Abstract
Interstrand cross-link (ICL) is a covalent modification of both strands of DNA, which prevents DNA strand separation during transcription and replication. Upon photoactivation 8-methoxypsoralen (8-MOP+UVA) alkylates both strands of DNA duplex at the 5,6-double bond of thymidines, generating monoadducts (MAs) and ICLs. It was thought that bulky DNA lesions such as MAs are eliminated only in the nucleotide excision repair pathway. Instead, non-bulky DNA lesions are substrates for DNA glycosylases and AP endonucleases which initiate the base excision repair (BER) pathway. Here we examined whether BER might be involved in the removal of psoralen–DNA photoadducts. The results show that in human cells DNA glycosylase NEIL1 excises the MAs in duplex DNA, subsequently the apurinic/apyrimidinic endonuclease 1, APE1, removes the 3′-phosphate residue at single-strand break generated by NEIL1. The apparent kinetic parameters suggest that NEIL1 excises MAs with high efficiency. Consistent with these results HeLa cells lacking APE1 and/or NEIL1 become hypersensitive to 8-MOP+UVA exposure. Furthermore, we demonstrate that bacterial homologues of NEIL1, the Fpg and Nei proteins, also excise MAs. New substrate specificity of the Fpg/Nei protein family provides an alternative repair pathway for ICLs and bulky DNA damage.
INTRODUCTION
Interstrand cross-links (ICLs) are highly lethal DNA lesions which block DNA transcription and replication by preventing strand separation. Due to their high cytotoxicity, ICL-inducing agents like mitomycin C (MMC), cisplatin and psoralens are widely used against hyperplasic diseases such as cancer and psoriasis. Furanocoumarins (psoralens) are naturally occurring secondary metabolites in plants, including many fruits and vegetables. Among several ICL-inducing agents, psoralens require UVA-photoactivation following DNA intercalation to chemically react with DNA. 8-methoxypsoralen (8-MOP) is a planar, tricyclic compound which intercalates into DNA duplex at pyrimidines, preferentially at 5′-TpA sites. Upon photoactivation, 8-MOP primarily photoalkylates DNA by cycloaddition to the 5,6-double bond of a thymidine, generating two types of monoadducts (MA) with either the 4′,5′-double bond of the furan (MAf) or the 3,4-double bond of the pyrone (MAp) side of the psoralen (1). A unique property of psoralen photochemistry is that the absorption of a second photon by the MAf leads to formation of a pyrone side 5,6-double bond adduct with a flanking thymine in the complementary strand, thus generating an ICL (2). Although the yield of psoralen MAs to pyrimidine bases is 3-fold higher than that of ICLs, the latter class of damage appears to have more severe biological effect (3).
Genetic and biochemical data indicate that elimination of ICLs is mainly linked to DNA replication and involves several linear repair pathways including structure-specific endonucleases, proteins required for homologous recombination-mediated double-strand break (DSB) repair and error-prone translesion DNA polymerases (4–9). The thirteen FANC proteins, mutated in the Fanconi anaemia cancer-prone syndrome, participate in coordination of excision repair of ICLs in order to reconstitute the genetic material with high fidelity. However, at present, the mechanism of coordination and biochemical activities of the FANC proteins involved in the excision of ICLs remains poorly defined. More importantly, it was believed that bulky DNA lesions such as thymine–psoralen adducts can be eliminated only by the nucleotide excision repair (NER) pathway (10). However, with the exception of XPF/ERCC1-deficient cells, cells lacking critical NER components only show a moderate sensitivity to ICL-inducing agents exposure which induces both MA and ICL (11). These observations hint us at the existence of an alternative NER-independent repair pathway for ICLs and/or bulky DNA adducts.
Non-bulky endogenous oxidative DNA lesions generated by reactive oxygen species (ROS) are substrates for two overlapping pathways: base excision repair (BER) and nucleotide incision repair (NIR). The BER pathway is initiated by a DNA glycosylase cleaving the N-glycosylic bond between the abnormal base and deoxyribose, leaving either an abasic site or a DNA single-strand break which in turn is repaired by an apurinic/apyrimidinic (AP) endonuclease (12). Interestingly, it has been shown that mammalian polynucleotide kinase (PNK) can substitute AP endonuclease in removal of 3′-blocking phosphates produced by certain DNA glycosylases (13,14). In human cells, four DNA glycosylases/AP lyases OGG1, NTH1, NEIL1 and NEIL2, excise the majority of oxidized bases (14–17). Alternatively, in the NIR pathway, the major human AP endonuclease 1, APE1, incises oxidatively damaged DNA in a DNA glycosylase-independent manner (18). These two pathways can work in concert to cleanse genomic DNA from 7,8-dihydro-8-oxoguanine (8oxoG) residues, one of the major ROS-induced DNA lesions (19). Previously, it was shown that antisense APE1-RNA human cell transfectants exhibited hypersensitivity to MMC, suggesting a potential involvement of APE1 in ICLs repair (20). Furthermore, similar to FA cells, Ape1-depleted cells were sensitive to hyperoxia. APE1 is involved in both BER and NIR suggesting that these pathways may be involved in repair of ICLs.
Human homologues of the Escherichia coli oxidized base-specific DNA glycosylase endonuclease VIII (Nei) were identified by database search of the genome and named NEIL1, 2 and 3 (human Nei-like proteins 1, 2 and 3) (16,21). The Nei-like proteins share significant sequence similitude with the Fpg protein, an 8oxoG-DNA glycosylase found in E. coli. Fpg/Nei/NEIL are structurally and catalytically a distinct family of bi-functional DNA glycosylases endowed with an AP lyase activity that incises DNA at abasic sites by a β,δ-elimination mechanism and leaving single-strand break carrying a phosphate residue at the 3′ and 5′-terminus (22,23). Escherichia coli mutants deficient in both DNA glycosylases, endonuclease III (nth) and nei show spontaneous mutator phenotype and are hypersensitive to the lethal effects of oxidizing agents (24,25). Moreover, mouse embryonic stem cell lines deficient in NEIL1 protein were twice more sensitive than control cells to low doses of γ-irradiation (26). Indeed, NEIL1 initiates base excision repair of adenine-derived formamidopyrimidines and 5S,6R stereoisomers of thymine glycol (Tg) which are not excised by OGG1 and NTH1 (21,27). Recently, it has been demonstrated that NEIL1 can excise an oxidized base located in the proximity to single-strand break suggesting its involvement in repair of clustered DNA lesions induced by ionizing radiation (28,29). Interestingly, S phase specific expression of NEIL1 and excision of base lesions in single-stranded and bubble-structured DNA substrates point to its possible involvement in replication-dependent repair (21,30).
The purpose of this study was to search for alternative repair pathway(s) involved in removal of ICLs and bulky psoralen–DNA photoadducts. Using a short cross-linked oligonucleotide duplex, we demonstrated that human DNA glycosylase and AP endonuclease initiate alternative repair pathway for bulky psoralen–DNA photoadducts which sought to be substrates for the NER machinery. The potential biological importance of these findings is discussed.
MATERIALS AND METHODS
Reagents and oligonucleotides
Chemical reagents including 8-MOP were purchased from Sigma-Aldrich. All oligonucleotides were purchased from Eurogentec (Seraing, Belgium), including regular oligonucleotides, siRNAs and those containing Uracil, 5,6-dihydrouracil (DHU), Tg and 3′-terminal phosphate (3′-P). A 21-mer oligonucleotide GFP1, d(GCTCTCGTCTGXACACCGAAG), where X is either T, U or Tg, and complementary ones GFP2, d(GCTCTTCGGTGTACAGACGAG) and GFP3, d(CTTCGGTGTACAGACGAGagc) were hybridized to obtain duplexes referred to as GFP1·2 and GFP1·3, respectively (Figure 1C). These sequence contexts were previously used to study the repair of psoralen-induced ICLs in human cells (31). The following oligonucleotides were used to measure 3′-phosphatase activity: 3′P-Exo10, d(TGACTGCATAp), and complementary 30-mer d(ATGCACATCGTCTACATGCGTATGCAGTCA). Classic AP site substrate was prepared by treating uracil containing oligonucleotide with E. coli Uracil-DNA glycosylase (Ung). Oligonucleotides were either 5′ or 3′-end labelled and annealed as previously described (18). Oligonucleotides carrying 3′-P residue were 5′-end labelled using 3′-phosphatase-free T4 polynucleotide kinase from Roche Diagnostics GmbH (Meylan, France).
Figure 1.
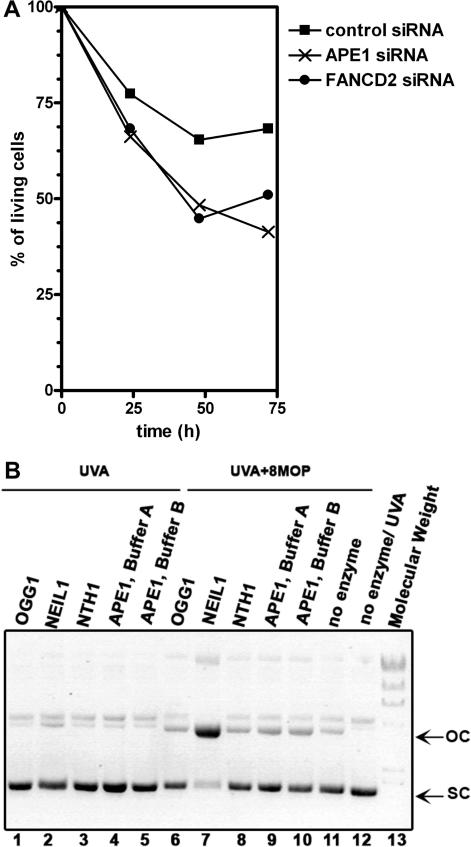
Role of the BER pathway in counteracting the lethal effect of 8MOP+UVA exposure. (A) Sensitivity of siRNA-treated HeLa cells to DNA damaging agents. HeLa cells were transfected with control, APE1 and FA-D2 siRNAs. Seventy-two hour after transfection, cells were exposed with 8MOP+UVA and cell viability was determined 24, 48 and 72 h after treatment. The figure represents the mean of three independent experiments. The results are expressed as the percentage of untreated cells transfected with the control siRNA 24 h after stimulation. (B) Excision of the 8-MOP+UVA induced DNA damage by purified DNA repair enzymes. The pUC18 plasmid DNA was irradiated in the presence or without 8-MOP and then incubated with APE1 in buffer containing 5 mM MgCl2 (lanes 4 and 9) or 0.5 mM MgCl2 (lanes 5 and 10) and DNA glycosylases OGG1, NEIL1, NTH1 in buffer containing 1 mM EDTA. Lanes 1–5, 12, UVA-irradiated plasmid; lanes 6–11, 8-MOP+UVA treated plasmid; lane 13, lambda DNA-Hind III digest. ‘oc’ indicates open circular plasmid and ‘sc’ supercoiled plasmid DNA.
The sequences of siRNA used to decrease APE1 in cells were previously described (32). Sequences of control siRNA were 5′ (ACUAUGUAUAGGAGUACGCTT)3′ and 3′ (TTUGAUACAUAUCCUCAUGCG)5′. Human FANCD2 and NEIL1 smart pool siRNAs were obtained from Dharmacon (Perbio Science, France).
Bacterial strains and proteins
Escherichia coli strains AB 1157 (IeuB6 thr-1 Δ(gpt-proA2) hisG4 argE3 lacY1 gaIK2 ara-14 mtl-1 xyl-5 thi-1 tsx-33 rpsL31 supE44 rac) (WT) and its isogenic derivatives BH20 (fpg::kanR) and BH110 (nfo::kanR [Δ(xth-pncA)90 X::Tn10]) were from the laboratory stock. DNA glycosylase deficient E. coli strains were constructed by insertional mutagenesis as described (33). Briefly, to construct fpg nei double mutant, the spectinomycine resistance cassette (SpcR) was inserted into NEI gene and transferred to the E. coli BH20 chromosome by recombination. Cells acquiring the selectable marker were selected and the presence of SpcR gene insertion was further confirmed by PCR of the genomic DNA. Single clone containing inactivated nei::SpcR and fpg::KanR genes (MS2000) was chosen for further work. Strains AB2480 (isogenic to AB1157 except uvrA6 recA13) and SW2-8 (isogenic to BW35 (KL16) except nei::camR) were gifts from Dr A. K. McCullough (Oregon Health & Science University, Portland, OR) and Dr S. S. Wallace (University of Vermont, Burlington, VT), respectively.
T4 DNA ligase was purchased from Roche Diagnostics GmbH. The purified E. coli Ung, Nfo, Fpg, Nth, AlkA and human OGG1, ANPG and NTH1 proteins were from laboratory stock. Human Ape1 protein was expressed and purified from E.coli BH110 (DE3) strain to avoid cross-contamination of bacterial AP endonucleases as described (34). The expression vectors phNEIL1 and phNEIL2 (27), the purified E. coli Nei protein (35) and human DNA polymerase β (36) were generously provided by Dr Hiroshi Ide (Hiroshima University, Japan), Dr Dmitry Zharkov (ICBFM, Novosibirsk, Russia) and Dr Grigory Dianov (MRC, Harwell, Oxfordshire, UK), respectively. The full-length native NEIL1 and NEIL2 were purified as described previously (27).
Preparation of psoralen mono- and diadducts
Five microgram of plasmid DNA pUC18 or 10 pmol of 32P-labelled GFP1·2 were incubated for 15 min in the dark with 0.1 mM 8-MOP in 50 µl of 100 mM Tris-HCl, pH 7.5, 5 mM EDTA and 50 mM NaCl, then irradiated at 365 nm and 240 kJ/m2 at room temperature. Plasmid DNA was purified by ethanol precipitation and the presence of ICLs was verified by denaturing agarose gel electrophoresis. GFP1·2 was desalted by spin-down columns filled with water equilibrated Sephadex G-50. Cross-linked and non-cross-linked oligonucleotides were separated by denaturing 20% PAGE. The oligonucleotides were eluted from the gel strips in 2 M LiClO4 and then acetone precipitated. To obtain MAp residue, the purified cross-linked GFP1·2 was treated with hot alkali (37).
DNA glycosylase assays
The standard reaction mixture (20 µl) for DNA glycosylase activity contained either 0.1 µg of pUC19 or 10 nM of 5′-[32P] or 3′-[32P]ddAMP-labelled GFP1·2, 25 mM HEPES-KOH, pH 7.6, 100 mM KCl, 1 mM EDTA, 5 mM 2-mercaptoethanol and 6% glycerol, unless otherwise stated. The assay mixture for the Fpg, Nei and OGG1 proteins contained 25 mM HEPES (pH 7.6), 100 mM KCl, 2 mM EDTA, 5 mM 2-mercaptoethanol and 6% glycerol. For Nfo and AlkA, the assay mixture was 20 mM HEPES-KOH, pH 7.6, 50 mM KCl, 0.1 mM EDTA, 0.1 mg/ml BSA, 1 mM DTT, for Nth and NTH1 the same but with 1 mM EDTA, for ANPG 70 mM HEPES-KOH, pH 7.6, 50 mM KCl, 1 mM EDTA, 0.1 mg/ml BSA and 5 mM 2-mercaptoethanol. APE1 assay conditions vary depending on the DNA repair pathway studied as described (18). Assays were performed with 5–20 nM of pure protein at 37°C for 10 min, unless otherwise stated. Reaction with pUC18 was stopped by adding 5 µl of 0.25% bromophenol blue, 50% glycerol and 10 mM EDTA and the products were analysed by 0.8% agarose gel electrophoresis (0.5× TBE). Reactions with the oligonucleotides were stopped by adding 10 µl of 0.5% SDS and 5 mM EDTA and analysed as previously described (18). Purified reaction products were heated at 65°C for 3 min and separated by electrophoresis in denaturing 20% (w/v) polyacrylamide gels (7 M Urea, 0.5× TBE). Gels were exposed to a Fuji FLA-3000 Phosphor Screen and analysed using Image Gauge V3.12 software.
For the reconstitution of the repair pathway of MAs in vitro, 20 nM of GFP1·3 with a single MAp was incubated in the presence of 20 nM NEIL1, 20 nM APE1, 10 nM DNA polymerase β and 2 U T4 DNA ligase in buffer (20 µl) containing 5 µCi of [α-32P]dTTP, 20 mM HEPES-KOH, pH 7.6, 50 mM KCl, 0.1 mg/ml BSA, 1 mM DTT, 2 mM ATP and 5 mM MgCl2 for 10 min at 37°C and then 20 min at 30°C. Reaction products were analysed as described above.
Formation of enzyme–DNA covalent complexes
The OGG1, NTH1 and NEIL1 proteins (200 nM) were incubated with 0.2 pmol of 8-MOP+UVA-treated 5-[32P]-labelled GFP1•2 at 37°C for 30 min in 20 µl of the standard reaction mixture but in the presence of 50 mM NaBH4 (a 2 M NaBH4 stock solution in water was prepared immediately prior to use) or 100 mM NaCl as a control. The reaction was stopped by adding 5 µl of buffer containing 10% SDS, 30% glycerol, 25% 2-mercaptoethanol, 0.1% bromophenol blue and 300 mM Tris-HCl, pH 6.8 and heating 10 min at 60°C. The reaction products were separated by 15% SDS-PAGE and analysed as described above.
Cell culture, drug treatment and survival assay
All strains of E. coli were grown with shaking at 37°C to 3 × 108 cells/ml in LB medium. Cells were centrifuged and resuspended in M9 medium at a density 5 × 108 cells/ml before irradiation. When appropriate, 8-MOP was added to a final concentration of 5 µM, and the suspension was allowed to stand on ice for 10 min in the dark before irradiation. Samples were irradiated at room temperature with HPW125 Philips lamp with a pyrex water filter. The fluence through the sample was 1 mW/cm2. All experiments were carried out at least three times.
HeLa cells were routinely grown at 37°C in 5% CO2 in Dulbecco minimal essential medium supplemented with 10% foetal calf serum, 2 mM glutamine, 100 U/ml of penicillin and 100 mg/ml of streptomycin. To decrease the target protein level, HeLa cells were plated in 6-well plates at the concentration of 180 000 cells/well and 24 h later were transfected with 100 nM of appropriate siRNA using Oligofectamine (Invitrogen) as described (38).
Cells were seeded into 96-well plate at the concentration of 104 cells/well in complete medium 72 h after siRNA transfection. ICL induction by photoactivated psoralen was achieved by incubating cells with 10 μM of 8-MOP for 20 min followed by exposure to 10 kJ/m2 UVA, afterwards cells were cultivated at 37°C in complete medium for increasing periods of time. Cell viability was assessed using Cell Proliferation Kit II (XTT) (Roche Diagnostics GmbH) according to the manufacturer instructions.
For immunoblotting cells were sonicated in Laemmli loading buffer. Total proteins were quantified by Bradford assay, and separated by SDS-PAGE, then electroblotted onto a nitrocellulose membrane (BA85, Schleicher & Schull). The membranes were saturated in 5% non-fat dry milk in PBS for 1 h at room temperature before adding primary antibodies for overnight at 4°C. The following antibodies were used: NEIL1 (1:600; Abcam), actin (1:500), APE1 (1:5000; Eurogentec, Belgium). Binding of the primary antibodies was detected with HRP-coupled secondary antibodies followed by enhanced chemiluminescence detection (GE Healthcare) and visualized with a GeneGnome gel documentation system (Syngene, MD, USA).
RESULTS
Role of the base excision repair proteins in the cellular response to DNA cross-linking agents
APE1−/− cells are sensitive to MMC, however, this cross-linking agent generates a variety of oxidative and non-oxidative DNA lesions and only 10% of them are ICLs (6). Consequently, to examine the potential involvement of APE1 in ICLs processing, we used exposure to photoactivated 8-MOP that generates distinct, well-characterized and easily detectable DNA adducts. The sensitivity of HeLa cells with reduced expression of APE1 and FANCD2 towards several genotoxins was examined. In agreement with the previous observations, HeLa cells transfected with Ape1-specific siRNA were highly sensitive to ionizing radiation (IR) and MMC but not to UVC treatments (data not shown). Strikingly, the cells with decreased Ape1 level were also sensitive to 8-MOP+UVA exposure, like cells with reduced expression of FANCD2 protein, suggesting a potential involvement of Ape1 in the processing of psoralen–DNA adducts (Figure 1A).
The sensitivity of APE1-depleted HeLa cells to cross-linking agents suggest that APE1-catalysed activities may remove psoralen-DNA adducts and thus perform function similar to that of the structure-specific endonuclease XPF-ERCC1 (8). In fact, in the NIR pathway APE1 can incise the DNA sugar-phosphate backbone 5′ next to bulky lesions such as 3,N4-benzetheno-2′-°deoxynucleotides (39), thus providing a back-up function to NER pathway. Furthermore, in the BER pathway, APE1 acts downstream of various DNA glycosylases to eliminate genotoxic repair intermediates. Thus depletion of APE1 removes the major AP and NIR endonuclease and 3′-repair diesterase activities in human cells, severely disabling two repair pathways. The first approach was to study whether the purified BER proteins cleave DNA cross-links and for this purpose 8-MOP+UVA treated supercoiled (sc) plasmid DNA was incubated with APE1 and human DNA glycosylases NTH1, NEIL1 and OGG1. Upon cleavage at the site of damage by a DNA repair enzyme, the sc form is converted to an open circular (oc) form and these two forms are identified by electrophoresis in agarose gel (40). As shown in Figure 1B, only NEIL1 converts the sc form to oc one, indicating that it specifically recognizes and incises photoactivated psoralen-induced DNA damage (lane 7).
Oxidative DNA glycosylase, NEIL1, excises psoralen-thymine MAs present in duplex DNA
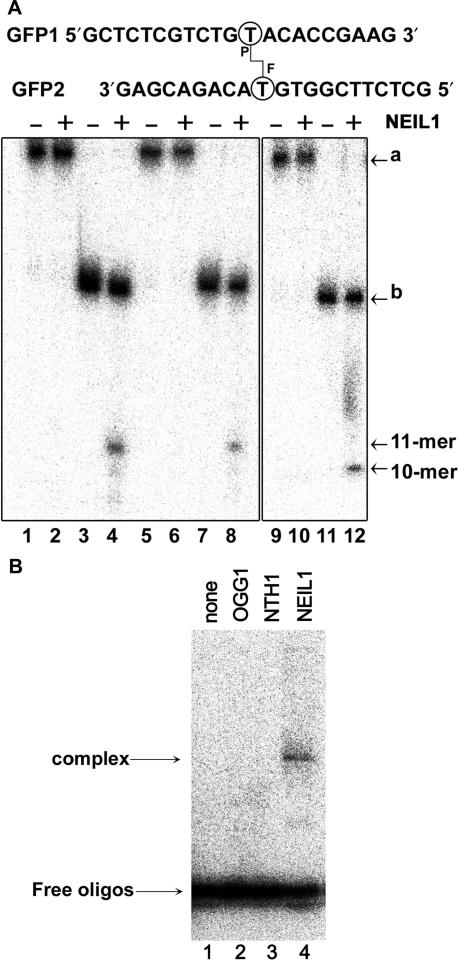
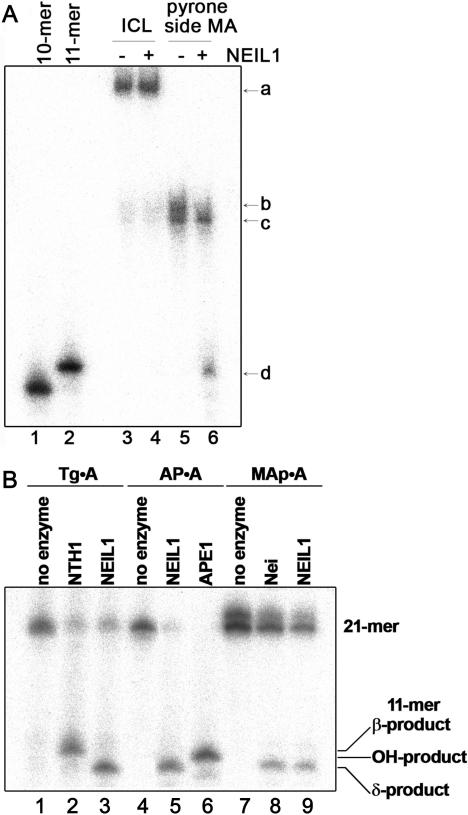
To further examine whether NEIL1 excises MAs or ICLs or both, the short 21-mer 5′-[32P] and 3′-[32P]-dCMP-labelled oligonucleotide duplexes, GFP1·2, were treated with 8-MOP+UVA and the cross-linked oligonucleotides were separated from non-cross-linked ones by denaturing PAGE and used as DNA substrates. Interestingly, NEIL1 cleaves only non-cross-linked GFP1·2 (Figure 2A, lanes 4, 8 and 12). The 5′ and 3′-labelled cleavage fragments migrating at position corresponding to an 11-mer (lanes 4 and 8) and 10-mer (lane 12), respectively, indicate excision of a psoralen–thymine MA at the position 12 of the 21-mer oligonucleotide. Consistent with the established mechanism of action of bi-functional DNA glycosylases/AP lyases, NEIL1 generates a Schiff base intermediate that can be characterized by the covalent trapping of the enzyme–substrate complex by reduction with NaBH4 (Figure 2B, lane 4) (41). To characterize the substrate specificity of NEIL1, an oligonucleotide containing single MAp was constructed by incubating cross-linked GFP1·2 under hot-alkali conditions (42). As expected, base-catalysed reversal of the cross-link yields two closely migrating DNA fragments, the ‘faster migrating’ native and ‘slow migrating’ MAp-containing oligonucleotides (Figure 3A, lane 5). Appearance of an 11-mer fragment (lane 6) upon incubation with NEIL1 was accompanied by the loss of MAp-containing fragment indicating that NEIL1 excises MAp residues present in duplex DNA. Furthermore, NEIL1 cleaves GFP1·2 irradiated at 405 nm in the presence of 8-MOP suggesting that it also excises MAfs (data not shown).
Figure 2.
The BER pathway removes psoralen-induced monoadducts. (A) NEIL1 is active on 8-MOP+UVA-induced MAs. 5′-[32P] (lanes 1–4) and 3′-[32P]-dCMP-labelled GFP1 (lanes 9–12) and 5′-[32P]-labelled GFP2 (lanes 5–8) were treated with 8-MOP+UVA. Cross-linked GFP1·2 (lanes 1, 2, 5, 6, 9 and 10) was purified from non-cross-linked GFP1·2 (lanes 3, 4, 7, 8, 11 and 12) and incubated or not with NEIL1. ‘a’ indicates cross-linked oligonucleotide duplex, ‘b’ non-cross-linked oligonucleotide duplex containing single MA. (B) The AP lyase activity of NEIL1 on GFP1·2 containing psoralen MA. Purified non-cross-linked 5′-[32P]-labelled GFP1·2 was incubated with 200 nM of OGG1, NTH1 and NEIL1 in presence of NaBH4 at 37°C for 30 min before separation of the DNA–protein covalent complex and free DNA by 10% SDS-PAGE.
Figure 3.
DNA glycosylase/AP lyase activities of NEL1 on various DNA substrates. (A) NEIL1 excises MAp. Interstrand cross-linked 5′-[32P]-labelled GFP2 oligonucleotide was reversed by hot alkaline treatment and then incubated with NEIL1. Lane 1, size marker 10-mer; lane 2, 11-mer; lane 3, oligonucleotide duplex containing ICL; lane 4, as 3 but incubated with NEIL1; lane 5, oligonucleotide containing MAp; lane 6, as 5 but incubated with NEIL1. ‘a’ indicates cross-linked oligonucleotide duplex, ‘b’ oligonucleotide containing single MAp, ‘c’ oligonucleotide without MAp and ‘d’ NEIL1-incision product. (B) Nature of 3′-termini of the single-strand breaks generated by NEL1. ‘β product’ indicate 11-mer fragment carrying 3′-α,β-unsaturated aldehyde moiety, ‘OH product’ 11-mer fragment carrying 3′-OH group and ‘δ product’ 11-mer fragment carrying 3′-P residue.
To further characterize the substrate specificities of the NEIL1 protein, the kinetic constants for the excision of MAp and DHU·G were measured. Comparison of the kinetic constants (Table 1) shows that MAp is the preferred substrate for NEIL1 (KM = 6.3′nM, kcat/KM = 3.9 min−1 µM−1) which is recognized with high specificity.
Table 1.
Kinetic parameters of NEIL1 for the excision of MAp and DHU residues present in oligonucleotide duplex
| DNA substrate | KM (nM) | kcat (min−1) | kcat/KM (min−1·µM−1) |
|---|---|---|---|
| GFP1·2 | 6.3 ± 1.4 | 0.025 ± 0.002 | 3.9 |
| DHU·G | 22 ± 6.3 | 0.102 ± 0.008 | 4.6 |
NEIL1-generated DNA 3′-phosphate termini are removed by APE1
To examine the nature of 3′-termini generated by NEIL1-catalysed excision of MAp, 5′-[32P] labelled GFP1·2 duplexes containing an AP site, MAp or Tg residues were used. As shown in Figure 3B, when excising a Tg, AP site and MAp NEIL1 generates 11-mer DNA fragments migrating the same distance a result that is consistent with β,δ-elimination mechanism of action of this enzyme (lanes 3, 5 and 9). In agreement with this observation, DNA cleavage fragments generated by the NEIL1 and E. coli Nei proteins (lanes 8–9) migrate faster than products generated by NTH1 and APE1 containing 3′-α,β-unsaturated aldehyde and 3′-hydroxyl residues, respectively (lanes 2 and 6). Taken together, these results demonstrate that with all DNA substrate tested NEIL1 and Nei generate products carrying a phosphate at the 3′-terminus.
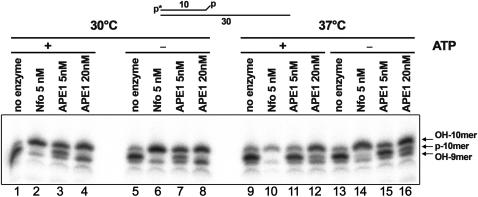
The catalytic rate of human APE1 acting on 3′-phosphoglycolates and 3′-Ps is nearly 200-fold lower than its AP endonuclease function (43). Recently, Wiederhold and colleagues (13) showed that polynucleotide kinase and not APE1 is primarily involved in the NEIL1-initiated BER pathway. To examine whether 3′-Ps are removed by APE1 under reaction conditions used, we incubated 5′-[32P] labelled 10-mer DNA fragment carrying 3′-P residue with the E. coli Nfo and varying amounts of APE1 proteins at 30 and 37°C. As shown in Figure 4, at 30°C 5 nM APE1 removes more than 50% of 3′-P (lanes 3 and 7) whereas 20 nM APE1 and 5 nM Nfo eliminate all 3′-P residues generating 10-mer fragment containing 3′-hydroxyl with lower mobility as compared to the substrate DNA (lanes 2, 4, 6 and 8). Addition of 1 mM ATP did not affect activity of enzymes. 3′-Phosphatase activity of APE1 but not that of Nfo is strongly inhibited at 37°C (lanes 10–12 and 14–16). Interestingly, APE1 at higher protein concentration exhibits significant 3′→5′ exonuclease activity generating 9-mer DNA fragment (lanes 4 and 8). These results show that at physiologically relevant protein concentration, APE1 exhibits efficient 3′-phosphatase activity.
Figure 4.
Electrophoretic mobility of DNA fragments generated by Nfo and APE1. 3′-phosphate removal by AP endonucleases was measured under various reaction conditions. The substrate (p-10-mer) and the products (OH-10 and OH-9-mer) are indicated.
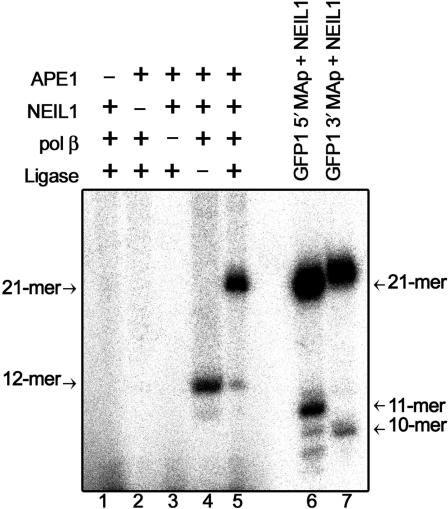
In vitro reconstitution of the BER pathway for bulky psoralen–DNA photoadducts
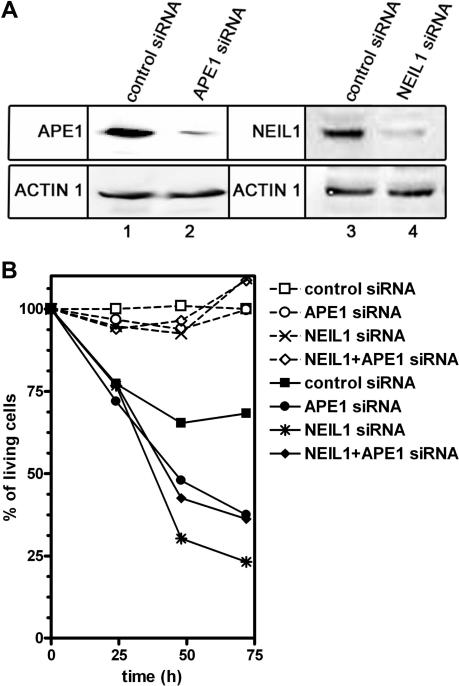
As we showed above, NEIL1-catalysed excision of MA generates a single-strand break with 3′-P terminus which has to be eliminated. Hence, we have reconstituted in vitro the BER pathway for MAps using purified proteins (Figure 5). Incubation of a 21-mer GFP1·3 containing a single MAp in the presence of NEIL1, APE1, DNA polymerase β and [α-32P]-TTP generated a labelled 12-mer DNA fragment (lane 4). The results indicate that APE1 removes 3′-P residues generated by NEIL1 and allows DNA polymerase β to insert one nucleotide. Addition of DNA ligase completes the restoration of the full-length 21-mer GFP1·3 (lane 5). These data demonstrate that 8-MOP-induced MAs are processed by the short-patch BER pathway. In agreement with biochemical data, the siRNA-transfected cells with decreased NEIL1 level were hypersensitive to 8-MOP+UVA exposure (Figure 6). It should be stressed that we did not observe significant additive increase in the sensitivity when cells were depleted for both APE1 and NEIL1. This result indicates that both proteins participate to a same pathway to cope with psoralen-induced DNA damage. These in vitro and in vivo data imply new functions of NEIL1 and APE1 in the removal of DNA adducts generated by cross-linking agents.
Figure 5.
NEIL1 and APE1 are indispensable for the Pol β-dependent repair of psoralen-induced MAp. In vitro reconstitution of the repair was performed in presence of [α-32P]dTTP with GFP1•3 containing single MAp (lanes 1–5). Assay was conducted with various combinations of proteins. Size markers were generated by incubating 5′-[32P] and 3′-[32P]-dCMP-labelled GFP1 containing MAp with NEIL1 (lanes 6 and 7, respectively).
Figure 6.
Sensitivity of NEIL1 and/or APE1-depleted HeLa cells to 8-MOP+UVA exposure. (A) Expression of the APE1 and NEIL1 proteins in HeLa cells after siRNA transfection. Western blot of HeLa cells transfected with APE1-siRNA, NEIL1-siRNA or unrelated control-siRNA. (B) Cells survival 72 h after transfection. Open symbols, control non-irradiated cells; filled symbols, cells exposed to UVA+8-MOP. Living cells were measured by XTT assay at 24, 48 and 72 h as described in Materials and Methods section. The data represent the mean of three independent experiments.
Activity of various E. coli and human DNA repair proteins on oligodeoxynucleotides containing MAp
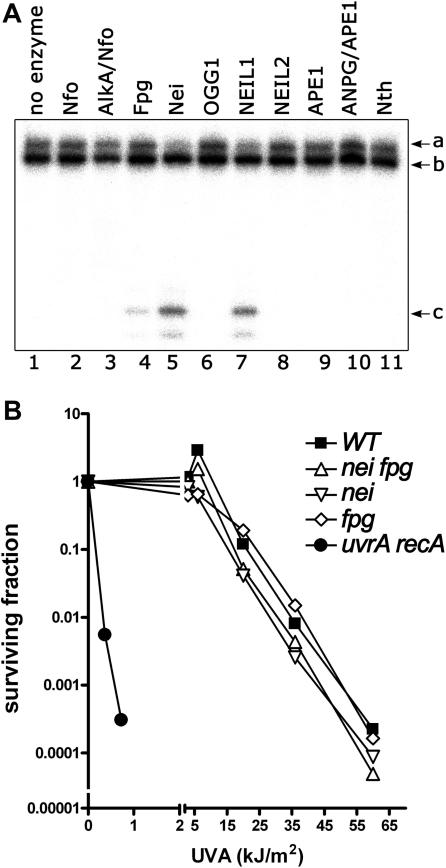
To further investigate the excision of psoralen MA in DNA, we examined whether this lesion was a substrate for previously characterized BER enzymes from E. coli and human. We challenged a 5′-[32P]-labelled oligonucleotide duplex containing single MAp with a variety of highly purified AP endonucleases and DNA glycosylases. Since not all DNA glycosylases possess AP-nicking activity, the assays were made in the presence of AP endonuclease in order to cleave DNA duplex at the potential abasic sites generated by the base excision. When the various E. coli and human enzymes were tested on GFP1·2 (Figure 7A), only incubation with Fpg, Nei and NEIL1 led to the cleavage of the labelled oligonucleotide at the position of the psoralen–thymine MAp. Interestingly, the excision of MAp by the Nei-like enzymes was more efficient than that of Fpg (lanes 5 and 7 versus 4). Despite being used in excess amount, Nfo, AlkA, Nth, OGG1, NEIL2, APE1, ANPG and NTH1 proteins did not act on GFP1·2 (lanes 2–3, 6, 8–11 and data not shown).
Figure 7.
Repair of psoralen-induced DNA damage in bacteria. (A) The E. coli Fpg and Nei proteins excise pyrone-side psoralen monoadduct. The 5′-[32P]-labelled cross-linked GFP1•2 was reversed by hot alkaline treatment and then incubated 10 min at 37°C in the presence of 10 nM Nfo, Alka+Nfo, Fpg, Nei, hOGG1, NEIL1, NEIL2, APE1, ANPG+APE1 and Nth. ‘a’ indicates oligonucleotide with MAp, ‘b’ oligonucleotide without MAp and ‘c’ NEIL1-incision product. (B) Survival of E. coli mutant strains exposed to photoactivated psoralen. AB1157 wild-type strain (filled square), MS2000 fpg nei strain (open triangle), SW2-8 nei strain (open square), BH20 fpg strain (open diamond) and AB2480 uvrA recA strain (filled circle) as a function of exposure to UVA in presence of 5 µM 8MOP. The data represent the mean of three independent experiments.
Photoactivated psoralen sensitivity of DNA glycosylase-deficient E. coli mutants
Genetic and biochemical studies of E. coli have established that both the RecA-dependent recombination and the NER pathways participate in removal of psoralen-induced cross-links (44,45). Subsequently, it was demonstrated that psoralen MAs are removed by UvrABC nuclease in vitro with high efficiency (46). To address the physiological relevance of the Nei and Fpg-catalysed excision of psoralen MAs, we assessed the sensitivity of the E. coli mutant strains to 8-MOP+UVA exposure. As shown in Figure 7B, control E. coli uvrB recA strain was extremely sensitive to the photoactivated psoralen, whereas E. coli nei and fpg single and nei fpg double mutants were not particularly sensitive to treatment. These results suggest that in bacteria, the NER system is a major pathway for removal of psoralen MAs present in DNA.
DISCUSSION
Repair of ICLs and other complex DNA lesions in mammalian cells is linked to replication and required successive involvement of several distinct repair pathways in coordinated manner. At present, the underlying mechanism of this coordination is poorly understood. The results obtained in this study reveal that psoralen–thymine MAs present in duplex DNA are substrate for human oxidative DNA glycosylase, NEIL1. Analysis of the kinetic data demonstrates that NEIL1 can efficiently backup the NER pathway to repair psoralen-induced MAs. It has been shown that NEIL1 catalyses the β,δ-elimination at AP site after oxidized base excision, leaving a 3′-P at the resulting single-strand break (16). Here, we demonstrated using size-markers generated by Nei, NTH1 and APE1 that NEIL1 in fact generates 3′-P termini while removing the psoralen adduct. Based on the fact that PNK has a much higher 3′-phosphatase activity than APE1, it has been proposed that in mammalian cells 3′-P generated by NEIL1 and NEIL2 is removed by PNK in the AP endonuclease-independent BER pathway (13). Although, human major AP endonuclease is a weak 3′-phosphatase, we demonstrated that at 20 nM protein concentration APE1 efficiently repairs oligonucleotide fragment containing 3′-P residue. In agreement with this result, we have fully reconstituted in vitro the repair of MAs using purified APE1, NEIL1, DNA polymerase β and DNA ligase. Following NEIL1 excision APE1 removes 3′-P residue and allows subsequent gap filling and ligation. Consistent with the biochemical data, HeLa cells lacking APE1 and/or NEIL1 become hypersensitive to 8-MOP+UVA exposure. Thus, we added one arm of the BER pathway to the cellular defence used to cope with the genotoxic effects of cross-linking agents. These results demonstrate that NEIL1 is not simply a backup DNA glycosylase for OGG1, NTH1 and NEIL2 but has distinct substrate specificity towards a specific class of bulky DNA adducts.
NEIL1, a 44 kDa protein, was initially characterized as a DNA glycosylase specific for oxidized, saturated and ring-fragmented bases. Several lines of evidence argue for a biological role of NEIL1 in counteracting oxidative DNA damage. Oxidative stress could transiently increase NEIL1 level in human colon carcinoma cells (47). Analysis of human polymorphic and mutant variants of NEIL1 revealed that their low DNA glycosylase activities and reduced expression may be involved in the pathogenesis in a subset of gastric cancers and increased risk of metabolic syndrome (48,49). Cells deficient in the Werner syndrome protein (WRN), a member of the RecQ family of DNA helicases, are hypersensitive to psoralen induced ICLs (50,51). Lately, it has been shown that WRN interacts with and stimulates NEIL1 in excision of oxidative lesions from bubble DNA substrates suggesting that NEIL1–WRN complex participates in the same repair pathway (52). Recently, Vartanian and colleagues (53) generated knockout (KO) mice with a frameshift deletion and insertion in the NEIL1 gene. The steady-state mitochondrial DNA damage and deletions from liver tissues of NEIL1−/− KO mice were significantly increased compared to wild-type controls. The mice develop spontaneously severe obesity, dyslipidemia and fatty liver disease, suggesting that accumulation of spontaneous DNA damage in cells lacking NEIL1 may lead to the diseases associated with the metabolic syndrome. Interestingly, dietary consumption or handling of psoralen-containing plants have been shown to cause adverse effects to human health (54). In light of a new function of NEIL1, it is tempting to speculate that NEIL1−/− KO mice may be more sensitive to psoralen-rich diet.
Since human DNA glycosylase can excise bulky MAp, we investigated whether this lesion was a substrate for previously characterized E. coli Nfo, AlkA, Nth, Fpg and Nei and human APE1, OGG1, NTH1, NEIL2 and ANPG proteins. The results show that in addition to NEIL1, the bacterial Fpg and Nei proteins can also excise psoralen MAp when present in duplex oligonucleotides. In E. coli, psoralen MAs are good substrates for UvrABC nuclease (46) however both NER and homologous recombination systems are required to remove the psoralen-induced ICLs (55). Evolutionary conservation of the new substrate specificity in bacterial Nei-like DNA glycosylases suggests its possible biological role. Therefore, we examined whether in E. coli Fpg/Nei-catalysed excision of MAp provides an alternative pathway to classic NER. Surprisingly, E. coli DNA glycosylase-deficient mutants, in contrast to uvrA recA mutant, were not sensitive to 8-MOP+UVA exposure indicating that in E. coli NER is a major pathway to remove psoralen MAs.
Three-dimensional crystal structures of the Nei–DNA complex and free NEIL1 protein show the same overall fold for both DNA glycosylases (35,56). Both proteins consist of two domains connected by a linker, this module structure giving rise to a DNA-binding cleft between domains. Based on molecular modelling it was suggested that Nei binds to DNA and flips out damaged base in a shallow hydrophobic pocket (35). At present, structural information for Nei and NEIL1 complexed with DNA duplex containing an oxidized base is not yet available. A bulky psoralen MA presents topological constraint for its accommodation in the active sites of Nei-like DNA glycosylases. A 3D crystal structure of the complex of T4 endonuclease V bound to a DNA substrate containing a bulky cyclobutane thymine dimer showed that the enzyme kinks the DNA helix by about 60° and flips out the opposing adenine base complementary to thymine dimer out of the DNA base stack (57). Based on these observations we may hypothesize that the mechanism of lesion recognition by Nei-like enzymes is different from that of well-studied DNA glycosylases. To access the C1′ atom, most of DNA glycosylases bent the DNA and flip out an oxidized base into a specific pocket (35,58–60), while Nei and NEIL1 similar to T4 endonuclease V may access it by a drastic kink of the helix and flipping out of the complementary base opposite to psoralen MA. Insight into the structural basis for clustered and bulky DNA damage recognition by Nei-family DNA glycosylases will have to await further investigations.
In conclusion, although in vitro NEIL1 does not incise DNA containing single ICL, we hypothesized that it may participate in psoralen-induced ICLs removal after XPF-ERCC1-mediated unhooking. Alternatively, but not exclusively, NEIL1/APE1 action could significantly reduce ICLs formation by eliminating MAf residues.
ACKNOWLEDGEMENTS
We wish to thank D. Averbeck and J. -C. Ehrhart for helpful discussion; H. Ide for pNEIL1 expression construct; D. Zharkov and G. Dianov for the Nei and DNA polymerase β proteins; S. S. Wallace and A. K. McCullough for the E. coli mutant strains. S.C.-P. and G.M. were supported by postdoctoral fellowships from the European Community and the Institut Gustave Roussy, respectively. This research was supported by grants to M.K.S. from the FP6 Euroatom Grant RISC-RAD FI6R-CT-2003-508842, Association pour la Recherche sur le Cancer and National Center for Biotechnology, Kazakhstan and to F.R. from La Ligue Contre le Cancer ‘Equipe Labellisée 2006’ and Institut National du Cancer. Funding to pay the Open Access publication charges for this article was provided by Institut National du Cancer grant (to M.K.S.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Cole RS. Psoralen monoadducts and interstrand cross-links in DNA. Biochim. Biophys. Acta. 1971;254:30–39. doi: 10.1016/0005-2787(71)90111-0. [DOI] [PubMed] [Google Scholar]
- 2.Johnston BH, Hearst JE. Low-level psoralen – deoxyribonucleic acid cross-links induced by single laser pulses. Biochemistry. 1981;20:739–745. doi: 10.1021/bi00507a012. [DOI] [PubMed] [Google Scholar]
- 3.Cimino GD, Gamper HB, Isaacs ST, Hearst JE. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu. Rev. Biochem. 1985;54:1151–1193. doi: 10.1146/annurev.bi.54.070185.005443. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy RD, D'Andrea AD. The Fanconi Anemia/BRCA pathway: new faces in the crowd. Genes Dev. 2005;19:2925–2940. doi: 10.1101/gad.1370505. [DOI] [PubMed] [Google Scholar]
- 5.Niedernhofer LJ, Lalai AS, Hoeijmakers JH. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat. Res. 2001;486:217–247. doi: 10.1016/s0921-8777(01)00092-1. [DOI] [PubMed] [Google Scholar]
- 7.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mu D, Bessho T, Nechev LV, Chen DJ, Harris TM, Hearst JE, Sancar A. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell. Biol. 2000;20:2446–2454. doi: 10.1128/mcb.20.7.2446-2454.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Svoboda DL, Taylor JS, Hearst JE, Sancar A. DNA repair by eukaryotic nucleotide excision nuclease. Removal of thymine dimer and psoralen monoadduct by HeLa cell-free extract and of thymine dimer by Xenopus laevis oocytes. J. Biol. Chem. 1993;268:1931–1936. [PubMed] [Google Scholar]
- 11.Hoy CA, Thompson LH, Mooney CL, Salazar EP. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer Res. 1985;45:1737–1743. [PubMed] [Google Scholar]
- 12.Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu. Rev. Genet. 2004;38:445–476. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- 13.Wiederhold L, Leppard JB, Kedar P, Karimi-Busheri F, Rasouli-Nia A, Weinfeld M, Tomkinson AE, Izumi T, Prasad R, et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Das A, Wiederhold L, Leppard JB, Kedar P, Prasad R, Wang H, Boldogh I, Karimi-Busheri F, et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: Evidence for a repair complex in human cells. DNA Repair (Amst) 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boiteux S, Radicella JP. The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys. 2000;377:1–8. doi: 10.1006/abbi.2000.1773. [DOI] [PubMed] [Google Scholar]
- 16.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 17.Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: an alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res. 2002;30:4926–4936. doi: 10.1093/nar/gkf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gros L, Ishchenko AA, Ide H, Elder RH, Saparbaev MK. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004;32:73–81. doi: 10.1093/nar/gkh165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishchenko AA, Yang X, Ramotar D, Saparbaev M. The 3'->5' exonuclease of Apn1 provides an alternative pathway to repair 7,8-dihydro-8-oxodeoxyguanosine in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:6380–6390. doi: 10.1128/MCB.25.15.6380-6390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker LJ, Craig RB, Harris AL, Hickson ID. A role for the human DNA repair enzyme HAP1 in cellular protection against DNA damaging agents and hypoxic stress. Nucleic Acids Res. 1994;22:4884–4889. doi: 10.1093/nar/22.23.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc. Natl Acad. Sci. USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Connor TR, Laval J. Physical association of the 2,6-diamino-4-hydroxy-5N-formamidopyrimidine-DNA glycosylase of Escherichia coli and an activity nicking DNA at apurinic/apyrimidinic sites. Proc. Natl Acad. Sci. USA. 1989;86:5222–5226. doi: 10.1073/pnas.86.14.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang D, Hatahet Z, Melamede RJ, Kow YW, Wallace SS. Characterization of Escherichia coli endonuclease VIII. J. Biol. Chem. 1997;272:32230–32239. doi: 10.1074/jbc.272.51.32230. [DOI] [PubMed] [Google Scholar]
- 24.Saito Y, Uraki F, Nakajima S, Asaeda A, Ono K, Kubo K, Yamamoto K. Characterization of endonuclease III (nth) and endonuclease VIII (nei) mutants of Escherichia coli K-12. J. Bacteriol. 1997;179:3783–3785. doi: 10.1128/jb.179.11.3783-3785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang D, Hatahet Z, Blaisdell JO, Melamede RJ, Wallace SS. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2003;2:581–591. doi: 10.1016/s1568-7864(03)00025-9. [DOI] [PubMed] [Google Scholar]
- 27.Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J. Biol. Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 28.Parsons JL, Zharkov DO, Dianov GL. NEIL1 excises 3' end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res. 2005;33:4849–4856. doi: 10.1093/nar/gki816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons JL, Kavli B, Slupphaug G, Dianov GL. NEIL1 is the major DNA glycosylase that processes 5-hydroxyuracil in the proximity of a DNA single-strand break. Biochemistry. 2007;46:4158–4163. doi: 10.1021/bi0622569. [DOI] [PubMed] [Google Scholar]
- 30.Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Peterson CA, Zheng H, Nairn RS, Legerski RJ, Li L. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Mol. Cell. Biol. 2001;21:713–720. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang D, Luo M, Kelley MR. Human apurinic endonuclease 1 (APE1) expression and prognostic significance in osteosarcoma: enhanced sensitivity of osteosarcoma to DNA damaging agents using silencing RNA APE1 expression inhibition. Mol. Cancer Ther. 2004;3:679–686. [PubMed] [Google Scholar]
- 33.Jurado J, Maciejewska A, Krwawicz J, Laval J, Saparbaev MK. Role of mismatch-specific uracil-DNA glycosylase in repair of 3,N(4)-ethenocytosine in vivo. DNA Repair (Amst) 2004;3:1579–1590. doi: 10.1016/j.dnarep.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 34.Daviet S, Couve-Privat S, Gros L, Shinozuka K, Ide H, Saparbaev M, Ishchenko AA. Major oxidative products of cytosine are substrates for the nucleotide incision repair pathway. DNA Repair (Amst) 2007;6:8–18. doi: 10.1016/j.dnarep.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Zharkov DO, Golan G, Gilboa R, Fernandes AS, Gerchman SE, Kycia JH, Rieger RA, Grollman AP, Shoham G. Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J. 2002;21:789–800. doi: 10.1093/emboj/21.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parsons JL, Dianova II, Allinson SL, Dianov GL. DNA polymerase beta promotes recruitment of DNA ligase III alpha-XRCC1 to sites of base excision repair. Biochemistry. 2005;44:10613–10619. doi: 10.1021/bi050085m. [DOI] [PubMed] [Google Scholar]
- 37.Kumaresan KR, Hwang M, Thelen MP, Lambert MW. Contribution of XPF functional domains to the 5' and 3' incisions produced at the site of a psoralen interstrand cross-link. Biochemistry. 2002;41:890–896. doi: 10.1021/bi011614z. [DOI] [PubMed] [Google Scholar]
- 38.Pichierri P, Rosselli F. The DNA crosslink-induced S-phase checkpoint depends on ATR-CHK1 and ATR-NBS1-FANCD2 pathways. EMBO J. 2004;23:1178–1187. doi: 10.1038/sj.emboj.7600113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hang B, Chenna A, Fraenkel-Conrat H, Singer B. An unusual mechanism for the major human apurinic/apyrimidinic (AP) endonuclease involving 5' cleavage of DNA containing a benzene-derived exocyclic adduct in the absence of an AP site. Proc. Natl Acad. Sci. USA. 1996;93:13737–13741. doi: 10.1073/pnas.93.24.13737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ishchenko AA, Sanz G, Privezentzev CV, Maksimenko AV, Saparbaev M. Characterisation of new substrate specificities of Escherichia coli and Saccharomyces cerevisiae AP endonucleases. Nucleic Acids Res. 2003;31:6344–6353. doi: 10.1093/nar/gkg812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun B, Latham KA, Dodson ML, Lloyd RS. Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme-DNA imino intermediates. J. Biol. Chem. 1995;270:19501–19508. doi: 10.1074/jbc.270.33.19501. [DOI] [PubMed] [Google Scholar]
- 42.Jones BK, Yeung AT. Repair of 4,5',8-trimethylpsoralen monoadducts and cross-links by the Escherichia coli UvrABC endonuclease. Proc. Natl Acad. Sci. USA. 1988;85:8410–8414. doi: 10.1073/pnas.85.22.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen DS, Herman T, Demple B. Two distinct human DNA diesterases that hydrolyze 3'-blocking deoxyribose fragments from oxidized DNA. Nucleic Acids Res. 1991;19:5907–5914. doi: 10.1093/nar/19.21.5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoakum GH, Cole RS. Role of ATP in removal of psoralen cross-links from DNA of Escherichia coli permeabilized by treatment with toluene. J. Biol. Chem. 1977;252:7023–7030. [PubMed] [Google Scholar]
- 45.Cole RS. Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl Acad. Sci. USA. 1973;70:1064–1068. doi: 10.1073/pnas.70.4.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Houten B, Gamper H, Hearst JE, Sancar A. Analysis of sequential steps of nucleotide excision repair in Escherichia coli using synthetic substrates containing single psoralen adducts. J. Biol. Chem. 1988;263:16553–16560. [PubMed] [Google Scholar]
- 47.Das A, Hazra TK, Boldogh I, Mitra S, Bhakat KK. Induction of the human oxidized base-specific DNA glycosylase NEIL1 by reactive oxygen species. J. Biol. Chem. 2005;280:35272–35280. doi: 10.1074/jbc.M505526200. [DOI] [PubMed] [Google Scholar]
- 48.Shinmura K, Tao H, Goto M, Igarashi H, Taniguchi T, Maekawa M, Takezaki T, Sugimura H. Inactivating mutations of the human base excision repair gene NEIL1 in gastric cancer. Carcinogenesis. 2004;25:2311–2317. doi: 10.1093/carcin/bgh267. [DOI] [PubMed] [Google Scholar]
- 49.Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M, Lloyd RS. Human polymorphic variants of the NEIL1 DNA glycosylase. J. Biol. Chem. 2007:15790–15798. doi: 10.1074/jbc.M610626200. [DOI] [PubMed] [Google Scholar]
- 50.Poot M, Gollahon KA, Emond MJ, Silber JR, Rabinovitch PS. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 2002;16:757–758. doi: 10.1096/fj.01-0906fje. [DOI] [PubMed] [Google Scholar]
- 51.Bohr VA, Souza Pinto N, Nyaga SG, Dianov G, Kraemer K, Seidman MM, Brosh, R M., Jr DNA repair and mutagenesis in Werner syndrome. Environ. Mol. Mutagen. 2001;38:227–234. doi: 10.1002/em.1076. [DOI] [PubMed] [Google Scholar]
- 52.Das A, Boldogh I, Lee JW, Harrigan JA, Hegde ML, Piotrowski J, de Souza-Pinto N, Ramos W, Greenberg MM, et al. The human we rner syndrome protein stimulates repair of oxidative DNA base damage by the DNA glycosylase Neil1. J. Biol. Chem. 2007:1–31. doi: 10.1074/jbc.M703343200. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 53.Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, et al. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc. Natl Acad. Sci. USA. 2006;103:1864–1869. doi: 10.1073/pnas.0507444103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watson DH, Hones SM. Carcinogenic higher plant metabolites in the human diet in temperate countries: a review. Food Addit. Contam. 1985;2:25–32. doi: 10.1080/02652038509373523. [DOI] [PubMed] [Google Scholar]
- 55.Cheng S, Sancar A, Hearst JE. RecA-dependent incision of psoralen-crosslinked DNA by (A)BC excinuclease. Nucleic Acids Res. 1991;19:657–663. doi: 10.1093/nar/19.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doublie S, Bandaru V, Bond JP, Wallace SS. The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity. Proc. Natl Acad. Sci. USA. 2004;101:10284–10289. doi: 10.1073/pnas.0402051101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vassylyev DG, Kashiwagi T, Mikami Y, Ariyoshi M, Iwai S, Ohtsuka E, Morikawa K. Atomic model of a pyrimidine dimer excision repair enzyme complexed with a DNA substrate: structural basis for damaged DNA recognition. Cell. 1995;83:773–782. doi: 10.1016/0092-8674(95)90190-6. [DOI] [PubMed] [Google Scholar]
- 58.Guan Y, Manuel RC, Arvai AS, Parikh SS, Mol CD, Miller JH, Lloyd S, Tainer JA. MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol. 1998;5:1058–1064. doi: 10.1038/4168. [DOI] [PubMed] [Google Scholar]
- 59.Bruner SD, Norman DP, Verdine GL. Structural basis for recognition and repair of the endogenous mutagen 8–oxoguanine in DNA. Nature. 2000;403:859–866. doi: 10.1038/35002510. [DOI] [PubMed] [Google Scholar]
- 60.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]