Abstract
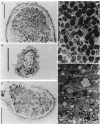
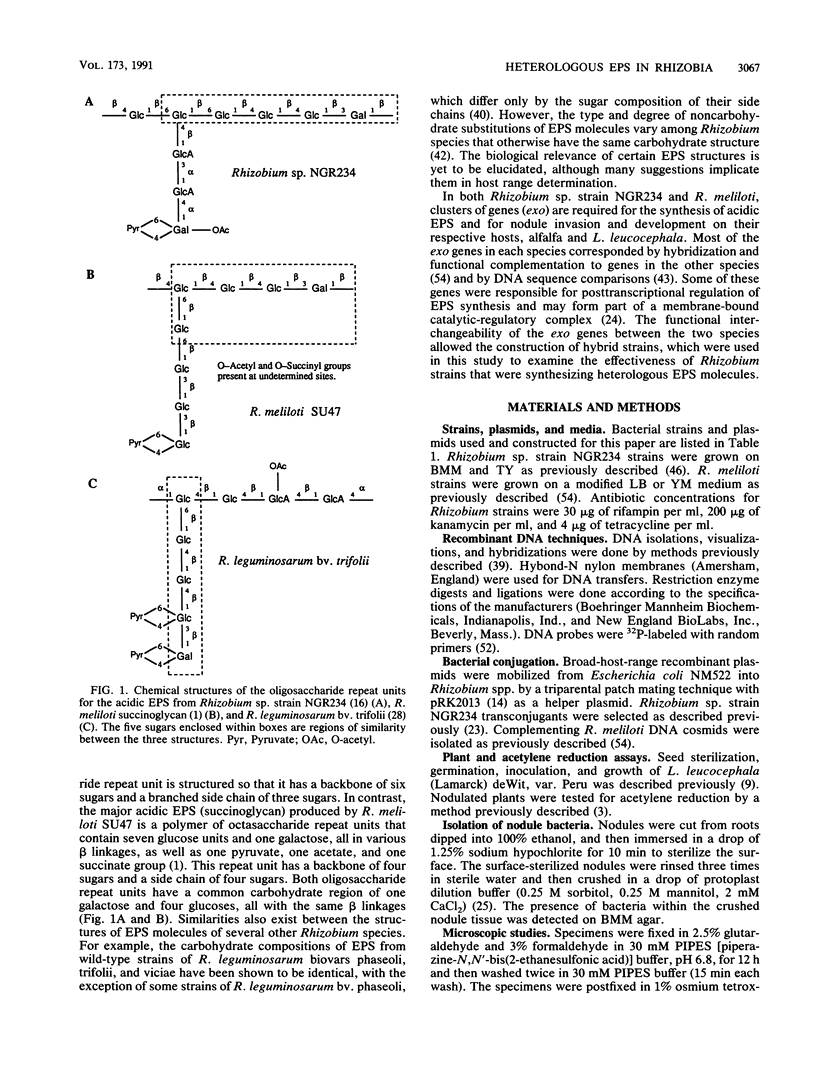
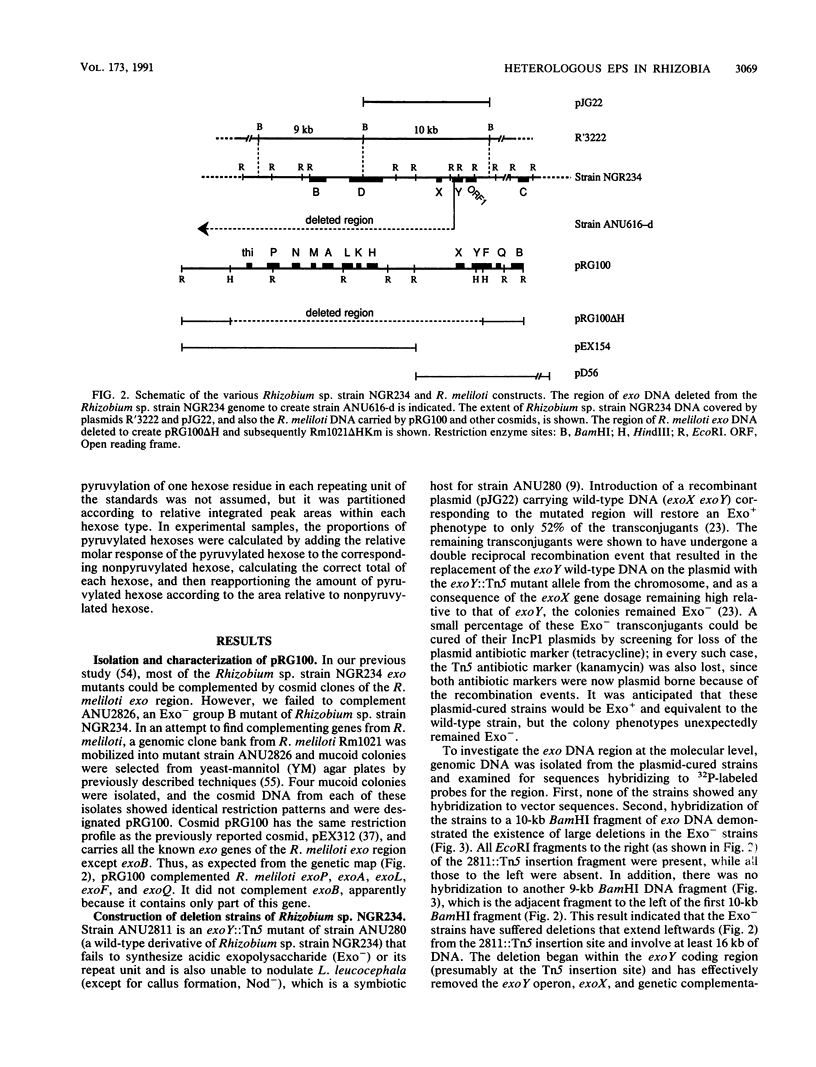
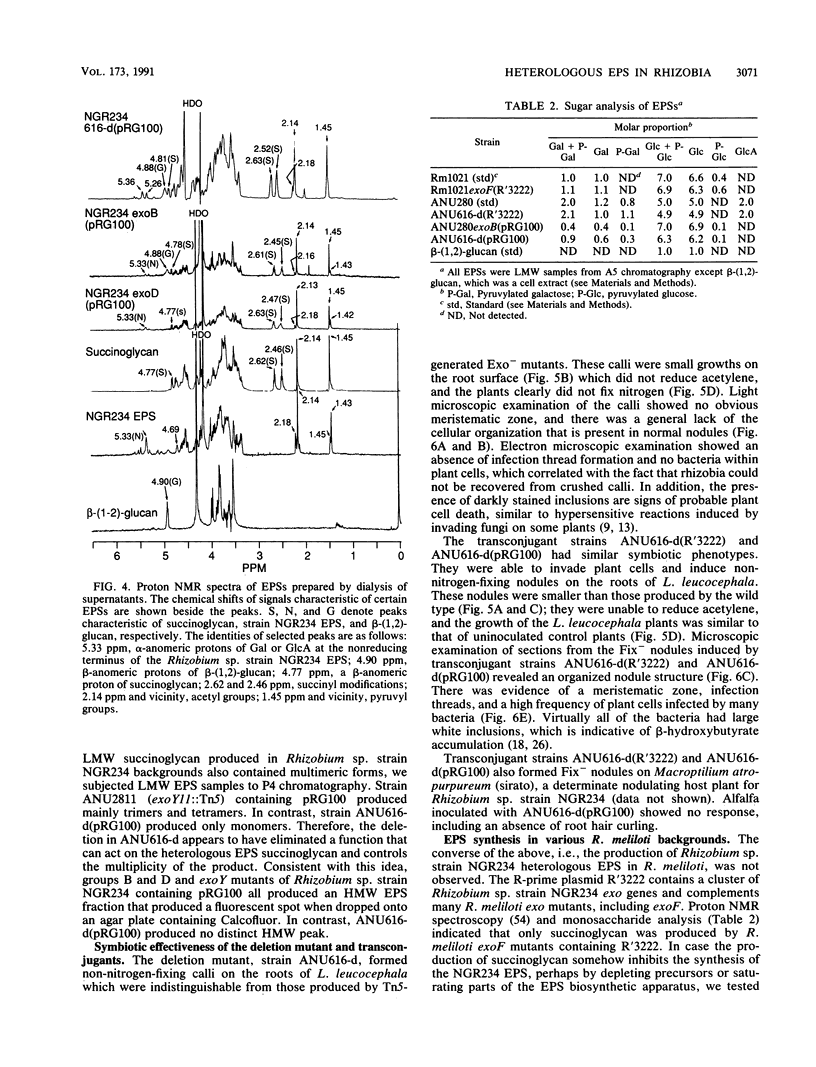
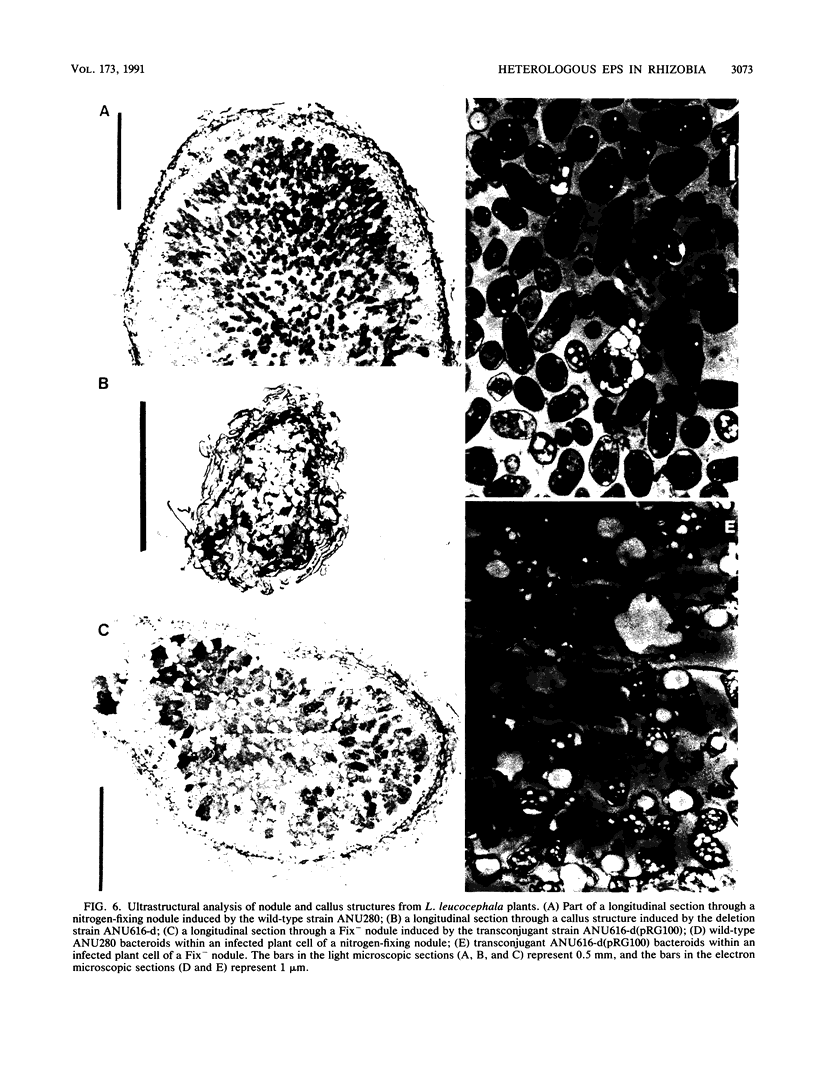
Rhizobium sp. strain NGR234 produces large amounts of acidic exopolysaccharide. Mutants that fail to synthesize this exopolysaccharide are also unable to nodulate the host plant Leucaena leucocephala. A hybrid strain of Rhizobium sp. strain NGR234 containing exo genes from Rhizobium meliloti was constructed. The background genetics and nod genes of Rhizobium sp. strain NGR234 are retained, but the cluster of genes involved in exopolysaccharide biosynthesis was deleted. These exo genes were replaced with genes required for the synthesis of succinoglycan exopolysaccharide from R. meliloti. As a result of the genetic manipulation, the ability of these hybrids to synthesize exopolysaccharide was restored, but the structure was that of succinoglycan and not that of Rhizobium sp. strain NGR234. The replacement genes were contained on a cosmid which encoded the entire known R. meliloti exo gene cluster, with the exception of exoB. Cosmids containing smaller portions of this exo gene cluster did not restore exopolysaccharide production. The presence of succinoglycan was indicated by staining with the fluorescent dye Calcofluor, proton nuclear magnetic resonance spectroscopy, and monosaccharide analysis. Although an NGR234 exoY mutant containing the R. meliloti exo genes produced multimers of the succinoglycan repeat unit, as does the wild-type R. meliloti, the deletion mutant of Rhizobium sp. strain NGR234 containing the R. meliloti exo genes produced only the monomer. The deletion mutant therefore appeared to lack a function that affects the multiplicity of succinoglycan produced in the Rhizobium sp. strain NGR234 background. Although these hybrid strains produced succinoglycan, they were still able to induce the development of an organized nodule structure on L. leucocephala. The resulting nodules did not fix nitrogen, but they did contain infection threads and bacteroids within plant cells. This clearly demonstrated that a heterologous acidic exopolysaccharide structure was sufficient to enable nodule development to proceed beyond the developmental barrier imposed on mutants of Rhizobium sp. strain NGR234 that are unable to synthesize any acidic exopolysaccharide.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Borthakur D., Barker R. F., Latchford J. W., Rossen L., Johnston A. W. Analysis of pss genes of Rhizobium leguminosarum required for exopolysaccharide synthesis and nodulation of peas: their primary structure and their interaction with psi and other nodulation genes. Mol Gen Genet. 1988 Jul;213(1):155–162. doi: 10.1007/BF00333413. [DOI] [PubMed] [Google Scholar]
- Canter Cremers H. C., Batley M., Redmond J. W., Eydems L., Breedveld M. W., Zevehuizen L. P., Pees E., Wijffelman C. A., Lugtenberg B. J. Rhizobium leguminosarum exoB mutants are deficient in the synthesis of UDP-glucose 4'-epimerase. J Biol Chem. 1990 Dec 5;265(34):21122–21127. [PubMed] [Google Scholar]
- Chakravorty A. K., Zurkowski W., Shine J., Rolfe B. G. Symbiotic nitrogen fixation: molecular cloning of Rhizobium genes involved in exopolysaccharide synthesis and effective nodulation. J Mol Appl Genet. 1982;1(6):585–596. [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic S. P., Chen H., Batley M., Redmond J. W., Rolfe B. G. Nitrogen fixation ability of exopolysaccharide synthesis mutants of Rhizobium sp. strain NGR234 and Rhizobium trifolii is restored by the addition of homologous exopolysaccharides. J Bacteriol. 1987 Jan;169(1):53–60. doi: 10.1128/jb.169.1.53-60.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M. Genetic and physical analyses of group E exo- mutants of Rhizobium meliloti. J Bacteriol. 1988 Jan;170(1):474–477. doi: 10.1128/jb.170.1.474-477.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan T. M., Hirsch A. M., Leigh J. A., Johansen E., Kuldau G. A., Deegan S., Walker G. C., Signer E. R. Symbiotic mutants of Rhizobium meliloti that uncouple plant from bacterial differentiation. Cell. 1985 Apr;40(4):869–877. doi: 10.1016/0092-8674(85)90346-0. [DOI] [PubMed] [Google Scholar]
- Glazebrook J., Walker G. C. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell. 1989 Feb 24;56(4):661–672. doi: 10.1016/0092-8674(89)90588-6. [DOI] [PubMed] [Google Scholar]
- Gough J. A., Murray N. E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983 May 5;166(1):1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- Gray J. X., Djordjevic M. A., Rolfe B. G. Two genes that regulate exopolysaccharide production in Rhizobium sp. strain NGR234: DNA sequences and resultant phenotypes. J Bacteriol. 1990 Jan;172(1):193–203. doi: 10.1128/jb.172.1.193-203.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray J. X., Rolfe B. G. Exopolysaccharide production in Rhizobium and its role in invasion. Mol Microbiol. 1990 Sep;4(9):1425–1431. doi: 10.1111/j.1365-2958.1990.tb02052.x. [DOI] [PubMed] [Google Scholar]
- Halverson L. J., Stacey G. Signal exchange in plant-microbe interactions. Microbiol Rev. 1986 Jun;50(2):193–225. doi: 10.1128/mr.50.2.193-225.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth R. I., Dazzo F. B., Hallenga K., Musselman B. The complete structure of the trifoliin A lectin-binding capsular polysaccharide of Rhizobium trifolii 843. Carbohydr Res. 1988 Jan 15;172(1):97–112. doi: 10.1016/s0008-6215(00)90845-0. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Klein S., Hirsch A. M., Smith C. A., Signer E. R. Interaction of nod and exo Rhizobium meliloti in alfalfa nodulation. Mol Plant Microbe Interact. 1988 Feb;1(2):94–100. doi: 10.1094/mpmi-1-094. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Lee C. C. Characterization of polysaccharides of Rhizobium meliloti exo mutants that form ineffective nodules. J Bacteriol. 1988 Aug;170(8):3327–3332. doi: 10.1128/jb.170.8.3327-3332.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Reed J. W., Hanks J. F., Hirsch A. M., Walker G. C. Rhizobium meliloti mutants that fail to succinylate their calcofluor-binding exopolysaccharide are defective in nodule invasion. Cell. 1987 Nov 20;51(4):579–587. doi: 10.1016/0092-8674(87)90127-9. [DOI] [PubMed] [Google Scholar]
- Leigh J. A., Signer E. R., Walker G. C. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J. C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990 Apr 19;344(6268):781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- Long S. R. Rhizobium-legume nodulation: life together in the underground. Cell. 1989 Jan 27;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- Long S., Reed J. W., Himawan J., Walker G. C. Genetic analysis of a cluster of genes required for synthesis of the calcofluor-binding exopolysaccharide of Rhizobium meliloti. J Bacteriol. 1988 Sep;170(9):4239–4248. doi: 10.1128/jb.170.9.4239-4248.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip-Hollingsworth S., Hollingsworth R. I., Dazzo F. B. Host-range related structural features of the acidic extracellular polysaccharides of Rhizobium trifolii and Rhizobium leguminosarum. J Biol Chem. 1989 Jan 25;264(3):1461–1466. [PubMed] [Google Scholar]
- REYNOLDS E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963 Apr;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky M. E., Staneloni R. J., Leloir L. F. Lipid-bound saccharides in Rhizobium meliloti. J Biol Chem. 1982 Jun 25;257(12):6751–6757. [PubMed] [Google Scholar]
- Uttaro A. D., Cangelosi G. A., Geremia R. A., Nester E. W., Ugalde R. A. Biochemical characterization of avirulent exoC mutants of Agrobacterium tumefaciens. J Bacteriol. 1990 Mar;172(3):1640–1646. doi: 10.1128/jb.172.3.1640-1646.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Whitfeld P. L., Seeburg P. H., Shine J. The human pro-opiomelanocortin gene: organization, sequence, and interspersion with repetitive DNA. DNA. 1982;1(2):133–143. doi: 10.1089/dna.1.1982.1.133. [DOI] [PubMed] [Google Scholar]
- Zhan H. J., Gray J. X., Levery S. B., Rolfe B. G., Leigh J. A. Functional and evolutionary relatedness of genes for exopolysaccharide synthesis in Rhizobium meliloti and Rhizobium sp. strain NGR234. J Bacteriol. 1990 Sep;172(9):5245–5253. doi: 10.1128/jb.172.9.5245-5253.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan H. J., Levery S. B., Lee C. C., Leigh J. A. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc Natl Acad Sci U S A. 1989 May;86(9):3055–3059. doi: 10.1073/pnas.86.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]