Abstract
The toxR gene of Vibrio cholerae encodes a transcriptional activator required for the expression of the cholera toxin genes (ctxAB) and more than 15 other genes encoding secreted or membrane proteins. The latter group includes virulence genes involved in the biogenesis of the TCP pilus, the accessory colonization factor, and such ToxR-activated genes as tagA, mutations in which cause no detectable virulence defect in the suckling mouse model. To analyze the regulation of expression and the structure of tagA, we have cloned and sequenced about 2 kb of DNA upstream from a tagA::TnphoA fusion. While the portion of the tagA gene product examined presented no extensive similarity to any known protein, the amino acid sequence deduced from an open reading frame (designated aldA) located upstream from and in opposite orientation to tagA was highly similar to the sequences of eukaryotic aldehyde dehydrogenases. An assay of aldehyde dehydrogenase activity in extracts of a wild-type V. cholerae strainand an aldA mutant confirmed that aldA encodes an aldehyde dehydrogenase. Expression of the aldA gene was studied together with that of tagA in both V. cholerae and Escherichia coli. The expression of both tagA and aldA was environmentally regulated and dependent on a functional toxR gene in V. cholerae, but neither promoter was activated by ToxR in E. coli, suggesting that expression of tagA and aldA requires an additional transcriptional activator besides ToxR. The aldA gene is the first example of a gene encoding a cytoplasmic protein that is under the control of ToxR, and this suggests that metabolic enzymes may constitute novel members of virulence regulons in bacteria.
Full text
PDF

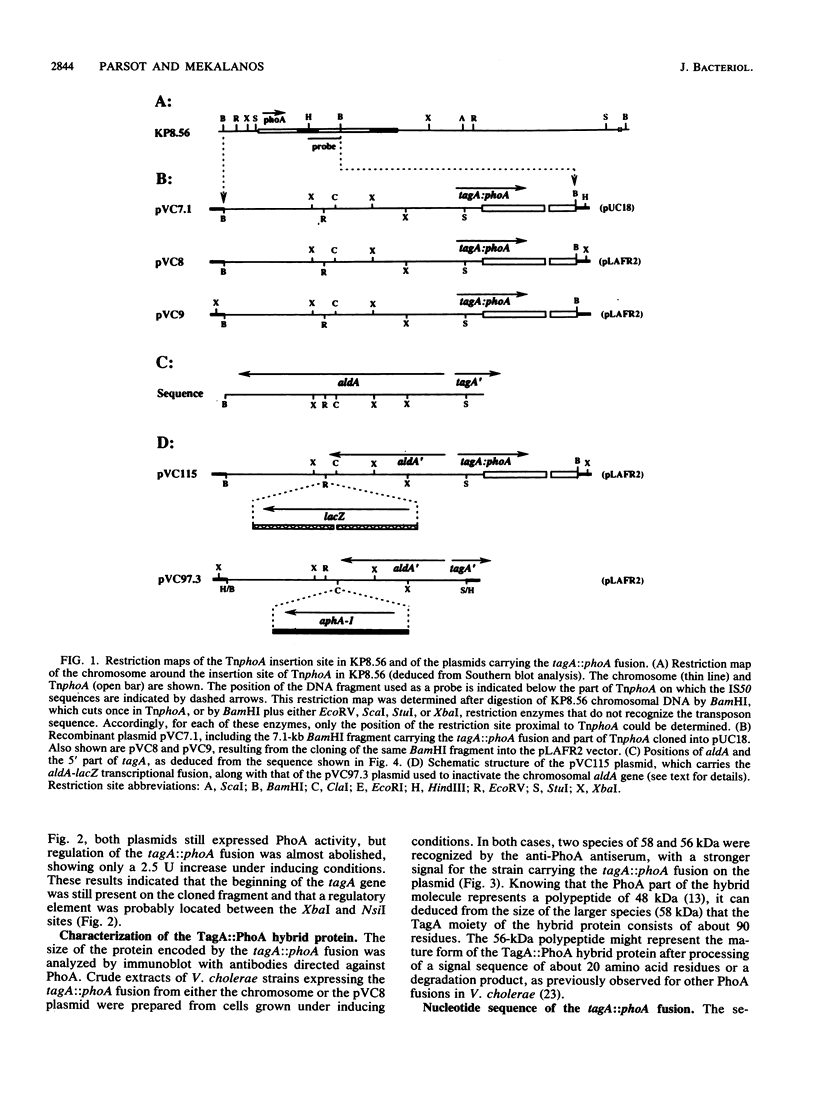
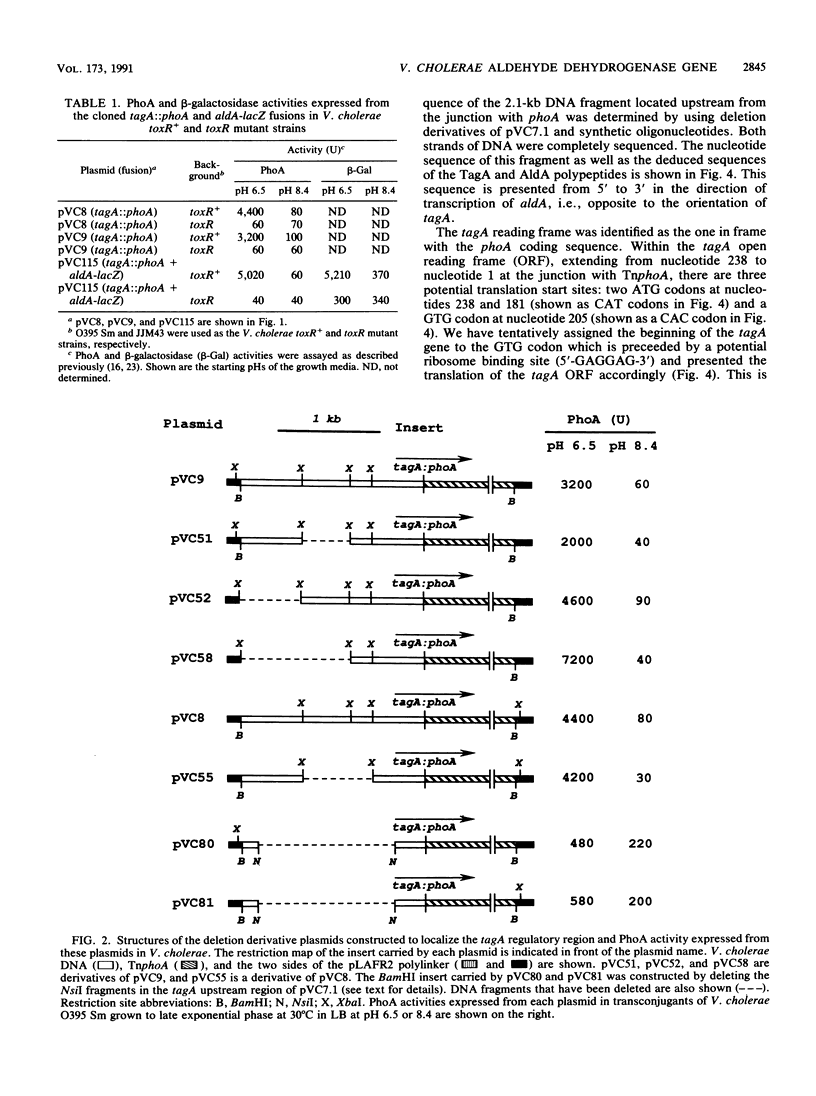






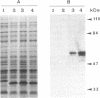
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Braun T., Bober E., Singh S., Agarwal D. P., Goedde H. W. Isolation and sequence analysis of a full length cDNA clone coding for human mitochondrial aldehyde dehydrogenase. Nucleic Acids Res. 1987 Apr 10;15(7):3179–3179. doi: 10.1093/nar/15.7.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freter R., O'Brien P. C., Macsai M. S. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect Immun. 1981 Oct;34(1):234–240. doi: 10.1128/iai.34.1.234-240.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Hempel J., Pietruszko R., Fietzek P., Jörnvall H. Identification of a segment containing a reactive cysteine residue in human liver cytoplasmic aldehyde dehydrogenase (isoenzyme E1). Biochemistry. 1982 Dec 21;21(26):6834–6838. doi: 10.1021/bi00269a032. [DOI] [PubMed] [Google Scholar]
- Hempel J., von Bahr-Lindström H., Jörnvall H. Aldehyde dehydrogenase from human liver. Primary structure of the cytoplasmic isoenzyme. Eur J Biochem. 1984 May 15;141(1):21–35. doi: 10.1111/j.1432-1033.1984.tb08150.x. [DOI] [PubMed] [Google Scholar]
- Jones P. W., Turner J. M. A model for the common control of enzymes of ethanolamine catabolism in Escherichia coli. J Gen Microbiol. 1984 Apr;130(4):849–860. doi: 10.1099/00221287-130-4-849. [DOI] [PubMed] [Google Scholar]
- Jones P. W., Turner J. M. Interrelationships between the enzymes of ethanolamine metabolism in Escherichia coli. J Gen Microbiol. 1984 Feb;130(2):299–308. doi: 10.1099/00221287-130-2-299. [DOI] [PubMed] [Google Scholar]
- Krzywicki K. A., Brandriss M. C. Primary structure of the nuclear PUT2 gene involved in the mitochondrial pathway for proline utilization in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Dec;4(12):2837–2842. doi: 10.1128/mcb.4.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekalanos J. J. Cholera toxin: genetic analysis, regulation, and role in pathogenesis. Curr Top Microbiol Immunol. 1985;118:97–118. doi: 10.1007/978-3-642-70586-1_6. [DOI] [PubMed] [Google Scholar]
- Mekalanos J. J., Swartz D. J., Pearson G. D., Harford N., Groyne F., de Wilde M. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature. 1983 Dec 8;306(5943):551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- Miller V. L., DiRita V. J., Mekalanos J. J. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol. 1989 Mar;171(3):1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988 Jun;170(6):2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Mekalanos J. J. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3471–3475. doi: 10.1073/pnas.81.11.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller V. L., Taylor R. K., Mekalanos J. J. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987 Jan 30;48(2):271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- Parsot C. Evolution of biosynthetic pathways: a common ancestor for threonine synthase, threonine dehydratase and D-serine dehydratase. EMBO J. 1986 Nov;5(11):3013–3019. doi: 10.1002/j.1460-2075.1986.tb04600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsot C., Saint-Girons I., Cohen G. N. Enzyme specialization during the evolution of amino acid biosynthetic pathways. Microbiol Sci. 1987 Sep;4(9):258, 260-2. [PubMed] [Google Scholar]
- Parsot C., Taxman E., Mekalanos J. J. ToxR regulates the production of lipoproteins and the expression of serum resistance in Vibrio cholerae. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1641–1645. doi: 10.1073/pnas.88.5.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson K. M., Mekalanos J. J. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun. 1988 Nov;56(11):2822–2829. doi: 10.1128/iai.56.11.2822-2829.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett M., Gwynne D. I., Buxton F. P., Elliott R., Davies R. W., Lockington R. A., Scazzocchio C., Sealy-Lewis H. M. Cloning and characterization of the aldA gene of Aspergillus nidulans. Gene. 1987;51(2-3):217–226. doi: 10.1016/0378-1119(87)90310-6. [DOI] [PubMed] [Google Scholar]
- Roof D. M., Roth J. R. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988 Sep;170(9):3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch K. L., Beckwith J. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1576–1580. doi: 10.1073/pnas.85.5.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. K., Miller V. L., Furlong D. B., Mekalanos J. J. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A. 1987 May;84(9):2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. C., Tokunaga M. Biogenesis of lipoproteins in bacteria. Curr Top Microbiol Immunol. 1986;125:127–157. doi: 10.1007/978-3-642-71251-7_9. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Bahr-Lindström H., Hempel J., Jörnvall H. The cytoplasmic isoenzyme of horse liver aldehyde dehydrogenase. Relationship to the corresponding human isoenzyme. Eur J Biochem. 1984 May 15;141(1):37–42. doi: 10.1111/j.1432-1033.1984.tb08152.x. [DOI] [PubMed] [Google Scholar]