SUMMARY
Detection and discrimination of chemical compounds in potential foods are essential sensory processes when animals feed. The fruit fly Drosophila melanogaster employs 68 different gustatory receptors (GRs) for the detection of mostly non-volatile chemicals that include sugars, a diverse group of toxic compounds present in many inedible plants and spoiled foods, and pheromones [1–6]. With the exception of a trehalose (GR5a) and a caffeine (GR66a) receptor [7–9], the functions of GRs involved in feeding are unknown. Here, we show that the Gr64 genes encode receptors for numerous sugars. We generated a fly strain that contained a deletion for all six Gr64 genes (ΔGr64) and showed that these flies exhibit no or a significantly diminished proboscis extension reflex (PER) response when stimulated with glucose, maltose, sucrose and several other sugars. The only considerable response was detected when Gr64 mutant flies were stimulated with fructose. Interestingly, response to trehalose is also abolished in these flies, even though they contain a functional Gr5a gene, which has been previously shown to encode a receptor for this sugar [8, 9]. This observation indicates that two or more Gr genes are necessary for trehalose detection, suggesting that GRs function as multimeric receptor complexes. Finally, we present evidence that some members of the Gr64 gene family are transcribed as a polycistronic mRNA, providing a mechanism for co-expression of multiple sugar receptors in the same taste neurons.
INTRODUCTION
In fruit flies and probably most other insects, non-volatile compounds, most notably many food chemicals, are thought to be recognized by seven transmembrane receptors which are expressed in taste neurons located on the labial palps (the equivalent of the mammalian tongue), the legs and wings [10, 11]. The Drosophila gustatory receptor (Gr) gene family is comprised of 68 relatively poorly conserved genes, with amino acid sequence similarity in the range of 8 to 20 % between most pairs [12] (Figure S1). Gr orthologs are found in other insects, but they are absent in vertebrates, C. elegans and more primitive organisms, such as yeast or bacteria. The large differences in gene number - the honeybee has only 12 GRs, while Drosophila has almost 70 [13]- and their poor conservation suggests that this gene family is subject to rapid adaptation driven by the vastly different ecological niches these insect species occupy.
Due to the dispersed location of taste sensilla throughout the body – flies and many other insects harbor taste sensilla not only on the labellum, but also on legs and wings - and the overall low abundance of Gr mRNAs in gustatory receptor neurons (GRNs), expression analyses of Gr genes has been performed mainly with the use of the Gal4/UAS system [1–4, 14, 15]. These studies have led to the identification of two distinct sets of GRNs, which are characterized by the mutually exclusive expression of different Gr genes. The first group is composed of about 22 GRNs – a single taste neuron of each of the 22 I- and S-type sensilla – and expresses Gr66a [1, 2]. However, all these neurons express additional, but distinct Gr genes, and hence, each neuron is defined by a unique Gr expression code. Functional studies revealed that Gr66a-expressing neurons detect bitter compounds, most notably caffeine, as flies in which these cells are impaired show significantly reduced sensitivity to such chemicals [1, 2]. Thus, it is generally assumed that the Gr genes expressed in these 22 I- and S-type sensilla encode receptors for harmful, noxious and toxic (and to humans, bitter tasting) compounds. The second group of GRNs is currently represented by a single Gr gene, Gr5a, and ablation/inactivation of these neurons lead to reduced sensitivity for this sugar, and to a weaker extent to some other sugars as well [1, 2]. Importantly, Gr5a and Gr66a are expressed in distinct, non-overlapping sets of GRNs, and mediate distinct behavioral responses, feeding and avoidance, respectively [1, 2].
The specific molecular roles of all but a few Gr genes are currently unknown. The only two GRs expressed in gustatory receptor neurons (GRNs) with known ligands are GR5a and GR66a, which detect the sugar trehalose and the bitter compound caffeine, respectively [7–9]. Gr5a is a member of a gene subfamily comprised of seven additional members, Gr61a and Gr64a-f, which evolved through recent gene duplication events [12] and therefore share relatively high similarity to each other (41 to 73 % at the amino acid level; Figure S1). Here, we investigate the role of Gr64a-f and show that they encode receptors for many sugars. Moreover, our data suggests an uncommon mode of co-expression of these genes, being transcribed as multi-cistronic mRNA(s), which would provide an efficient and elegant strategy to express these receptors in the same taste neurons.
RESULTS AND DISCUSSION
Generating a Drosophila strain that lacks six putative sugar receptor genes
To gain a basic understanding of sugar perception in Drosophila, we performed a reverse genetic analysis of the six Gr64 genes, which are tightly clustered on the left arm of chromosome 3. We used FRT mediated trans-recombination [16, 17] to create a 25 kb deletion of the region containing the Gr64a gene cluster (ΔGr64), and we confirmed the expected molecular nature of this deletion using genomic PCR and DNA sequencing from the trans-recombined chromosome (see Experimental Procedures; Figure 1). In addition to the six Gr64 genes, this trans-recombination event also removed five additional genes on either side of the cluster, resulting in a homozygous lethal mutation, presumably because some of the neighboring genes have essential functions required for viability. We therefore cloned two genomic DNA constructs containing the two genes proximal (R1) or the three genes distal (R2) to the Gr64 locus, respectively, into a transformation vector and generated corresponding transgenic Drosophila lines (see Experimental Procedures; Figure 1). When R1 was crossed into the ΔGr64 mutant strain, viability was completely restored, indicating that at least one of the two proximally located genes provides a life-essential function. Even though unlikely, it is possible that some genes on R1 and/or R2 may have functions related to taste perception, and we used ΔGr64/ΔGr64 flies that carried a copy of each of these rescue constructs for all behavioral experiments (see below).
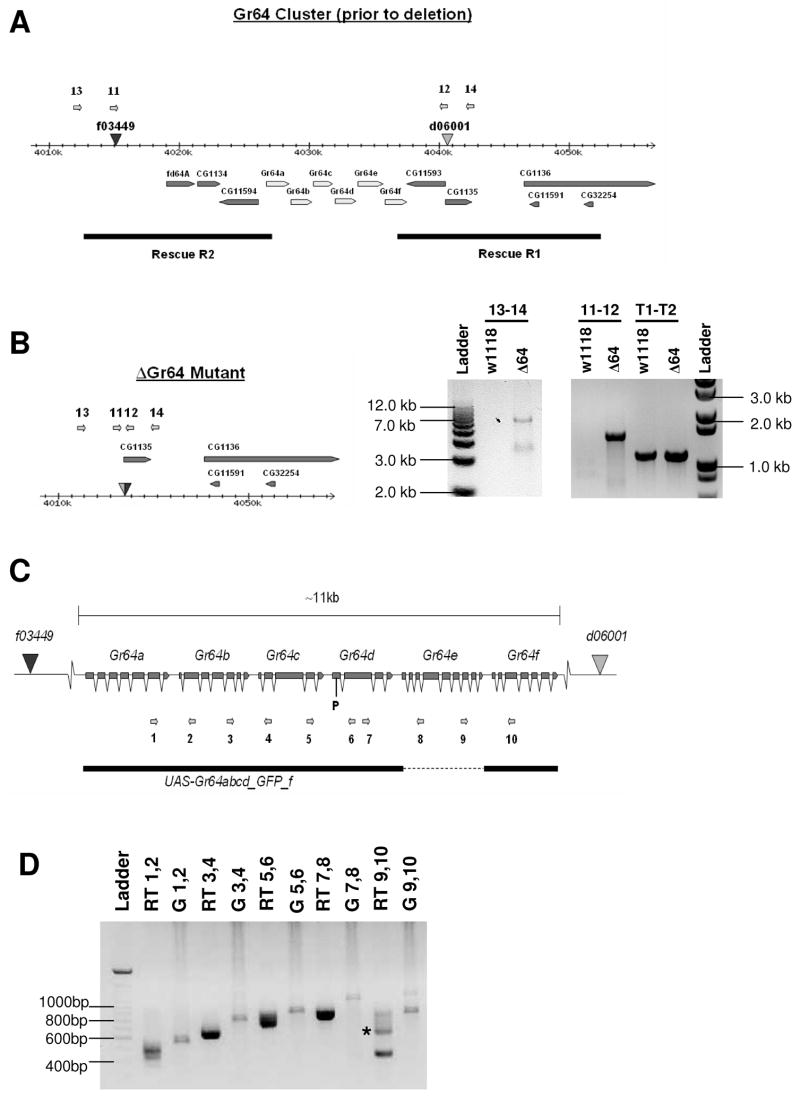
Figure 1. Generation of a Gr64 mutant strain (ΔGr64) and RT-PCR expression analysis of the six Gr64 genes.
(A) Diagram of the Gr64 gene cluster. The positions of the piggyBac transposons are indicated by triangles. The diagram shows the Gr64 cluster prior to generation of the deletion line. The numbered arrows indicate the positions of the primers used for PCR analysis of the deletion. The black bars represent the rescue constructs for the genes flanking the Gr64 cluster.
(B) Molecular analysis of ΔGr64 mutant strain. The diagram shows the structure of the Gr64 deletion (ΔGr64) after trans-recombination. Genomic DNA from w1118 flies was also analyzed for comparison. Expected band sizes are as follows: 1.1 kb for primers T1 and T2, 1.5 kb for primers 11 and 12, and 6.9 kb for primers 13 and 14. Relevant band sizes from the ladder are marked along the sides of the gel. The 1.5 and 6.9 kb products were cloned and sequenced to further confirm the presence of the deletion. The 1.1 kb product is derived from the tubulin gene and serves as a control for DNA integrity.
(C) Exon-intron structure of the Gr64 cluster. Exons are represented by boxes and introns by v-shaped lines. The numbered arrows show the positions of the primers used for RT-PCR analysis. The black bar indicates the rescue construct (UAS-Gr64abcd_GFP_f) in which Gr64e was replaced by EGFP (indicated by the dashed line).
(D) RT-PCR of total RNA from fly heads indicates the presence of polycistronic transcripts in the Gr64 cluster. RNA was extracted from wild-type ORE-R flies. Each pair of primers spans at least one intron in each of the two genes being investigated. For each pair of primers used in an RT-PCR reaction, a corresponding PCR reaction was performed on genomic DNA to provide a size comparison. RT-PCR products were isolated for each primer pair, cloned and sequenced to confirm integrity of appropriately spliced cDNA products. RT-PCR from leg tissue showed similar results (data not shown). Lanes marked “RT” represent RT-PCR products, while lanes marked “G” represent PCR products from genomic DNA.
The Gr64 genes encode sugar receptors
We first investigated whether the Gr64 genes were required for the detection of six sugars by generating ΔGr64 homozygous mutant flies that contained one copy of each of the rescue constructs (R1/+;R2/+;ΔGr64/ΔGr64). We determined the behavioral response of these and control flies to sucrose, glucose, trehalose, fructose, arabinose and maltose using the proboscis extension reflex (PER; Figure 2). As controls, we tested flies that were heterozygous for each of the two piggyBac elements used to generate the ΔGr64 mutation (Figure 2), as well as flies with an intact Gr64 cluster, but containing a copy of R1 and R2 to rule out a dominant phenotype of these transgenes (Figure S2). PER is a robust indicator of a fly’s attraction and motivation to eat a given chemical compound [18]. If taste neurons in labial palps or the forelegs are stimulated with a solution containing sugars, the fly extends its proboscis to attempt feeding. Indeed, we find that both control strains responded with high probability of a PER, ranging from 42 to 97% when stimulated with 500 mM solution of various sugars (Figure 2A). Even at a five-fold lower concentration (100 mM), both types of control flies responded to all sugars, albeit with a reduced PER (Figure 2B, Figure S2). In contrast, R1/+;R2/+;ΔGr64/ΔGr64 flies showed a drastic reduction in PER for all sugars at both 500 and 100 mM, except for fructose. In most cases, the reduction was at least 10 fold, while sensitivity for sucrose was reduced only by about three-fold (Figure 2A and 2B). However, PER response to fructose was the same in control flies and R1/+;R2/+;ΔGr64/ΔGr64 mutant flies at 100mM and reduced by only about 35% at 500mM, suggesting that a high affinity fructose receptor is present in flies lacking all six Gr64 genes.
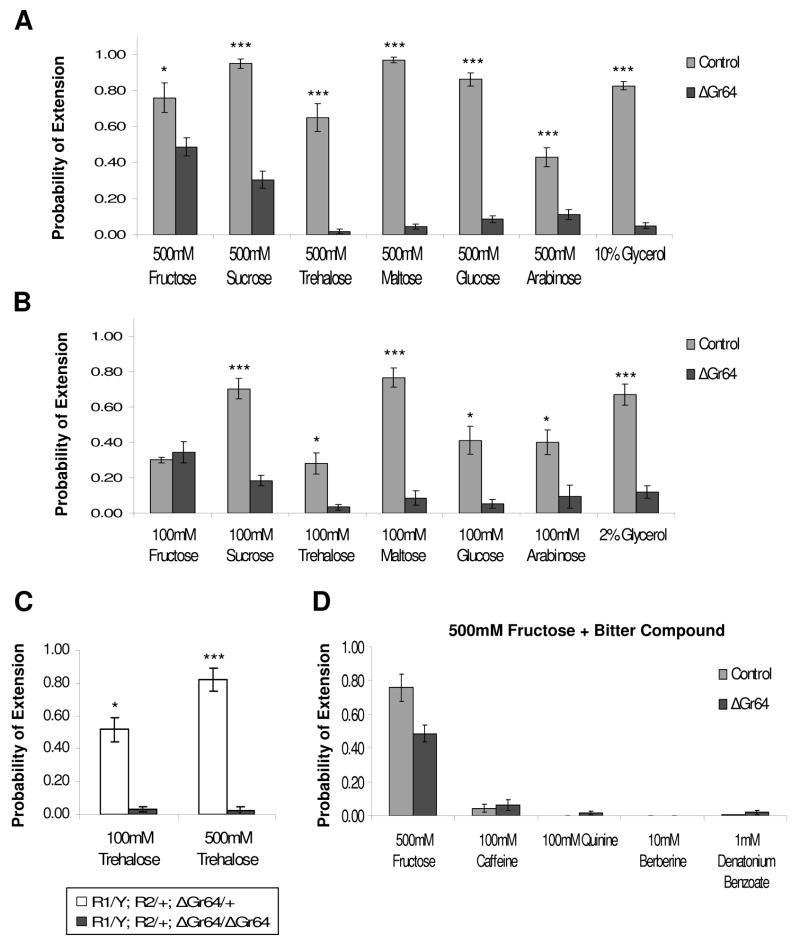
Figure 2. ΔGr64 mutants are severely deficient in the perception of most sugars Proboscis Extension Response (PER) of ΔGr64 mutants and isogenic control flies to 500mM (A) and 100mM (B) sugar solutions.
The genotype of ΔGr64 mutants is R1/+;R2/+;ΔGr64/ΔGr64 and that of the control flies is f03449/d06001. “Probability of Extension” represents the number of times flies from a given strain extended their proboscis when presented with a tastant, divided by the total number of times that the tastant was presented. For all data shown in Figure 2, each graph is the average of 4–15 experiments +/− SEM (3–11 flies per experiment, 20–105 flies total for each strain and tastant tested). Asterisks indicate a significant difference between the mutant and control strains, as determined by Student’s t-test (* indicates p<0.05, *** indicates p<0.0001). Glycerol was used as 10% or 2% solutions in water.
(C) The Gr5a gene is functional in R1/Y;R2/+;ΔGr64/ΔGr64 flies. Flies heterozygous for ΔGr64, but containing the same X chromosome (i.e. the same Gr5a) as the homozygous ΔGr64 flies, show normal and robust response to trehalose at both 100 mM and 500 mM concentrations.
(D) PER response of ΔGr64 mutant and control strains to various bitter tastants in the presence of 500mM fructose. The response to 500mM fructose alone is shown for comparison. There was no significant difference between mutants and controls for any of the bitter solutions by Student’s t-test.
It was recently shown that flies exhibit a behavioral feeding response to glycerol, a linear triol, and indeed, glycerol was shown to stimulate sugar-sensitive neurons [19]. We wondered whether glycerol detection is also mediated by some of the GR64 receptors and therefore examined the PER response in control and ΔGr64 mutant flies. Indeed we observed almost a 6 and 16 fold reduction of PER to 2% and 10% glycerol, respectively, in mutants when compared to controls.
The loss of behavioral response to trehalose in ΔGr64 mutant flies is surprising, since these flies contain presumably a wild type Gr5a gene, which encodes a receptor for this sugar [8, 9]. Therefore, the perception of trehalose appears to require at least two Gr genes, Gr5a and one or more members of the Gr64 gene cluster, suggesting that insect sugar receptors might function as dimers or multimers. To rule out the possibility that the loss of trehalose responses in these flies is caused by a defective Gr5a gene, we tested the PER response of flies heterozygous for ΔGr64, but containing the same X chromosome (i.e. the same Gr5a) as the homozygous ΔGr64 flies (Figure 2C). These flies showed indeed a robust response to trehalose, indicating that a second receptor in the Gr64 locus is necessary for the detection of this sugar. Thus, trehalose, and possibly sugars in general, are detected by mulimeric receptors composed of two or more GRs. There are precedents for insect chemoreceptors as multisubunit transmembrane receptors in the olfactory system: In most olfactory sensory neurons (OSNs), receptors for volatile chemicals appear to function as dimers consisting of the widely expressed OR83b protein and the single OR expressed in a given OSN [20, 21]. Furthermore, a distinct subset of CO2 – sensitive OSNs expresses the two gustatory receptors Gr63a and Gr21a, both of which are required for sensing this gas, a stress pheromone in flies [22–24].
Homozygous ΔGr64 mutant flies show as robust a response to 100 mM fructose as control flies, indicating that a functional fructose receptor does not contain any of the GR64 proteins. A receptor for this sugar might therefore be comprised of a heterodimer between GR61a and GR5a or a homodimer of either one of these two proteins. But other compositions are possible as well, such as heterodimers involving one of these subunits along with another GR proteins. Any such dimer may also serve as a low affinity receptor for non-fructose sugars and therefore be responsible for the residual PER responses to glucose, trehalose, maltose and arabinose in homozygous ΔGr64 mutant flies (Figure 2).
To assess whether lack of the Gr64 gene affects the behavioral responses to other chemicals, we tested PER response to four chemically diverse, bitter-tasting compounds. Such compounds, which are known to inhibit feeding, reduce PER responses if they are mixed with sugars solutions [1, 2]. Therefore, we tested PER responses to 500 mM fructose solutions that included caffeine, denatonium benzoate, berberine or quinine (Figure 2D). Both strains showed a similar decrease in PER response when stimulated with these solutions, suggesting that the Gr64 genes are not required for the detection (and avoidance) of bitter compounds. Taken together, our data suggest that the six Gr64 genes are necessary specifically for the detection of most sugars.
Gr64 genes are co-expressed as a poly-cistronic mRNA
The Gal4/UAS expression system [14] has been used very successfully to identify GRNs that express specific Gr genes [1–4, 15]. We generated four Gr64-Gal4 driver constructs and combined these with the UAS-gfp reporters, but we did not observe expression in the main taste organs with any of them (Thorne, Slone and Amrein; unpublished data), even though RNAs for all six Gr64 genes are detected by RT-PCR (see below). This suggested that crucial transcriptional regulatory elements are located upstream and/or downstream of the cluster and/or within introns of the Gr64 genes. Further support for an unusual arrangement of regulatory elements of the Gr64a genes is apparent from the dense genomic clustering of the six open reading frames (ORFs). Assuming at least 50 nucleotides of 5′ and 3′ UTR for each gene, the intergenic, non-transcribed regions harboring putative promoters are extremely short (<100 nt) and lack transcription termination signals (AAUAAA), which are present in most Drosophila genes (Table 1). These observations prompted us to test whether the Gr64 genes might be transcribed as a poly-cistronic mRNA. We isolated mRNA from heads and performed RT-PCR analysis across the whole cluster using primer pairs of adjacent genes (Figure 1C and 1D). To discriminate between products from spliced RNA and residual genomic DNA, primers were chosen such that the amplified fragments would represent spliced transcripts that lack at least one intron (Figure 1C). In each case, RT-PCR readily amplified a spliced RNA product composed of cDNAs corresponding to adjacent ORFs separated by the intergenic sequence (Figure 1D). The same result was obtained when RNA isolated from leg tissue was used (data not shown). These results suggest that coding sequences of adjacent Gr64 genes are present on the same mRNA and, by inference, that possibly all six ORFs may be transcribed as a large polycistronic mRNA.
Table 1. Distances between adjacent genes in selected Gr gene clusters.
Intergenic distances are indicted and refer to the number of nucleotides from stop codon to start codon. The nucleotide position of the polyadenylation signal (AAUAAA), if present in the intergenic region, is indicated and represents the number of nucleotides between the stop codon and the first nucleotide of the polyadenylation signal. The last column indicates the position of the AAUAAA after the last gene in each cluster. Gr59a/b and Gr59c/d are two separate gene clusters and have therefore been separated on this table. The intergenic regions highlighted in bold were shown to have no obvious promoter sequences in a bioinformatic analysis performed by another group (28).
| Gene Cluster | a to b | AAUAAA | b to c | AAUAAA | c to d | AAUAAA | d to e | AAUAAA | e to f | AAUAAA | AAUAAA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gr22 | 399 | None | 233 | None | 255 | None | -- | -- | -- | -- | 107 |
| Gr93 | -- | -- | 230 | 74 | 188 | 123 | -- | -- | -- | -- | 111 |
| Gr98 | -- | -- | 554 | 39 | 215 | None | -- | -- | -- | -- | 293 |
| Gr28 | 838 | 164 | -- | -- | -- | -- | -- | -- | -- | -- | 67 |
| Gr64 | 200 | None | 191 | None | 136 | None | 183 | None | 180 | None | 30 |
| Gr59a,b | 477 | 8 | -- | -- | -- | -- | -- | -- | -- | -- | 178 |
| Gr59c,d | -- | -- | -- | -- | 207 | None | -- | -- | -- | -- | 9 |
| Gr10 | 309 | 128 | -- | -- | -- | -- | -- | -- | -- | -- | 219 |
| Gr36 | 859 | 23 | 273 | None | 305 | None | -- | -- | -- | -- | 1098 |
With the exception of nematodes, polycistronic transcripts are not thought to be common in higher eukaryotes. In C. elegans, however, a significant number of genes (~15 %) are co-transcribed as operons, and independently trans-spliced to the abundantly expressed spliced leader (SL2) RNA [25]. However, it has recently become apparent that operon-like gene organizations and polycistronic mRNAs do exist in Drosophila; at least two transcripts initially postulated to be non-coding RNAs were shown to encode multiple, albeit redundant peptides, with functions necessary in early development [26, 27]. Examples more similar to the Gr64 genes were described for four pairs of Drosophila Or genes and the Drosophila CheB42a-llz locus [28, 29]. The basic translation mechanism of poly-cistronic mRNAs of the Or gene pairs is unknown; however the CheB42a-llz dicistronic transcript is subsequently cleaved into two mRNAs that appear to be translated separately [29]. While a polyadenylation signal is present after the upstream gene (CheB42a) in this case, no putative promoter sequences were identified for the intergenic region in the CheB42a-llz locus [29]. A genomic survey by these authors for closely clustered genes lacking promoter sequences in the intergenic region identified almost 1400 Drosophila gene pairs, suggesting that operon-like gene structures may be much more common in eukaryotes than generally assumed [29]. Not surprisingly several Or and Gr gene pairs were found to lack such promoter sequences, including the five downstream genes in the Gr64 gene cluster (see also Table 1).
Transgene rescue of sugar phenotypes
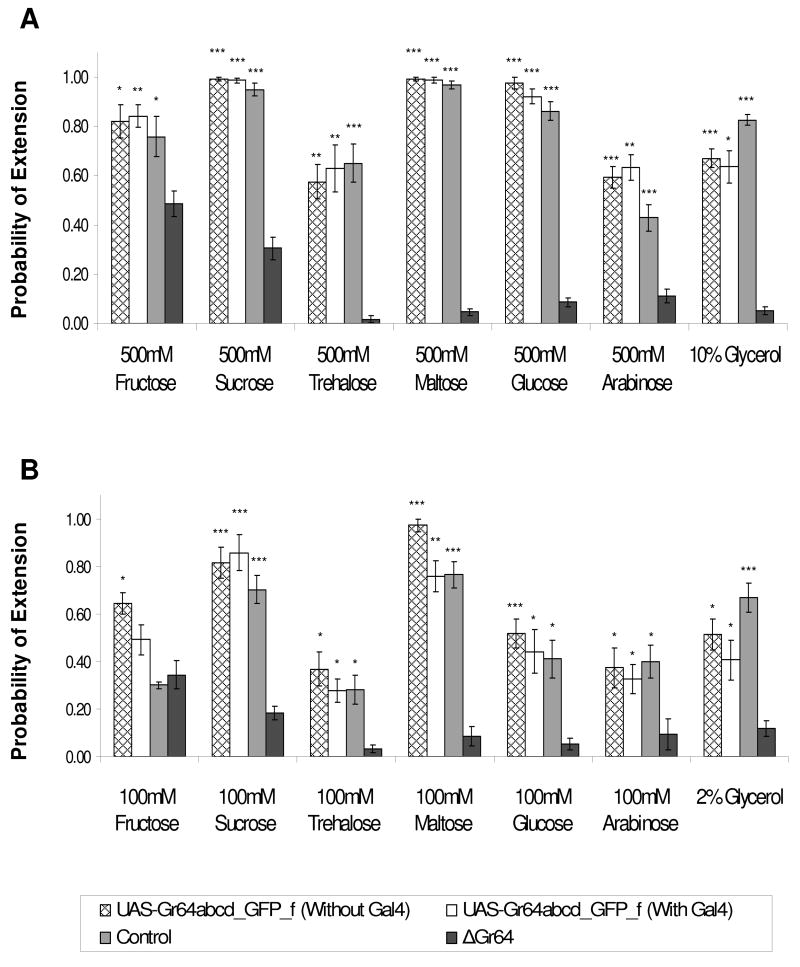
To conclusively prove that the Gr64 genes indeed encode sugar receptors, we performed transgene rescue experiments. We cloned a genomic fragment containing five of the six Gr64 genes into the UAS reporter (UAS-Gr64abcd_GFP_f; Gr64e was replaced by GFP) and generated two types of R1/+;R2/+;ΔGr64/ΔGr64 flies, the first containing the UAS-Gr64abcd_GFP_f rescue construct (see Figure 1C), and the second containing the same rescue construct as well as Gr5a-Gal4 driver. The Gr5a-Gal4 driver is expressed in sugar-sensitive neurons of both the labellum and the legs [2] and should confer such expression on the rescue construct. At both 100mM and 500mM concentration, the UAS-Gr64abcd_GFP_f reporter rescued the PER response to similar levels as observed in the control strain (Figure 3). Surprisingly, this rescue was independent of the GAL4 driver, indicating that intragenic regulatory elements confer sufficient expression onto the Gr64 genes. This expression was confirmed using RT-PCR analysis, which showed that regardless of whether the Gr5a-Gal4 was present or not, the Gr64 transcripts were readily amplified (Figure S3). We note that a second Gr64abcd_GFP_f reporter integrated in a different genomic location provided only partial rescue (data not shown).
Figure 3. Rescue of ΔGr64 mutant phenotype.
PER response of ΔGr64 mutants (R1/+;R2/+;ΔGr64/ΔGr64) carrying one copy of the UAS-Gr64abcd_GFP_f reporter, with or without Gr5a-Gal4 driver. Sugars were tested at 500mM (A) and 100mM (B) concentration. PER response of flies with the rescue construct is similar to that of the control flies, regardless of whether or not the Gr5a-Gal4 driver is present. Each graph is the average of 4–15 experiments +/− SEM (3–11 flies per experiment, 20–105 flies total for each strain and tastant tested). Asterisks indicate a significant difference between the mutant and control strains, as determined by Student’s t-test (* indicates p<0.05, ** indicates p<0.001, *** indicates p<0.0001).
CONCLUSION
Sugars are essential dietary compounds for many insects, including most Drosophila species. While a single trehalose receptor has been identified previously [8, 9], the molecular genetic basis for the perception of sugars in general was unknown. Here we showed that the six Gr64 genes encode receptors for the detection of most sugars: sucrose, glucose, maltose, trehalose, and arabinose. Our data also suggests that the Drosophila taste receptors, similar to insect olfactory [20, 21] and CO2 receptors [22, 24] and mammalian sweet taste receptors [30, 31], function as dimers (or possibly multimers), since detection of trehalose requires, in addition to GR5a, at least one of the six receptors encoded by the Gr64 genes. However, in contrast to mammals, which use T1R2/T1R3 heterodimers for the detection of all sugars [30, 31], Drosophila appears to use distinct combinations of GRs for the detection of different sugars. As GRs have been difficult to express in cell or heterologous systems, the availability of a mutant lacking the six Gr64 genes should help elucidate the molecular nature of dimeric (or multimeric) sugar receptors in insects.
RT-PCR analysis of Gr64 transcripts suggests that the six Gr64 genes are transcribed as a polycistronic mRNA, a rare mode of gene expression in eukaryotes other than C. elegans. Of the few known examples of operon-like genes in Drosophila, the CheB42a-llz locus is the best-characterized case of a polycistronic mRNA. The CheB42a-llz RNA, which encodes two proteins, is subsequently cleaved into two transcripts, which appear to be translated independently by a cap-independent process [29]. Elucidating how the Gr64 transcripts are processed post-transcriptionally and by which mechanism they are translated will be a challenging undertaking, especially if such processing only takes place in the correct cellular context (i.e. taste neurons). In any case, polycistronic, operon-like transcription of the Gr64 genes would provide an elegant solution for their coordinated expression in the same subset of sugar-responsive taste neurons.
EXPERIMENTAL PROCEDURES
Deletion of the Gr64 gene cluster
Insertion lines f03449 and d06001 were obtained from the Bloomington and Harvard stock centers, and used to generate the Gr64 deletion using an approach previously described by Parks et al. (2004). Briefly, insertion line f03449 was crossed to a line carrying a hsp:FLP insertion, and the resulting progeny were crossed to the d06001 insertion line. Then, the hsp:FLP insertion was activated in the progeny of the second cross by heat-shock, which resulted in deletion of the Gr64 genes through recombination between the FRT sites contained in the piggyBac insertions. Deletions were detected using a pair of primers that anneal within the piggyBac elements, and then confirmed using a second pair of primers that anneal within the genomic sequences flanking the site of the deletion. Both PCR products were sequenced and confirmed that the end points of the deletion coincided with the insertions sites of f03449 and d06001. The primers used for the detection of Gr64 deletions, and a control DNA (tubulin gene) were as follows:
| Primer11 | GACGCATGATTATCTTTTACGTGAC |
| Primer12 | AATGATTCGCAGTGGAAGGCT |
| Primer13 | GGAAAGTGGCGGCGGTGGGTGGAGGCC |
| Primer14 | CTGATCGCAACTAGTTGAGGGGATTCG |
| PrimerT1 | CCTTGTCGCGTGTGAAACACTTCC |
| PrimerT2 | GATAGCCTCGTTGTCGACCATGAA |
Transgenic Constructs
The genes flanking the Gr64 cluster that were also deleted during the excision process were rescued using genomic fragments digested out of BAC clone RPCI98-9C2 (Roswell Park Cancer Institute Drosophila BAC Library). A 14.2 kb EcoRI fragment containing fd64A, CG1134, and CG11594 was cloned into pUAST (Construct R2), and a 15.3 kb NsiI fragment containing CG11593 and CG1135 was cloned into the PstI site of pCaspR4 (Construct R1). In addition, the Gr64 rescue construct was derived from the same BAC clone and subcloned into the NotI and Acc65I sites of pUAST. The coding sequence of Gr64e was replaced with that of egfp (PEGFP-N3; Clontech Inc.) by recombinant PCR. All constructs were injected into w1118 embryos, and transgenic flies were recovered according to standard procedures.
RT-PCR
PCR from genomic DNA was performed using the TAKARA LA PCR kit. RT-PCR was performed on DNase-treated RNA samples using the One-Step RT-PCR kit (Invitrogen), according to the manufacturer’s protocol. Each RT-PCR product derived from total head RNA was cloned into the TOPO cloning vector and sequenced in order to prove that the PCR product represents a spliced cDNA product. In all cases, the PCR products were either completely or partially spliced. The relative position of each primer used for RT-PCR is shown in Figure 3A. The nucleotide sequence of each primer is as follows:
| Primer1 | CCACACAGACAGTGGCACTTTC |
| Primer2 | GGAAGCAGGCCGTAGATTTGTG |
| Primer3 | CGAAAGATTGTCACAGCCTTGAGG |
| Primer4 | GATAGCGCACAGTCCACGATG |
| Primer5 | CCCACTGAGTTTTGGTGCGTG |
| Primer6 | GCGCTGTTTCCCGGATGATATG |
| Primer7 | GTTGTCTGGACTGATTCTGGTCTGC |
| Primer8 | GCTTGATGGCTTCCTGGAAAGATC |
| Primer9 | GATGAGTCCAAGCGACCACTGG |
| Primer10 | CCTCCTTATCGCTACGAGACAGC |
Behavioral Experiments and Statistical Analysis
Proboscis Extension Response (PER) assays were performed using a modified version of the protocol described in Wang et al. (2004). Flies were collected on the day of eclosion, and allowed to feed in food vials for 2–5 days. Flies were starved for approximately 28–30 hours at 22 to 24° C, and mounted the following afternoon. During the mounting process, flies were immobilized using ice rather than carbon dioxide. Mounted flies were allowed to recover from the ice treatment for two to four hours in a humidified chamber, and then tested for sensitivity to various tastants using the PER assay.
During the assay, each fly was first administered water and allowed to drink until satiation. Only flies that responded to water were used to calculate PER. Each fly was tested with a given tastant by briefly applying the taste solution to the fly’s labellum and recording whether or not the animal extended its proboscis. Each tastant was applied three times per fly, and the flies were given water between each application of a taste solution. Error bars represent +/− SEM, and statistical significance was calculated using Student’s t-test (assuming unequal variance).
Supplementary Material
Supplementary Figure S1: Sequence conservation of the sugar receptors in Drosophila melanogaster
(A) Alignment of the eight putative sugar receptors. Alignment was constructed with the Multiple Alignment feature of MacVector, using a BLOSUM series matrix and the default parameter settings.
(B) Evolutionary relationship between GR proteins. The phylogenetic tree was generated in MEGA4, using the Neighbor-Joining method [32]. Bootstrap values are indicated next to the branches. The eight sugar receptors are at the bottom (indicated by a bracket) and have bootstrap support of 100%.
Supplementary Figure S2: Flies with R1 and R2 transgenes show normal PER response. The graph shows PER responses for three different sugars of flies containing the two transgenes R1 and R2 and a wild type copy of the Gr64 gene (R1/+;R2/+;ΔGR64/+), in comparison to control flies (see Figure 2), as well as homozygous ΔGR64 mutants. R1/+;R2/+;ΔGR64/+ flies show normal response to these sugars, compared to the highly reduced or lost response in mutants. At 500 mM, the response of R1/+;R2/+;ΔGR64/+ flies is the same as that observed in the control strain, while at 100 mM, the response appears slightly higher. Asterisks indicate a significant difference between the mutant and control strains, as determined by Student’s t-test (* indicates p<0.05, *** indicates p<0.0001).
Supplementary Figure S3: Expression of the Gr64 genes in flies containing the rescue Gr64 construct
Flies containing the UAS-Gr64abcd_GFP_f transgene express the Gr64 genes, both in the absence (lanes 1) and presence (lanes 2) of the Gr5a-Gal4 driver. In lanes 3, RNA from homozygous ΔGr64a mutants flies (R1/+;R2/+;ΔGR64/ΔGR64) was loaded. RT-PCR analysis of RNA isolated from heads and legs (not shown) was carried out for the first four genes. Integrity of cDNA was confirmed using primers against the tubulin gene. The same primers were used as in Figure 1D (1–6, T1 and TR2).
Acknowledgments
This work was supported by a grant from the NIH (GM-DC05606-01) to HA. JS was supported by an NSF graduate research fellowship. We thank Anupama Dahanukar for communicating results prior to publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wang Z, Singhvi A, Kong P, Scott K. Taste representations in the Drosophila brain. Cell. 2004;117:981–991. doi: 10.1016/j.cell.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Thorne N, Chromey C, Bray S, Amrein H. Taste perception and coding in Drosophila. Current Biology. 2004;14:1065–1079. doi: 10.1016/j.cub.2004.05.019. [DOI] [PubMed] [Google Scholar]
- 3.Scott K, Brady R, Cravchik A, Morozov P, Rzhetsky A, Zuker C, Axel R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell. 2001;104:661–673. doi: 10.1016/s0092-8674(01)00263-x. [DOI] [PubMed] [Google Scholar]
- 4.Dunipace L, Meister S, McNealy C, Amrein H. Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Current Biology: Cb. 2001;11:822–835. doi: 10.1016/s0960-9822(01)00258-5. [DOI] [PubMed] [Google Scholar]
- 5.Clyne PJ, Warr CG, Carlson JR. Candidate taste receptors in Drosophila. Science. 2000;287:1830–1834. doi: 10.1126/science.287.5459.1830. [DOI] [PubMed] [Google Scholar]
- 6.Bray S, Amrein H. A putative Drosophila pheromone receptor expressed in male-specific taste neurons is required for efficient courtship. Neuron. 2003;39:1019–1029. doi: 10.1016/s0896-6273(03)00542-7. [DOI] [PubMed] [Google Scholar]
- 7.Moon SJ, Kottgen M, Jiao Y, Xu H, Montell C. A taste receptor required for the caffeine response in vivo. Curr Biol. 2006;16:1812–1817. doi: 10.1016/j.cub.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Ueno K, Ohta M, Morita H, Mikuni Y, Nakajima S, Yamamoto K, Isono K. Trehalose sensitivity in Drosophila correlates with mutations in and expression of the gustatory receptor gene Gr5a. Current Biology: Cb. 2001;11:1451–1455. doi: 10.1016/s0960-9822(01)00450-x. [DOI] [PubMed] [Google Scholar]
- 9.Dahanukar A, Foster K, van der Goes van Naters WM, Carlson JR. A Gr receptor is required for response to the sugar trehalose in taste neurons of Drosophila. Nat Neurosci. 2001;4:1182–1186. doi: 10.1038/nn765. [DOI] [PubMed] [Google Scholar]
- 10.Stocker RF. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell and Tissue Research. 1994;275:3–26. doi: 10.1007/BF00305372. [DOI] [PubMed] [Google Scholar]
- 11.Amrein H, Thorne N. Gustatory perception and behavior in Drosophila melanogaster. Curr Biol. 2005;15:R673–684. doi: 10.1016/j.cub.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Robertson HM, Warr CG, Carlson JR. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2003;100(Suppl 2):14537–14542. doi: 10.1073/pnas.2335847100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16:1395–1403. doi: 10.1101/gr.5057506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development (Cambridge, England) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 15.Hiroi M, Marion-Poll F, Tanimura T. Differentiated response to sugars among labellar chemosensilla in Drosophila. Zoolog Sci. 2002;19:1009–1018. doi: 10.2108/zsj.19.1009. [DOI] [PubMed] [Google Scholar]
- 16.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, Ryner L, Cheung LM, Chong A, Erickson C, Fisher WW, Greer K, Hartouni SR, Howie E, Jakkula L, Joo D, Killpack K, Laufer A, Mazzotta J, Smith RD, Stevens LM, Stuber C, Tan LR, Ventura R, Woo A, Zakrajsek I, Zhao L, Chen F, Swimmer C, Kopczynski C, Duyk G, Winberg ML, Margolis J. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 17.Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- 18.Dethier VG. The Hungry Fly. Cambridge, MA: Harvard University Press; 1976. [Google Scholar]
- 19.Koseki T, Koganezawa M, Furuyama A, Isono K, Shimada I. A specific receptor site for glycerol, a new sweet tastant for Drosophila: structure-taste relationship of glycerol in the labellar sugar receptor cell. Chem Senses. 2004;29:703–711. doi: 10.1093/chemse/bjh075. [DOI] [PubMed] [Google Scholar]
- 20.Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol. 2006;4:e20. doi: 10.1371/journal.pbio.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 22.Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature. 2007;445:86–90. doi: 10.1038/nature05466. [DOI] [PubMed] [Google Scholar]
- 23.Suh GS, Wong AM, Hergarden AC, Wang JW, Simon AF, Benzer S, Axel R, Anderson DJ. A single population of olfactory sensory neurons mediates an innate avoidance behaviour in Drosophila. Nature. 2004;431:854–859. doi: 10.1038/nature02980. [DOI] [PubMed] [Google Scholar]
- 24.Kwon JY, Dahanukar A, Weiss LA, Carlson JR. The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci U S A. 2007;104:3574–3578. doi: 10.1073/pnas.0700079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, Kim SK. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–854. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- 26.Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- 27.Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray A, van Naters WG, Shiraiwa T, Carlson JR. Mechanisms of odor receptor gene choice in Drosophila. Neuron. 2007;53:353–369. doi: 10.1016/j.neuron.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Shahar Y, Nannapaneni K, Casavant TL, Scheetz TE, Welsh MJ. Eukaryotic operon-like transcription of functionally related genes in Drosophila. Proc Natl Acad Sci U S A. 2007;104:222–227. doi: 10.1073/pnas.0609683104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. Mammalian sweet taste receptors. Cell. 2001;106:381–390. doi: 10.1016/s0092-8674(01)00451-2. [DOI] [PubMed] [Google Scholar]
- 31.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell. 2003;115:255–266. doi: 10.1016/s0092-8674(03)00844-4. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1: Sequence conservation of the sugar receptors in Drosophila melanogaster
(A) Alignment of the eight putative sugar receptors. Alignment was constructed with the Multiple Alignment feature of MacVector, using a BLOSUM series matrix and the default parameter settings.
(B) Evolutionary relationship between GR proteins. The phylogenetic tree was generated in MEGA4, using the Neighbor-Joining method [32]. Bootstrap values are indicated next to the branches. The eight sugar receptors are at the bottom (indicated by a bracket) and have bootstrap support of 100%.
Supplementary Figure S2: Flies with R1 and R2 transgenes show normal PER response. The graph shows PER responses for three different sugars of flies containing the two transgenes R1 and R2 and a wild type copy of the Gr64 gene (R1/+;R2/+;ΔGR64/+), in comparison to control flies (see Figure 2), as well as homozygous ΔGR64 mutants. R1/+;R2/+;ΔGR64/+ flies show normal response to these sugars, compared to the highly reduced or lost response in mutants. At 500 mM, the response of R1/+;R2/+;ΔGR64/+ flies is the same as that observed in the control strain, while at 100 mM, the response appears slightly higher. Asterisks indicate a significant difference between the mutant and control strains, as determined by Student’s t-test (* indicates p<0.05, *** indicates p<0.0001).
Supplementary Figure S3: Expression of the Gr64 genes in flies containing the rescue Gr64 construct
Flies containing the UAS-Gr64abcd_GFP_f transgene express the Gr64 genes, both in the absence (lanes 1) and presence (lanes 2) of the Gr5a-Gal4 driver. In lanes 3, RNA from homozygous ΔGr64a mutants flies (R1/+;R2/+;ΔGR64/ΔGR64) was loaded. RT-PCR analysis of RNA isolated from heads and legs (not shown) was carried out for the first four genes. Integrity of cDNA was confirmed using primers against the tubulin gene. The same primers were used as in Figure 1D (1–6, T1 and TR2).