Abstract
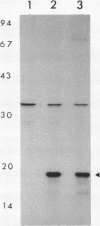
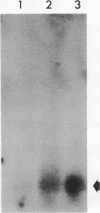
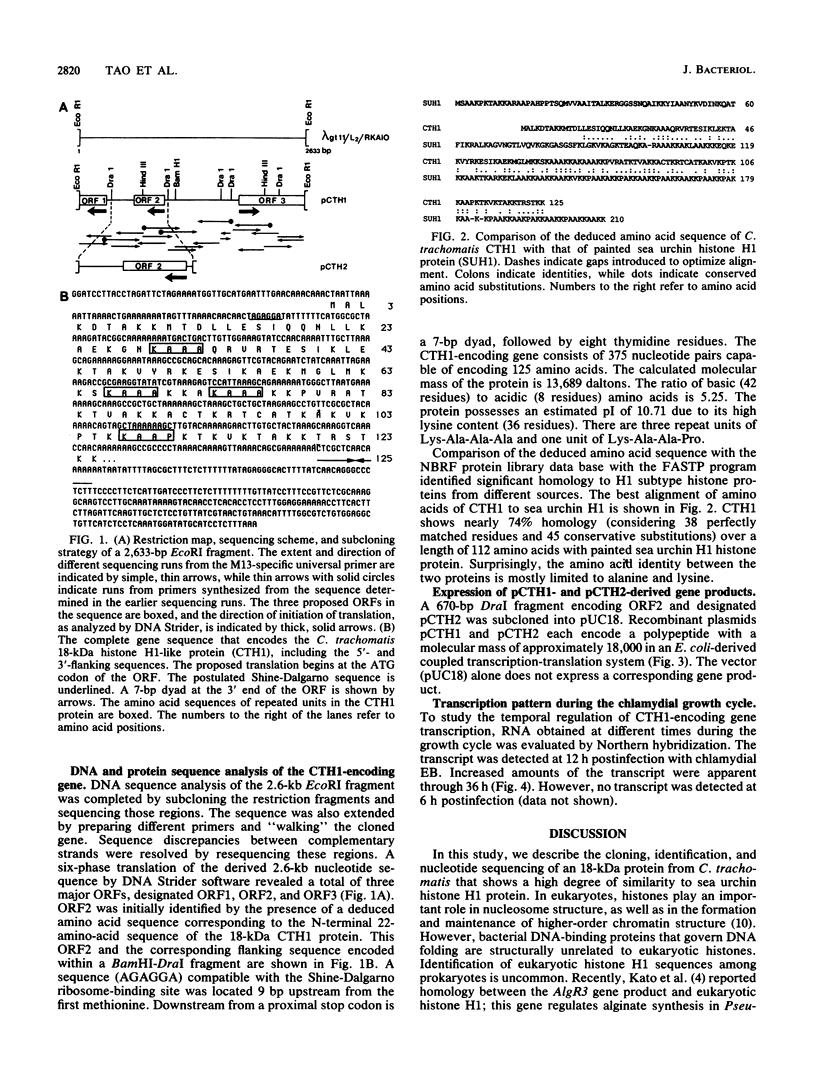
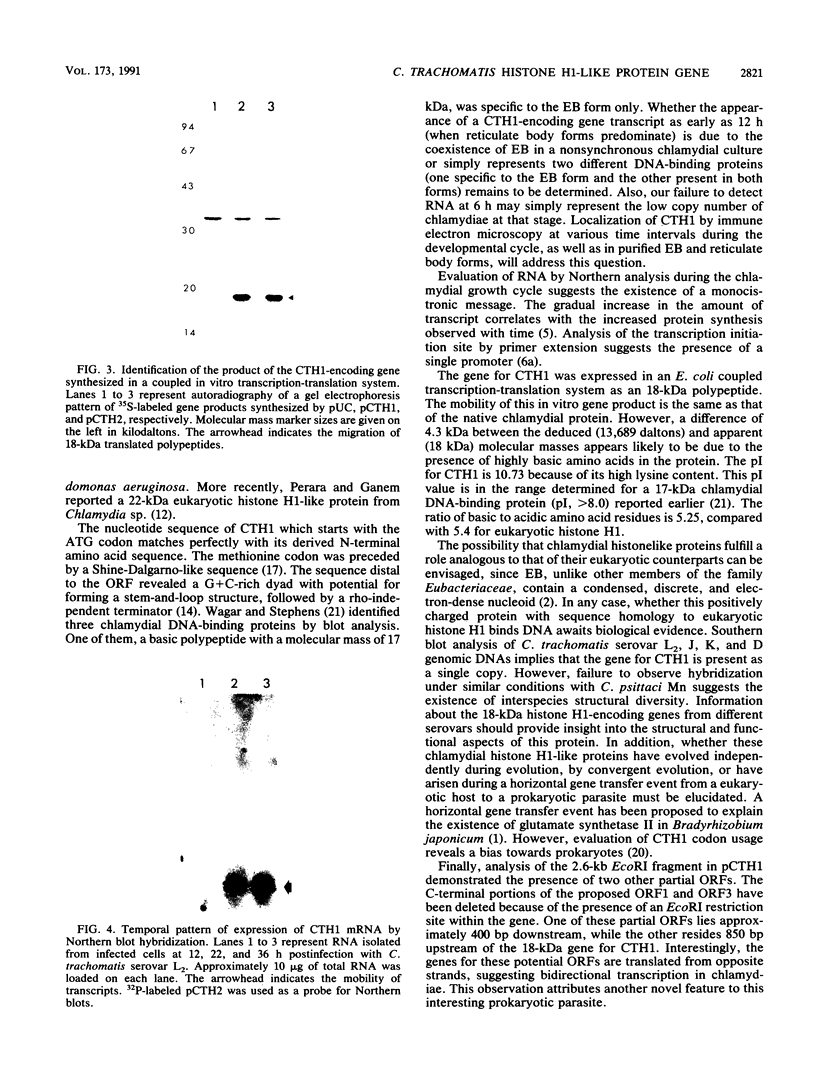
A lambda gt11 recombinant library of Chlamydia trachomatis serovar L2 chromosomal DNA was screened with a 29-mer synthetic oligonucleotide specific to the N-terminal amino acids of a predominant 18-kDa chlamydial protein. One recombinant clone, designated lambda gt11/L2/RKA10, was selected on the basis of its strong hybridization signal. Restriction endonuclease analysis and complete nucleotide sequencing of the recombinant revealed a 2,633-bp insert containing one complete open reading frame (ORF2) and two partial ORFs (ORF1 and ORF3). The deduced amino acid sequence of ORF2 matched perfectly at its N-terminal end with the derived amino acid sequence. The 375-bp ORF is capable of encoding a protein comprising 125 amino acids with a molecular mass of 13,689. A sequence compatible with a Shine-Dalgarno ribosome-binding site was located 9 bp upstream from the initiation codon, while the sequence distal to ORF2 revealed a rho-independent terminator. The protein, designated CTH1, possesses an estimated pI of 10.71 due to its high lysine content. This highly basic protein contains no tryptophan or phenylalanine. A protein data base search identified significant homology between CTH1 and painted sea urchin histone H1. Northern (RNA) blot analysis of Chlamydia-infected host cells demonstrated transcripts at 12 h postinfection. The recombinant plasmid encoding ORF2 expressed a gene product of approximately 18 kDa, similar to the native chlamydial protein as analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. This protein appears to represent one of the few eukaryotic histonelike proteins described to date in prokaryotes.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Costerton J. W., Poffenroth L., Wilt J. C., Kordová N. Ultrastructural studies of the nucleoids of the pleomorphic forms of Chlamydia psittaci 6BC: a comparison with bacteria. Can J Microbiol. 1976 Jan;22(1):16–28. doi: 10.1139/m76-003. [DOI] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Misra T. K., Chakrabarty A. M. AlgR3, a protein resembling eukaryotic histone H1, regulates alginate synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 1990 Apr;87(8):2887–2891. doi: 10.1073/pnas.87.8.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul R., Tao S., Wenman W. M. Cyclic AMP inhibits protein synthesis in Chlamydia trachomatis at a transcriptional level. Biochim Biophys Acta. 1990 Jun 12;1053(1):106–112. doi: 10.1016/0167-4889(90)90032-9. [DOI] [PubMed] [Google Scholar]
- Matsudaira P. Sequence from picomole quantities of proteins electroblotted onto polyvinylidene difluoride membranes. J Biol Chem. 1987 Jul 25;262(21):10035–10038. [PubMed] [Google Scholar]
- McGhee J. D., Felsenfeld G. Nucleosome structure. Annu Rev Biochem. 1980;49:1115–1156. doi: 10.1146/annurev.bi.49.070180.005343. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E. Structure and properties of the bacterial nucleoid. Cell. 1982 Oct;30(3):667–669. doi: 10.1016/0092-8674(82)90269-0. [DOI] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wada K., Aota S., Tsuchiya R., Ishibashi F., Gojobori T., Ikemura T. Codon usage tabulated from the GenBank genetic sequence data. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2367–2411. doi: 10.1093/nar/18.suppl.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagar E. A., Stephens R. S. Developmental-form-specific DNA-binding proteins in Chlamydia spp. Infect Immun. 1988 Jul;56(7):1678–1684. doi: 10.1128/iai.56.7.1678-1684.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg W. G., Hatch T. P., Woese C. R. Eubacterial origin of chlamydiae. J Bacteriol. 1986 Aug;167(2):570–574. doi: 10.1128/jb.167.2.570-574.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenman W. M., Lovett M. A. Expression in E. coli of Chlamydia trachomatis antigen recognized during human infection. Nature. 1982 Mar 4;296(5852):68–70. doi: 10.1038/296068a0. [DOI] [PubMed] [Google Scholar]
- Wenman W. M., Meuser R. U. Chlamydia trachomatis elementary bodies possess proteins which bind to eucaryotic cell membranes. J Bacteriol. 1986 Feb;165(2):602–607. doi: 10.1128/jb.165.2.602-607.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]