. However, L-ribose-positive mutants could be isolated from strains of E. coli K-12 which contained a ribitol operon. L-ribose-positive strains of E. coli, isolated after 15 to 20 days, had a growth rate of 0.22 generation per h on L-ribose. Growth on L-ribose was found to induce the enzymes of the L-arabinose and ribitol pathways, but only ribitol-negative mutants derived from strains originally L-ribose positive lost the ability to grow on L-ribose, showing that a functional ribitol pathway was required. One of the mutations permitting growth on L-ribose enabled the mutants to produce constitutively an NADPH-linked reductase which converted L-ribose to ribitol. L-ribose is not metabolized by an isomerization to L-ribulose, as would be predicted on the basis of other pentose pathways in enteric bacteria. Instead, L-ribose was metabolized by the reduction of L-ribose to ribitol, followed by the conversion to D-ribulose by enzymes of the ribitol pathway.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ANDERSON R. L., WOOD W. A. Purification and properties of L-xylulokinase. J Biol Chem. 1962 Apr;237:1029–1033. [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkus J. M., Mortlock R. P. Isolation of a mutation resulting in constitutive synthesis of L-fucose catabolic enzymes. J Bacteriol. 1986 Mar;165(3):710–714. doi: 10.1128/jb.165.3.710-714.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Burleigh B. D., Rigby P. W., Hartley B. S. A comparison of wild-type and mutant ribitol dehydrogenases from Klebsiella aerogenes. Biochem J. 1974 Nov;143(2):341–352. doi: 10.1042/bj1430341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CRIBBS R., ENGLESBERG E. L-ARABINOSE NEGATIVE MUTANTS OF THE L-RIBULOKINASE STRUCTURAL GENE AFFECTING THE LEVELS OF L-ARABINOSE ISOMERASE IN ESCHERICHIA COLI. Genetics. 1964 Jan;49:95–108. doi: 10.1093/genetics/49.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DISCHE Z., BORENFREUND E. A new spectrophotometric method for the detection and determination of keto sugars and trioses. J Biol Chem. 1951 Oct;192(2):583–587. [PubMed] [Google Scholar]
- Doten R. C., Mortlock R. P. Characterization of xylitol-utilizing mutants of Erwinia uredovora. J Bacteriol. 1985 Feb;161(2):529–533. doi: 10.1128/jb.161.2.529-533.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOWARD-FLANDERS P., SIMSON E., THERIOT L. A LOCUS THAT CONTROLS FILAMENT FORMATION AND SENSITIVITY TO RADIATION IN ESCHERICHIA COLI K-12. Genetics. 1964 Feb;49:237–246. doi: 10.1093/genetics/49.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- LeBlanc D. J., Mortlock R. P. Metabolism of D-arabinose: a new pathway in Escherichia coli. J Bacteriol. 1971 Apr;106(1):90–96. doi: 10.1128/jb.106.1.90-96.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg J. Gene Recombination and Linked Segregations in Escherichia Coli. Genetics. 1947 Sep;32(5):505–525. doi: 10.1093/genetics/32.5.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Formation of merodiploids in matings with a class of Rec- recipient strains of Escherichia coli K12. Proc Natl Acad Sci U S A. 1968 May;60(1):160–167. doi: 10.1073/pnas.60.1.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M. S., Patterson R. A., Hartley B. S. Purification and properties of Klebsiella aerogenes D-arabitol dehydrogenase. Biochem J. 1979 Oct 1;183(1):31–42. doi: 10.1042/bj1830031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver E. J., Bisson T. M., LeBlanc D. J., Mortlock R. P. D-Ribulose production by a mutant of Aerobacter aerogens. Anal Biochem. 1969 Feb;27(2):300–305. doi: 10.1016/0003-2697(69)90036-0. [DOI] [PubMed] [Google Scholar]
- Ornston L. N., Parke D. The evolution of induction mechanisms in bacteria: insights derived from the study of the beta-ketoadipate pathway. Curr Top Cell Regul. 1977;12:209–262. doi: 10.1016/b978-0-12-152812-6.50011-1. [DOI] [PubMed] [Google Scholar]
- Reiner A. M. Genes for ribitol and D-arabitol catabolism in Escherichia coli: their loci in C strains and absence in K-12 and B strains. J Bacteriol. 1975 Aug;123(2):530–536. doi: 10.1128/jb.123.2.530-536.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosner J. L. Formation, induction, and curing of bacteriophage P1 lysogens. Virology. 1972 Jun;48(3):679–689. doi: 10.1016/0042-6822(72)90152-3. [DOI] [PubMed] [Google Scholar]
- Scher B. M., Horecker B. L. Pentose metabolism in Candida. 3. The triphosphopyridine nucleotide-specific polyol dehydrogenase of Candida utilis. Arch Biochem Biophys. 1966 Sep 26;116(1):117–128. doi: 10.1016/0003-9861(66)90020-8. [DOI] [PubMed] [Google Scholar]
- TREVELYAN W. E., PROCTER D. P., HARRISON J. S. Detection of sugars on paper chromatograms. Nature. 1950 Sep 9;166(4219):444–445. doi: 10.1038/166444b0. [DOI] [PubMed] [Google Scholar]
- WOOD W. A., McDONOUGH M. J., JACOBS L. B. Rihitol and D-arabitol utilization by Aerobacter aerogenes. J Biol Chem. 1961 Aug;236:2190–2195. [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]