Abstract
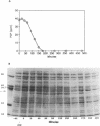
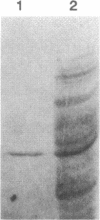
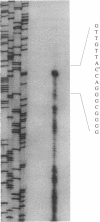
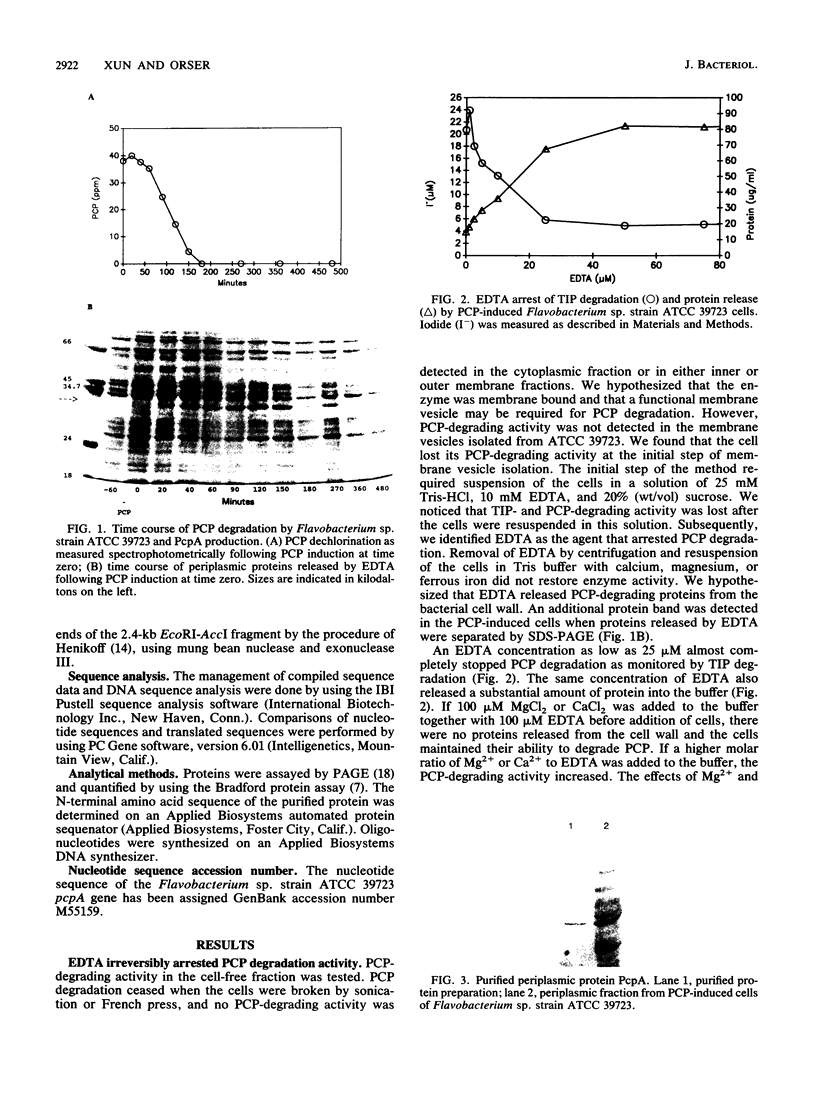
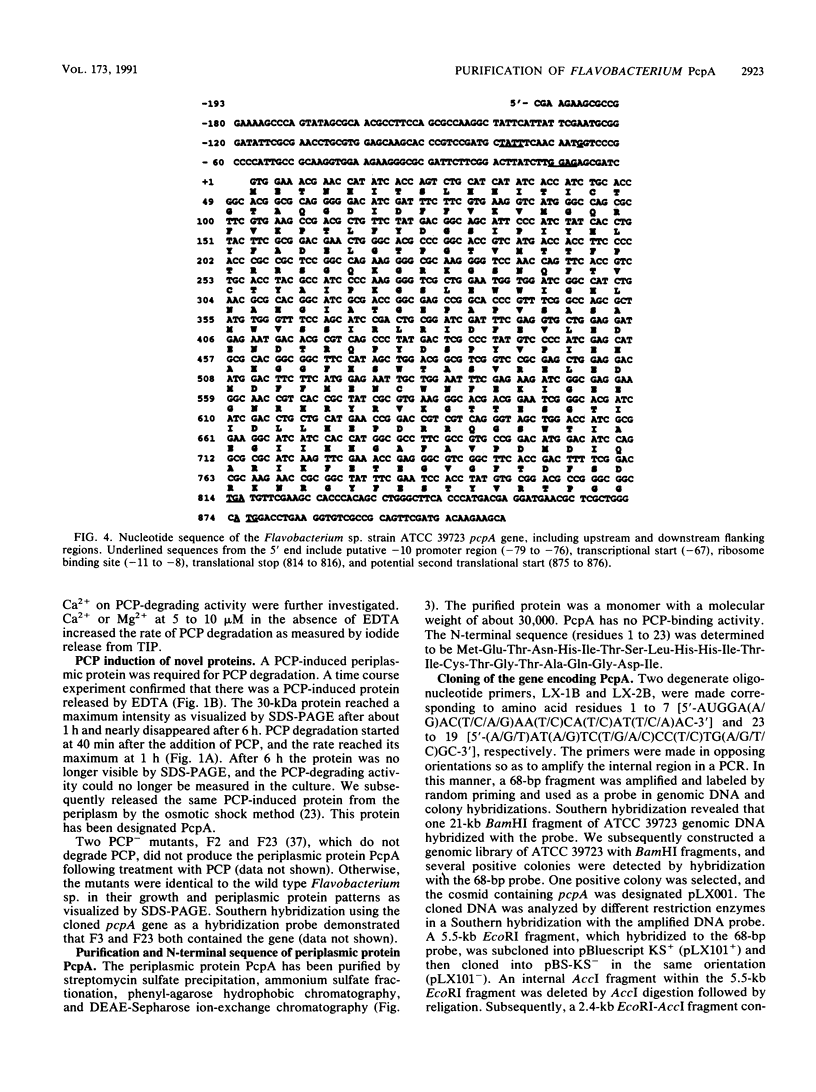
A pentachlorophenol (PCP)-induced periplasmic protein (PcpA) with a molecular weight of 30,000 has been identified and purified from Flavobacterium sp. strain ATCC 39723. Results suggest that PcpA may be involved in PCP mineralization. The induction of PcpA correlated with the induction of PCP-degrading activity. When PcpA was released from the periplasmic space by EDTA or osmotic shock treatment, PCP-degrading activity was arrested. Two PCP- mutants of ATCC 39723, which do not degrade PCP, did not produce PcpA following induction with PCP. The N terminus of PcpA was sequenced, and two degenerate primers were synthesized for use in DNA amplification of a 68-bp probe which hybridized to a genomic library clone containing pcpA. The nucleotide sequence of pcpA was determined and found to encode an open reading frame of 816 bp. pcpA was found to be part of a 1.35-kb transcript with a transcriptional start site 67 bp upstream of the translational start site. Data base search comparisons yielded no sequences of high similarity to pcpA or its protein product.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benson S. A., Hall M. N., Silhavy T. J. Genetic analysis of protein export in Escherichia coli K12. Annu Rev Biochem. 1985;54:101–134. doi: 10.1146/annurev.bi.54.070185.000533. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chu J. P., Kirsch E. J. Metabolism of pentachlorophenol by an axenic bacterial culture. Appl Microbiol. 1972 May;23(5):1033–1035. doi: 10.1128/am.23.5.1033-1035.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetzner S., Müller R., Lingens F. Degradation of 2-chlorobenzoate by Pseudomonas cepacia 2CBS. Biol Chem Hoppe Seyler. 1989 Nov;370(11):1173–1182. doi: 10.1515/bchm3.1989.370.2.1173. [DOI] [PubMed] [Google Scholar]
- Frantz B., Chakrabarty A. M. Organization and nucleotide sequence determination of a gene cluster involved in 3-chlorocatechol degradation. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4460–4464. doi: 10.1073/pnas.84.13.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III in DNA sequence analysis. Methods Enzymol. 1987;155:156–165. doi: 10.1016/0076-6879(87)55014-5. [DOI] [PubMed] [Google Scholar]
- Häggblom M. M., Janke D., Salkinoja-Salonen M. S. Hydroxylation and Dechlorination of Tetrachlorohydroquinone by Rhodococcus sp. Strain CP-2 Cell Extracts. Appl Environ Microbiol. 1989 Feb;55(2):516–519. doi: 10.1128/aem.55.2.516-519.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida N., Tawaragi Y., Inuzuka C., Sugita O., Kubota I., Nakazato H., Noguchi T., Sassa S. Four species of cDNAs for cytochrome P450 isozymes immunorelated to P450C-M/F encode for members of P450IID subfamily, increasing the number of members within the subfamily. Biochem Biophys Res Commun. 1988 Oct 31;156(2):681–688. doi: 10.1016/s0006-291x(88)80896-9. [DOI] [PubMed] [Google Scholar]
- Jones K. A., Yamamoto K. R., Tjian R. Two distinct transcription factors bind to the HSV thymidine kinase promoter in vitro. Cell. 1985 Sep;42(2):559–572. doi: 10.1016/0092-8674(85)90113-8. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levy S. B., Leive L. Release from Escherichia coli of a galactosyltransferase complex active in lipopolysaccharide synthesis. J Biol Chem. 1970 Feb 10;245(3):585–594. [PubMed] [Google Scholar]
- Markus A., Klages U., Krauss S., Lingens F. Oxidation and dehalogenation of 4-chlorophenylacetate by a two-component enzyme system from Pseudomonas sp. strain CBS3. J Bacteriol. 1984 Nov;160(2):618–621. doi: 10.1128/jb.160.2.618-621.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin H. J., ter Beest M. B., Witholt B. Release of outer membrane fragments from wild-type Escherichia coli and from several E. coli lipopolysaccharide mutants by EDTA and heat shock treatments. J Bacteriol. 1989 Oct;171(10):5262–5267. doi: 10.1128/jb.171.10.5262-5267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orser C., Staskawicz B. J., Panopoulos N. J., Dahlbeck D., Lindow S. E. Cloning and expression of bacterial ice nucleation genes in Escherichia coli. J Bacteriol. 1985 Oct;164(1):359–366. doi: 10.1128/jb.164.1.359-366.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn M. J., Gander J. E., Parisi E., Carson J. Mechanism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972 Jun 25;247(12):3962–3972. [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Prossnitz E., Gee A., Ames G. F. Reconstitution of the histidine periplasmic transport system in membrane vesicles. Energy coupling and interaction between the binding protein and the membrane complex. J Biol Chem. 1989 Mar 25;264(9):5006–5014. [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Saber D. L., Crawford R. L. Isolation and characterization of Flavobacterium strains that degrade pentachlorophenol. Appl Environ Microbiol. 1985 Dec;50(6):1512–1518. doi: 10.1128/aem.50.6.1512-1518.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk T., Müller R., Mörsberger F., Otto M. K., Lingens F. Enzymatic dehalogenation of pentachlorophenol by extracts from Arthrobacter sp. strain ATCC 33790. J Bacteriol. 1989 Oct;171(10):5487–5491. doi: 10.1128/jb.171.10.5487-5491.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stanlake G. J., Finn R. K. Isolation and characterization of a pentachlorophenol-degrading bacterium. Appl Environ Microbiol. 1982 Dec;44(6):1421–1427. doi: 10.1128/aem.44.6.1421-1427.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiert J. G., Crawford R. L. Catabolism of pentachlorophenol by a Flavobacterium sp. Biochem Biophys Res Commun. 1986 Dec 15;141(2):825–830. doi: 10.1016/s0006-291x(86)80247-9. [DOI] [PubMed] [Google Scholar]
- Summers W. C. A simple method for extraction of RNA from E. coli utilizing diethyl pyrocarbonate. Anal Biochem. 1970 Feb;33(2):459–463. doi: 10.1016/0003-2697(70)90316-7. [DOI] [PubMed] [Google Scholar]
- Suzuki T. Metabolism of pentachlorophenol by a soil microbe. J Environ Sci Health B. 1977;12(2):113–127. doi: 10.1080/03601237709372057. [DOI] [PubMed] [Google Scholar]
- Xun L., Orser C. S. Biodegradation of triiodophenol by cell-free extracts of a pentachlorophenol-degrading Flavobacterium sp. Biochem Biophys Res Commun. 1991 Jan 15;174(1):43–48. doi: 10.1016/0006-291x(91)90482-m. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]