Abstract
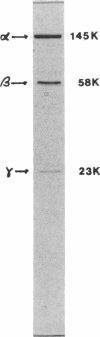
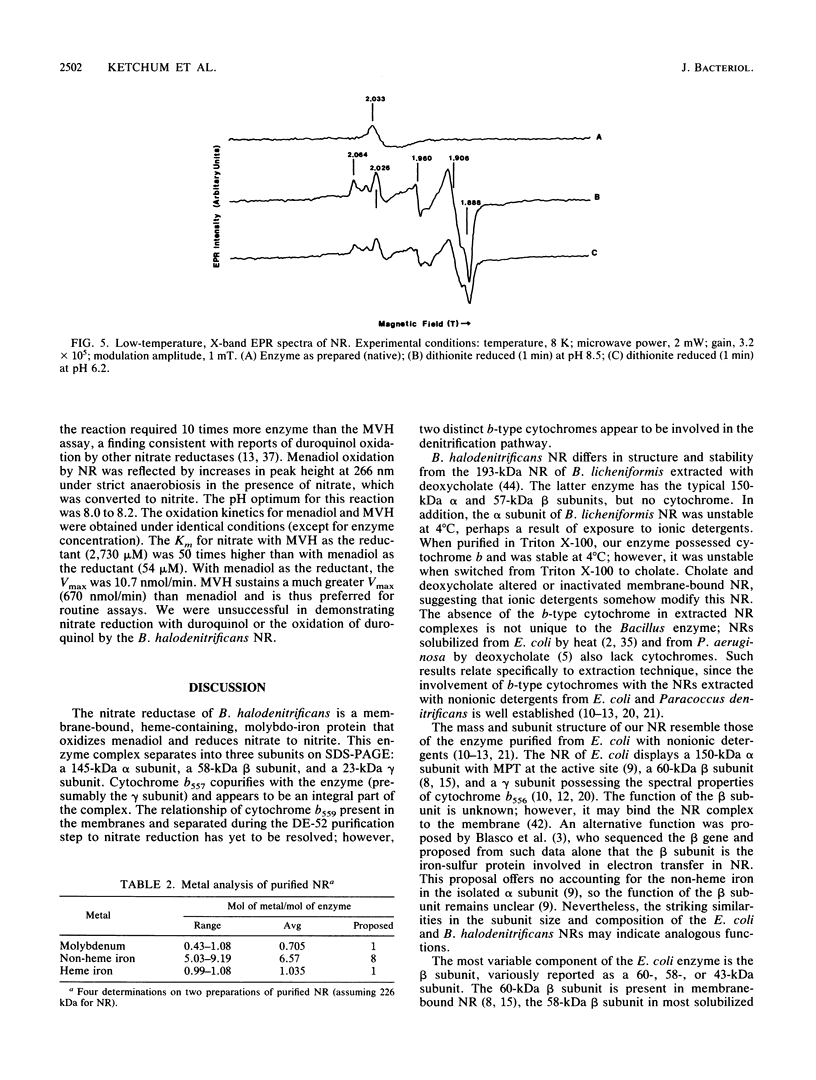
When grown anaerobically on nitrate-containing medium, Bacillus halodenitrificans exhibited a membrane-bound nitrate reductase (NR) that was solubilized by 2% Triton X-100 but not by 1% cholate or deoxycholate. Purification on columns of DE-52, hydroxylapatite, and Sephacryl S-300 yielded reduced methyl viologen NR (MVH-NR) with specific activities of 20 to 35 U/mg of protein that was stable when stored in 40% sucrose at -20 degrees C for 6 weeks. 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxypropone-1-sulfonat e (CHAPSO) and dodecyl-beta-D-maltoside stimulated enzyme activity three- to fourfold. Membrane extractions yielded purified NR that separated after electrophoresis into a 145-kDa alpha subunit, a 58-kDa beta subunit, and a 23-kDa gamma subunit. The electronic spectrum of dithionite-reduced, purified NR displayed peaks at 424.6, 527, and 557 nm, indicative of the presence of a cytochrome b, an interpretation consistent with the pyridine hemochrome spectrum formed. Analyses revealed a molybdenum-heme-non-heme iron ratio of 1:1:8 for the NR and the presence of molybdopterin. Electron paramagnetic resonance (EPR) signals characteristic of iron-sulfur centers were detected at low temperature. EPR also revealed a minor signal centered in the g = 2 region of the spectra. Upon reduction with dithionite, the enzyme displayed signals at g = 2.064, 2.026, 1.906, and 1.888, indicative of the presence of low-potential iron-sulfur centers, which resolve most probably as two [4Fe-4S]+1 clusters. With menadiol as the substrate for nitrate reduction, the Km for nitrate was 50-fold less than that seen when MVH was the electron donor. The cytochrome b557-containing enzyme from B. halodenitrificans is characterized as a menaquinol-nitrate:oxidoreductase.
Full text
PDF

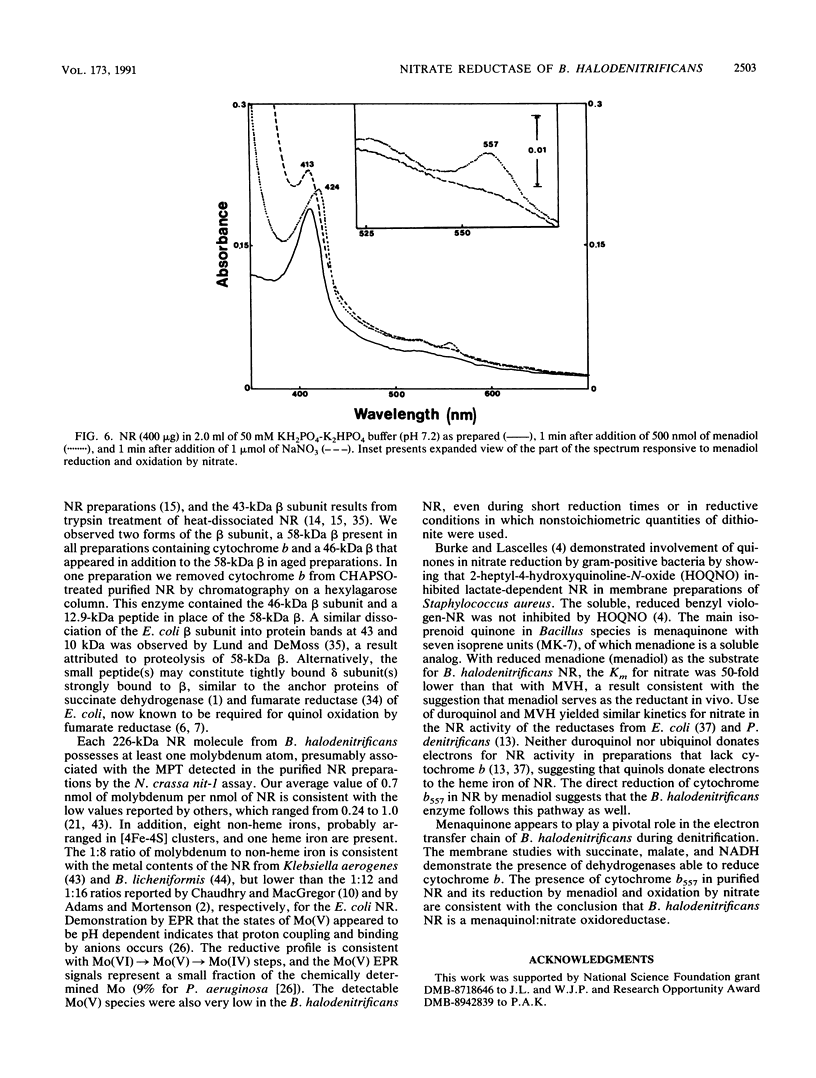


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackrell B. A., Ball M. B., Kearney E. B. Peptides from complex II active in reconstitution of succinate-ubiquinone reductase. J Biol Chem. 1980 Apr 10;255(7):2761–2769. [PubMed] [Google Scholar]
- Adams M. W., Mortenson L. E. The effect of cyanide and ferricyanide on the activity of the dissimilatory nitrate reductase of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):1791–1799. [PubMed] [Google Scholar]
- Blasco F., Iobbi C., Giordano G., Chippaux M., Bonnefoy V. Nitrate reductase of Escherichia coli: completion of the nucleotide sequence of the nar operon and reassessment of the role of the alpha and beta subunits in iron binding and electron transfer. Mol Gen Genet. 1989 Aug;218(2):249–256. doi: 10.1007/BF00331275. [DOI] [PubMed] [Google Scholar]
- Burke K. A., Lascelles J. Nitrate reductase system in Staphylococcus aureus wild type and mutants. J Bacteriol. 1975 Jul;123(1):308–316. doi: 10.1128/jb.123.1.308-316.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C. A., Ferguson L. P., Ingraham J. L. Properties of dissimilatory nitrate reductase purified from the denitrifier Pseudomonas aeruginosa. J Bacteriol. 1982 Jul;151(1):162–171. doi: 10.1128/jb.151.1.162-171.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini G., Ackrell B. A., Deshler J. O., Gunsalus R. P. Reconstitution of quinone reduction and characterization of Escherichia coli fumarate reductase activity. J Biol Chem. 1986 Feb 5;261(4):1808–1814. [PubMed] [Google Scholar]
- Cecchini G., Thompson C. R., Ackrell B. A., Westenberg D. J., Dean N., Gunsalus R. P. Oxidation of reduced menaquinone by the fumarate reductase complex in Escherichia coli requires the hydrophobic FrdD peptide. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8898–8902. doi: 10.1073/pnas.83.23.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry G. R., Chaiken I. M., MacGregor C. H. An activity from Escherichia coli membranes responsible for the modification of nitrate reductase to its precursor form. J Biol Chem. 1983 May 10;258(9):5828–5833. [PubMed] [Google Scholar]
- Chaudhry G. R., MacGregor C. H. Cytochrome b from Escherichia coli nitrate reductase. Its properties and association with the enzyme complex. J Biol Chem. 1983 May 10;258(9):5819–5827. [PubMed] [Google Scholar]
- Chaudhry G. R., MacGregor C. H. Escherichia coli nitrate reductase subunit A: its role as the catalytic site and evidence for its modification. J Bacteriol. 1983 Apr;154(1):387–394. doi: 10.1128/jb.154.1.387-394.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikwem J. O., Downey R. J. Detergent solubilization of the respiratory nitrate reductase of Bacillus stearothermophilus. Microbios. 1986;47(192-193):159–163. [PubMed] [Google Scholar]
- Clegg R. A. Purification and some properties of nitrate reductase (EC 1.7.99.4) from Escherichia coli K12. Biochem J. 1976 Mar 1;153(3):533–541. doi: 10.1042/bj1530533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craske A., Ferguson S. J. The respiratory nitrate reductase from Paracoccus denitrificans. Molecular characterisation and kinetic properties. Eur J Biochem. 1986 Jul 15;158(2):429–436. doi: 10.1111/j.1432-1033.1986.tb09771.x. [DOI] [PubMed] [Google Scholar]
- DeMoss J. A. Limited proteolysis of nitrate reductase purified from membranes of Escherichia coli. J Biol Chem. 1977 Mar 10;252(5):1696–1701. [PubMed] [Google Scholar]
- Demoss J. A., Fan T. Y., Scott R. H. Characterization of subunit structural alterations which occur during purification of nitrate reductase from Escherichia coli. Arch Biochem Biophys. 1981 Jan;206(1):54–64. doi: 10.1016/0003-9861(81)90065-5. [DOI] [PubMed] [Google Scholar]
- Downey R. J. Nitrate reductase and respiratory adaptation in Bacillus stearothermophilus. J Bacteriol. 1966 Feb;91(2):634–641. doi: 10.1128/jb.91.2.634-641.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The role of a novel cytochrome b-containing nitrate reductase and quinone in the in vitro reconstruction of formate-nitrate reductase activity of E. coli. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1234–1241. doi: 10.1016/s0006-291x(74)80416-x. [DOI] [PubMed] [Google Scholar]
- Forget P., Dervartanian D. V. The bacterial nitrate reductases: EPR studies on nitrate reductase A from Micrococcus denitrificans. Biochim Biophys Acta. 1972 Feb 28;256(2):600–606. doi: 10.1016/0005-2728(72)90089-8. [DOI] [PubMed] [Google Scholar]
- Furth A. J., Bolton H., Potter J., Priddle J. D. Separating detergent from proteins. Methods Enzymol. 1984;104:318–328. doi: 10.1016/s0076-6879(84)04098-2. [DOI] [PubMed] [Google Scholar]
- Garrett R. H., Nason A. Further purification and properties of Neurospora nitrate reductase. J Biol Chem. 1969 Jun 10;244(11):2870–2882. [PubMed] [Google Scholar]
- Godfrey C., Greenwood C., Thomson A. J., Bray R. C., George G. N. Electron-paramagnetic-resonance spectroscopy studies on the dissimilatory nitrate reductase from Pseudomonas aeruginosa. Biochem J. 1984 Dec 1;224(2):601–608. doi: 10.1042/bj2240601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes T. R., Bray R. C. Quantitative transfer of the molybdenum cofactor from xanthine oxidase and from sulphite oxidase to the deficient enzyme of the nit-1 mutant of Neurospora crassa to yield active nitrate reductase. Biochem J. 1984 Apr 15;219(2):481–493. doi: 10.1042/bj2190481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstein L. I., Tomlinson G. A. The enzymes associated with denitrification. Annu Rev Microbiol. 1988;42:231–261. doi: 10.1146/annurev.mi.42.100188.001311. [DOI] [PubMed] [Google Scholar]
- Ingledew W. J., Poole R. K. The respiratory chains of Escherichia coli. Microbiol Rev. 1984 Sep;48(3):222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. W., Garland P. B. Sites and specificity of the reaction of bipyridylium compounds with anaerobic respiratory enzymes of Escherichia coli. Effects of permeability barriers imposed by the cytoplasmic membrane. Biochem J. 1977 Apr 15;164(1):199–211. doi: 10.1042/bj1640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketchum P. A., Cambier H. Y., Frazier W. A., 3rd, Madansky C. H., Nason A. In vitro assembly of Neurospora assimilatory nitrate reductase from protein subunits of a Neurospora mutant and the xanthine oxidizing or aldehyde oxidase systems of higher animals. Proc Natl Acad Sci U S A. 1970 Jul;66(3):1016–1023. doi: 10.1073/pnas.66.3.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lemire B. D., Robinson J. J., Weiner J. H. Identification of membrane anchor polypeptides of Escherichia coli fumarate reductase. J Bacteriol. 1982 Dec;152(3):1126–1131. doi: 10.1128/jb.152.3.1126-1131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund K., DeMoss J. A. Association-dissociation behavior and subunit structure of heat-released nitrate reductase from Escherichia coli. J Biol Chem. 1976 Apr 25;251(8):2207–2216. [PubMed] [Google Scholar]
- Read S. M., Northcote D. H. Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal Biochem. 1981 Sep 1;116(1):53–64. doi: 10.1016/0003-2697(81)90321-3. [DOI] [PubMed] [Google Scholar]
- Smith L. Bacterial cytochromes and their spectral characterization. Methods Enzymol. 1978;53:202–212. doi: 10.1016/s0076-6879(78)53025-5. [DOI] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman A. R. Purification of membrane proteins: chloroplast cytochromes f and b559. Methods Enzymol. 1974;32:406–422. doi: 10.1016/0076-6879(74)32040-x. [DOI] [PubMed] [Google Scholar]
- Weber K., Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969 Aug 25;244(16):4406–4412. [PubMed] [Google Scholar]
- van 't Riet J., Wientjes F. B., van Doorn J., Planta R. J. Purification and characterization of the respiratory nitrate reductase of Bacillus licheniformis. Biochim Biophys Acta. 1979 Feb 26;576(2):347–360. doi: 10.1016/0005-2795(79)90410-0. [DOI] [PubMed] [Google Scholar]
- van Riet J., van Ed J. H., Wever R., van Gelder B. F., Planta R. J. Characterization of the respiratory nitrate reductase of Klebsiella aerogenes as a molybdenum-containing iron-sulfur enzyme. Biochim Biophys Acta. 1975 Oct 20;405(2):306–317. doi: 10.1016/0005-2795(75)90096-3. [DOI] [PubMed] [Google Scholar]