Abstract
We have determined the subcellular localization of the chromosome partition protein Spo0J of Bacillus subtilis by immunofluorescence microscopy and visualizing fluorescence of a Spo0J–GFP fusion protein. Spo0J was associated with a region of the nucleoid proximal to the cell pole, both in growing cells dividing symmetrically and in sporulating cells dividing asymmetrically. Additional experiments indicated that Spo0J was bound to sites in the origin-proximal third of the chromosome. These results show that the replicating chromosomes are oriented in a specific manner during the division cycle, with the Spo0J binding region positioned toward the cell poles. Experiments characterizing cells at different stages of the cell cycle showed that chromosome orientation is established prior to the initiation of cell division. Our results indicate that there is a mechanism for orienting the chromosomes and that the chromosome partition protein Spo0J might be part of a bacterial mitotic-like apparatus.
Chromosome segregation is a fundamental process necessary for propagation of all organisms. Although not fully understood, many components of the segregation machinery have been identified and characterized in eukaryotes, including, centromeres, centromere-binding proteins, and the mitotic apparatus (1, 2). In contrast, these components, or their analogues, have not been identified in bacteria.
The study of bacterial chromosome segregation has focused mostly on Escherichia coli and Bacillus subtilis (3–5). Both organisms have a circular chromosome with a single origin of replication. Segregation is divided into two steps: physical separation of replicated chromosomes and partitioning of these chromosomes to daughter cells. Topoisomerases and site-specific recombinases are involved in the decatenation and separation of the replicated chromosomes. Several gene products have been identified that are involved in the partitioning of chromosomes to dividing cells (e.g., mukA, mukB), but the molecular mechanisms by which these proteins function are not yet known (3, 5). While it is clear that the chromosomes are associated with the cell membrane, at least during part of the cell cycle, it is not known if or how this association is related to partitioning. In addition, it is not clear if the circular chromosome has a defined orientation in the cell (3–5).
In B. subtilis, the spo0J gene product is needed for the initiation of sporulation and for proper chromosome partitioning during vegetative growth (6) and sporulation (7). soj, the gene immediately upstream from and cotranscribed with spo0J, is a negative regulator of sporulation (6) but does not seem to be required for partitioning. Null mutations in soj bypass the need for spo0J in sporulation, but not partitioning (6, 7).
Soj and Spo0J are similar to a family of proteins involved in plasmid partitioning, including ParA/ParB of prophage P1 and SopA/SopB of F (3, 5, 8). ParB (SopB) binds to a centromere-like site, parS, located near the plasmid origin of replication, and by a mechanism that is not clear, mediates partitioning. Null mutations in parB (or sopB) or deletion of the binding site (parS or sopC) lead to a rate of plasmid loss ≈100-fold greater than that of wild type (9–11). The plasmid-encoded ParA proteins are also required for partitioning. ParA is an ATPase that binds DNA, regulates transcription, and interacts with ParB (3, 12, 13). Soj is a DNA binding protein (D.C.-H.L. and A.D.G., unpublished results), and, by analogy to ParA and ParB, is probably an ATPase that interacts with Spo0J.
In B. subtilis, null mutations in spo0J cause a defect in chromosome partitioning during vegetative growth. Approximately 1.5% of cells in a growing culture of a spo0J mutant are anucleate, a frequency ≈100-fold higher than that observed in wild-type cells (6). spo0J homologues have been found in several bacterial species, including Pseudomonas putida (8), Coxiella burnetii, Mycobacterium leprae, and Caulobacter crescentus (J. Gober, personal communication), and it is likely that these homologues are also involved in chromosome partitioning.
Spo0J is also involved in chromosome partitioning during sporulation (7). Sporulating cells divide asymmetrically to produce two cell types, the larger mother cell and the smaller forespore, each with an intact chromosome and a distinct pattern of gene expression. Immediately following asymmetric division, ≈30% of the chromosome, centered around the origin of replication, is positioned in the forespore (14). The remainder of the chromosome is translocated into the forespore in a process that requires the spoIIIE gene product (14, 15). In the absence of SpoIIIE, the oriC-proximal 30% of the chromosome is “trapped” in the forespore while the other 70% remains in the mother cell. Spo0J is involved in positioning the origin-proximal part of the chromosome in the forespore (7).
We have determined the subcellular location of Spo0J using immunofluorescence microscopy with anti-Spo0J antibodies, and using a fusion of Spo0J to the Aequorea victoria green fluorescent protein (GFP) (16). Spo0J was associated with the regions of the nucleoid located near the cell poles, both during symmetric division in growing cells, and asymmetric division in sporulating cells. Experiments with a spoIIIE mutant indicated that Spo0J colocalized with the 30% of the origin-proximal part of the chromosome that is trapped in the forespore. These results demonstrate that the chromosome is oriented in a specific manner such that the Spo0J binding region is placed toward a cell pole, and suggest that Spo0J is directly involved in chromosome partitioning.
MATERIALS AND METHODS
Plasmids.
spo0J-(his)6 was constructed in pET21 (Novagen). An XhoI site was introduced by site directed mutagenesis (17) at the 3′ end of spo0J, in place of the stop codon. An EcoRI to XhoI fragment containing spo0J was cloned into pET21 to generate pDL3, containing spo0J-(his)6.
pDL50B contains the in-frame spo0J–gfp fusion in the vector pGEMcat. Briefly, spo0J-(his)6, with the XhoI site at the junction between spo0J and the (his)6 tag, was cloned into pGEMcat (18) to give pDL8. gfp was PCR amplified from plasmid pJK19–1 [gift from J. Kahana (19)] using PCR primers LIN13 (5′-GGAGATCTCGAGATGGCTAGCAAAGGAG-3′) and LIN14 (5′-GATCATGGCATGCACACCCGTCCTGTG-3′). The PCR product was digested with XhoI and SphI (introduced in the primers, underlined above) and ligated into pDL8 that had been digested with XhoI and SphI to produce plasmid pDL50B.
Strains.
Wild-type B. subtilis strains were JH642 (trp, phe) (20) and PY79 (prototroph) (21). The spoIIIE mutant was RL1259 [spoIIIE36 amyE::(sspE(2G)-lacZ tet)]. The spoIIIE36 mutation (15) and sspE(2G)–lacZ fusion (22) have been described. The spo0J–gfp fusion was introduced into B. subtilis strain AG1468 (Δspo0J::spc) (6) by single crossover at spo0J, selecting for chloramphenicol-resistance, to give strain DCL233. BL21 lambda DE3 (Novagen) was the E. coli strain used to overexpress Spo0J-(his)6.
Antibodies.
Antibodies against Spo0J-(his)6 were raised in rabbits (Babco, Richmond, CA) and antibodies against FtsZ were raised in chickens (Immuno-Dynamics, La Jolla, CA). Antibodies were affinity-purified essentially as described (23). We estimate that there are ≈200–400 molecules of Spo0J per cell during growth in minimal medium, and ≈500–1,000 molecules per cell during sporulation, based on immuno-blot analysis and comparison to known amounts of purified Spo0J protein (D.C.-H.L. and A.D.G., unpublished results).
Immunofluorescence Microscopy.
Cells were grown at 37°C in S750 minimal medium (24) with 0.1% glucose, 0.1% glutamate, and required amino acids (40 μg/ml), and samples were taken during exponential growth. Cells were induced to sporulate by the resuspension method (25, 26). Samples were taken at 0, 90, and 180 min after the onset of sporulation (the time of resuspension).
Cells were prepared for immunofluorescence microsocopy essentially as described (27, 28). Secondary antibodies coupled to fluorophores fluorescein isothiocyanate or Cy3 (Jackson ImmunoResearch) were used as indicated. Two different microscopes were used and the fluorescence of the Cy3 fluorophore appeared orange with one (see Fig. 1 B, F, and Q) and red with the other (Fig. 1 M and O). A spo0J null mutant had no detectable immunostaining with the anti-Spo0J antibodies (data not shown).
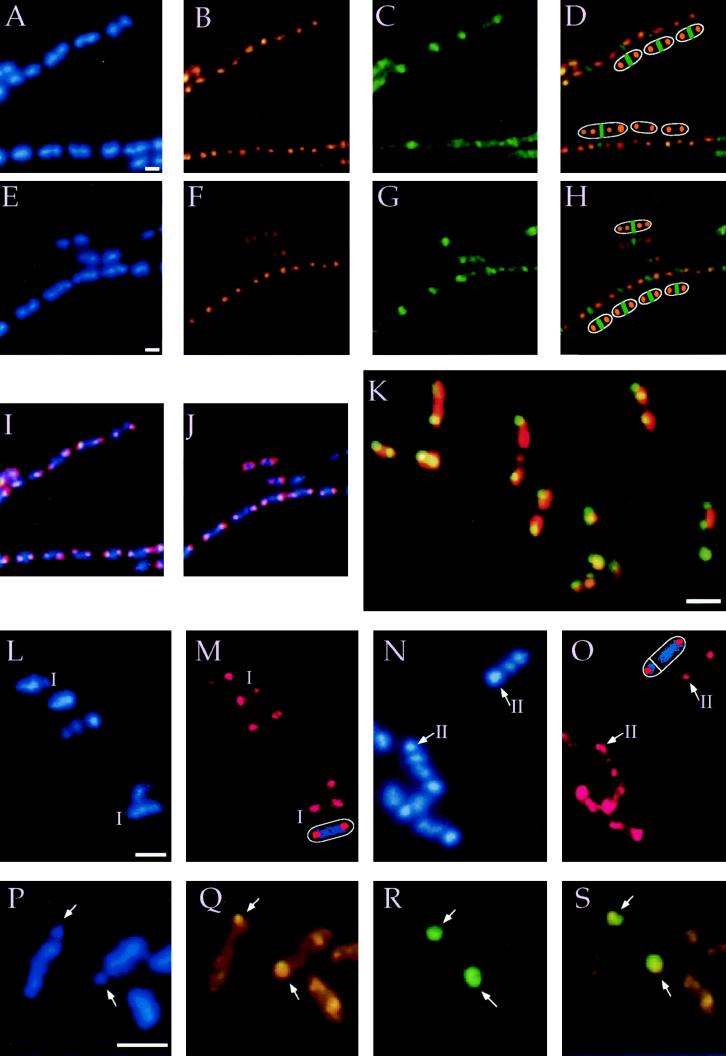
Figure 1.
Localization of Spo0J during growth and sporulation. (Bars ≈1 μm.) (A–J). Growth in minimal medium. A–D and I represent one field of cells, and E–H and J a different field of cells. (A and E) DAPI staining to visualize nucleoids (blue). (B and F) Immunostaining of Spo0J with affinity-purified rabbit antibodies and secondary antibodies coupled to the fluorophore Cy-3 (orange). (C and G) Immunostaining of FtsZ with affinity-purified chicken antibodies and secondary antibodies coupled to the fluorophore fluorescein isothiocyanate (green). (D) Overlay of exposures of Spo0J (B) and FtsZ (C). Cartoons of six cells are indicated. The three cells at the top of each panel have an FtsZ ring with a single site of Spo0J staining per nucleoid (class 3 of Fig. 2). Of the three cells at the bottom, the large cell to the left has an FtsZ ring with a nucleoid on each side and two sites of Spo0J per nucleoid (class 4 of Fig. 2). The other two cells have no FtsZ ring and a single nucleoid with two sites of Spo0J (class 2 of Fig. 2). (H) Overlay of exposures of Spo0J (F) and FtsZ (G). Cartoons of five cells are indicated; all have FtsZ rings. The large cell at the top has two sites of Spo0J per nucleoid (class 4 of Fig. 2), while all of the cells at the bottom have one site of Spo0J per nucleoid (class 3 of Fig. 2). (I) Overlay of exposures of DAPI (A) and Spo0J (B). (J) Overlay of exposures of DAPI (D) and Spo0J (E). (K) Spo0J–GFP localizes to the nucleoids in a bipolar manner. Living cells were stained with 7-amino-actinomycin D to visualize the DNA (red). Endogenous fluorescence from Spo0J–GFP appears yellow when it completely overlaps with the red from the DNA. (L–O) Localization of Spo0J during early stages of sporulation in wild-type cells. Cells were stained with DAPI (blue) to visualize DNA (L and N), and immuno-stained (red) to visualize Spo0J (M and O). (L and M) Three cells are at stage I, indicated by “I,” and a sketch of one of these is shown in M. (N and O) Arrows point to the condensed forespore nucleoids from two different stage II sporangia (II). A sketch is shown of one of these at the top of O. (P–S) Spo0J colocalizes with the origin-proximal 30% of the chromosome in the forespore of a spoIIIE mutant. Two sporangia of the spoIIIE mutant stained with DAPI (blue) to visualize DNA (P), and immunostained for Spo0J (orange) (Q), and β-galactosidase (green) (R). β-Galactosidase is produced from the sspE(2G)–lacZ fusion that is expressed only in the forespore. The fusion is in the origin-proximal part of the chromosome that gets trapped in the forespore in the spoIIIE mutant. (S) Overlay of the Spo0J and β-galactosidase staining. Arrows indicate the forespore. Note that in contrast to wild-type sporangia (N) that have highly condensed forespore nucleoids, the spoIIIE mutant forespore (P) has much less staining, reflecting the absence of a complete chromosome.
Visualization of Spo0J–GFP.
Cells containing the spo0J–gfp fusion (DCL233) were grown to confluence on an Luria-Bertani agar plate, scraped from the plate, resuspended in minimal salts (29), and adhered to a poly-l-lysine-coated slide. DNA was visualized by staining with 7-amino-actinomycin D (Molecular Probes). GFP was visualized essentially as described (30).
RESULTS
Localization of Spo0J During Vegetative Growth.
To identify the subcellular location of Spo0J, we performed immunofluorescence microscopy using antibodies against Spo0J. During vegetative growth in defined minimal medium (doubling time ≈45 min), Spo0J was associated with the nucleoid. Fig. 1 A–J shows the staining pattern from two different fields of cells (A–D and I representing one field and E–H and J the other). Nucleoids were visualized with 4′,6-diamidino-2-phenylindole (DAPI) and appear as bright blue bodies, often in chains, as B. subtilis tends to grow as septated filaments of cells (Fig. 1 A and E). The majority of nucleoid bodies (≥75%) had discrete immuno-staining of Spo0J, with at least one defined site of Spo0J staining per nucleoid (Fig. 1 A, B, and I; E, F, and J). Many of the nucleoids had two defined sites of Spo0J staining, usually with each site on opposite ends of the nucleoid and toward the cell poles (Fig. 1 A, B, and I; E, F, and J).
To determine the location of Spo0J at different times during the cell cycle and relative to the midcell, we compared the localization of Spo0J to that of the cell division protein FtsZ. FtsZ forms a ring structure at the midcell, with a nucleoid on each side, and marks the site of septation (31–34). In cells without an FtsZ ring, the nucleoid is usually in the middle of the cell. As the cell grows and the chromosome replicates, the nucleoid begins to separate into two discrete chromosomal bodies and the FtsZ ring forms at midcell. The FtsZ ring contracts and disappears during septation, then reforms prior to the next division (reviewed in refs. 35 and 36).
Several different patterns of staining were observed when comparing the localization of Spo0J (Fig. 1 B and F), FtsZ (Fig. 1 C and G), and the nucleoids (Fig. 1 A and I; E and J). A total of 421 cells that had discrete sites of Spo0J staining were counted and divided into four classes, based on the presence or absence of the FtsZ ring, and the number of sites of Spo0J staining per nucleoid (Fig. 2A). Classes 1 and 2 consist of cells with no FtsZ ring and a single nucleoid body with either a single site (class 1) or two sites (class 2) of Spo0J staining. Classes 3 and 4 consist of cells with an FtsZ ring and two nucleoid bodies, one on each side of the FtsZ ring and a single site (class 3) or two sites (class 4) of Spo0J staining per nucleoid. The four classes are described in more detail below.
Figure 2.
Analysis of Spo0J localization. Shaded blobs represent the nucleoids and black balls indicate Spo0J on the nucleoid. (A) Localization of Spo0J during vegetative growth (as in Fig. 1 A–J). In several different experiments, 75–90% of the cells had discrete sites of Spo0J staining and ≈60% of all cells had visible, well-defined FtsZ rings. The proportion of cells with discrete sites of Spo0J staining was similar in cells with and without FtsZ rings, indicating that the appearance of sites of Spo0J staining does not correlate with the presence or absence of FtsZ rings. (Classes 1 and 2) Cells with no FtsZ ring and a single nucleoid body with either a single site (class 1) or two sites (class 2) of Spo0J staining. (Classes 3 and 4) Cells with an FtsZ ring (indicated by the oval at midcell) and two nucleoid bodies, one on each side of the FtsZ ring and a single site (class 3) or two sites (class 4) of Spo0J staining per nucleoid. Of the 117 cells with an FtsZ ring and two sites of Spo0J per nucleoid, 7 cells actually had two sites on one nucleoid and only one site on the other nucleoid. We infer that in a small fraction of cells, reinitiation of replication or subsequent elongation did not occur synchronously on each chromosome. Alternatively, there were two Spo0J sites, but we could not resolve them. If these cells divide before the appearance of the second site of Spo0J staining, they would produce a few cells with a single nucleoid with a single site of Spo0J staining (class 1). For simplicity, cells in all four classes are drawn the same length. In fact, there is a rough correlation with the indicated class and cell length, with class 4 cells the longest and class 1 and 2 cells the shortest (see Fig. 1). (B) Localization of Spo0J during stage I of sporulation. (C) Localization of Spo0J during stage II of sporulation.
A small proportion of cells (4%) had a single nucleoid with one site of Spo0J staining and no FtsZ ring (Fig. 2A, class 1). (There are no examples of these in Fig. 1 A–J.) These are probably cells that have recently divided (no FtsZ ring), but that have not replicated enough of the chromosome to duplicate the Spo0J binding site.
Of the 165 cells with no FtsZ ring (18 class 1 plus 147 class 2), the majority (87%) had two distinct sites of Spo0J staining on a single nucleoid (Fig. 2A, class 2; Fig. 1 A–D and I). In these cells, the sites of Spo0J localization were near the ends of the nucleoid, toward the cell poles. These are probably cells that have replicated the Spo0J binding region and have recently divided (no FtsZ ring). The two distinct sites of Spo0J localization at the ends of the nucleoid indicate that chromosome orientation has already been established and that the newly replicated regions have been separated. Furthermore, the Spo0J binding region is likely to be in the part of the chromosome that replicates early (the origin region).
[The formal possibility exists that there are two Spo0J binding regions per chromosome. This seems highly unlikely based on the relatively large proportion of cells (see below) that have distinct nucleoids on each side of the FtsZ ring with a single site of Spo0J localization per nucleoid. In addition, if there were two distinct binding regions per chromosome, we would expect to see a significant number of nucleoids with three sites of Spo0J staining. This has not been observed.]
Approximately 60% of the cells had an FtsZ ring at midcell. [In contrast, >90% of cells growing in rich medium, with a rapid doubling time, have an FtsZ ring (32, 34)]. The cells that had FtsZ rings and discrete sites of Spo0J staining fell into two classes (Fig. 2A, classes 3 and 4), one class with a single site (class 3) and the other with two sites (class 4) of Spo0J staining per nucleoid. Fig. 1 D and H shows an overlay of the Spo0J (Fig. 1 B and F) and FtsZ (Fig. 1 C and G) staining and a sketch of several of the cells next to the image. Fig. 1 I and J shows an overlay of the DNA (A and E) and Spo0J (B and F) staining. Nucleoids with single sites and those with two sites of Spo0J staining are clearly visible and are illustrative of the way in which other cells were classified.
Of the 256 cells with an FtsZ ring (139 class 3 plus 117 class 4), 54% had a single site of Spo0J per nucleoid (Fig. 2A, class 3) while 46% had two sites of Spo0J per nucleoid (Fig. 2A, class 4). In the cells with a single site of Spo0J staining per nucleoid, Spo0J often appeared oriented toward the cell pole, away from the FtsZ ring at midcell.
A significant fraction (46%) of the cells with an FtsZ ring had two sites of Spo0J staining per nucleoid (Fig. 2A, class 4; Fig. 1 A–J). Each nucleoid had a site of Spo0J localization toward the cell pole and a second site of Spo0J localization closer to the FtsZ ring at midcell. We infer that these chromosomes have already begun a new round of DNA replication and that the Spo0J binding region has been duplicated, indicating again that the Spo0J binding region is probably near the origin of replication. In addition, since the FtsZ ring is still present and Spo0J is localized in a bipolar manner on each nucleoid, before cell division, the defined orientation and polarity of the chromosome is established for cell division in the next generation, before the current division has been completed. The machinery that separates the replicating chromosomes and establishes this polarity must be assembled and functional in this predivisional cell. After division, the young cells do not have an FtsZ ring and most have two sites of Spo0J (Fig. 2A, class 2) localized in the bipolar manner that was established before division.
Visualization of Spo0J–GFP.
To visualize the subcellular localization of Spo0J in living cells, in the absence of fixation, we constructed a Spo0J–GFP fusion and determined the localization based on endogenous fluorescence of GFP. The spo0J–gfp fusion produced a functional gene product as judged by the ability to complement a spo0J null mutation (data not shown). Spo0J–GFP was associated with the nucleoid and most cells had Spo0J clearly localized toward the poles of the nucleoid (Fig. 1K). These results are consistent with those from immunofluorescence and indicate that the bipolar localization of Spo0J was not caused by potential artifacts from fixation, permeablization, or any other immunostaining procedures.
Localization of Spo0J During Sporulation.
Because Spo0J is involved in chromosome partitioning during asymmetric division in sporulating cells (7), we determined the subcellular location of Spo0J during the early stages of sporulation. Cells entering stage I of sporulation have an axial filament, a single nucleoid body (containing at least two chromosomes) that stretches the length of the predivisional cell. Fig. 1L shows the DAPI staining of several cells, three of which are at stage I of sporulation (I). Fig. 1M is the same field of cells, but with Spo0J staining shown, and a sketch of one of the sporangia below the image. During sporulation, >90% of the cells that had an axial filament had discrete staining of Spo0J. In 86% of these cells, Spo0J was associated with both poles of the axial filament (as seen in Fig. 1 L and M; Fig. 2B), while in the remaining 14%, Spo0J was localized to one end of the axial filament (Fig. 2B).
At the beginning of stage II of sporulation, a polar septum is formed creating two cells of unequal size; the larger mother cell and the smaller forespore. The stage II sporangia (mother cell plus forespore) are distinguishable by DAPI staining as the forespore nucleoid is highly condensed and brightly staining while the mother cell nucleoid is more diffuse (37, 38). Fig. 1N shows the DAPI staining of several stage II sporangia, with arrows pointing to the condensed forespore nucleoid in two of these. Over 90% of the sporangia with a condensed forespore nucleoid (examples in Fig. 1 N and O) had discrete immunostaining of Spo0J. In 76% of these, Spo0J was still associated with the nucleoid in a bipolar manner (Fig. 1 N and O; Fig. 2C). That is, both the mother cell and forespore nucleoids had Spo0J staining, and in the mother cell, this staining was toward the cell pole away from the forespore. In the much smaller forespore, it is difficult to determine if Spo0J staining is to a localized part of the highly condensed nucleoid. At later times during sporulation, the chromosomes have already segregated and fewer sporangia had discrete localization of Spo0J (data not shown).
Spo0J Binds to the Region of the Chromosome Around the Origin of Replication.
When the polar septum forms at stage II to generate the two cell types, it bisects one end of the axial filament, trapping ≈30% of one chromosome in the smaller forespore and leaving ≈70% of the forespore chromosome in the larger mother cell. Mutations in spoIIIE prevent the translocation of the chromosome into the forespore, resulting in a forespore that contains only the origin-proximal 30% of the chromosome (14, 15).
We found that Spo0J was localized to the 30% of the chromosome that gets trapped in the forespore in a spoIIIE mutant. Fig. 1 P–S shows two sporangia from the spoIIIE mutant. The arrows point to the two forespore cells, each with a bit of the nucleoid (Fig. 1P). We visualized the forespore by immunostaining β-galactosidase (Fig. 1R, green) that was produced from a gene fusion [sspE(2G)–lacZ] that is in the origin-proximal 30% of the chromosome and that is expressed only in the forespore. In 53 sporangia examined that had β-galactosidase staining in the forespore, 48 sporangia had Spo0J staining (Fig. 1 Q and S) that colocalized with β-galactosidase staining (Fig. 1 R and S). This indicates that Spo0J binds to a region of the origin-proximal 30% of the chromosome that is trapped in the forespore in the spoIIIE mutant, consistent with our results from growing cells that also indicated the Spo0J binding region is in the origin-proximal part of the chromosome.
DISCUSSION
We favor a model in which Spo0J is a direct participant in chromosome partitioning, during both symmetric division in vegetatively growing cells and asymmetric division in sporulating cells. Evidence supporting this model includes the bipolar localization of Spo0J on the nucleoids, the chromosome partition defect during growth (6) and sporulation (7) caused by spo0J null mutations, and the homology of Spo0J to partition proteins of the ParB family (8). We propose that Spo0J assembles on a centromere-like site, probably in the origin region, and that this assembly interacts with as yet unidentified cellular machinery that drives the partitioning of the nucleoids to the daughter cells. While a specific binding site for Spo0J has not yet been identified, in vitro experiments indicate that Spo0J is a DNA binding protein with a high degree of cooperativity (D.C.-H.L. and A.D.G., unpublished results). We suspect that in vivo, Spo0J binding to specific sites helps to nucleate assembly of a large complex that contains many molecules of Spo0J.
The machinery that orients Spo0J in a bipolar manner on the nucleoid is established in the predivisional cell. Based on our results, it is clear that replicating chromosomes are in a defined orientation in the cell, with the Spo0J binding region of each chromosome oriented toward the cell poles during most of the cell cycle. Similar results have been obtained independently by Errington and coworkers (J. Errington, personal communication).
Webb et al. (39) used a different approach to visualize the orientation of the chromosome. The oriC region was visualized using a LacI–GFP fusion bound to an array of lac operators that had been integrated into the chromosome near oriC. Fluorescence from LacI–GFP was observed toward the cell poles (39), in a manner similar to the localization of Spo0J (Fig. 1). In contrast, in cells with the lac operator array near the terminus of replication, the fluorescence was observed closer to midcell (39). The results with LacI–GFP indicate that both the origin and terminus of replication can be oriented in a specific manner, at least during part of the B. subtilis cell cycle.
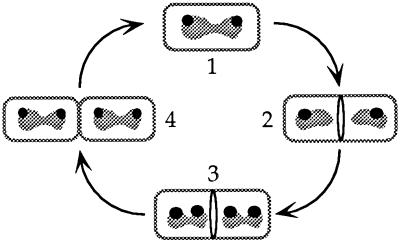
We envision the following sequence of events during the bacterial cell cycle (Fig. 3), keeping in mind that in rapidly growing cells (doubling time ≤ 60 min.) there is more than one replication fork per chromosome to compensate for the fact that chromosomal replication takes longer than cell division. (1) Immediately after cell division, there is a single, partly replicated nucleoid located at approximately midcell. Most of these cells have two sites of Spo0J localization, one at each end of the nucleoid, oriented toward a cell pole. (2) During cell growth and continued DNA replication, the nucleoid begins to separate into two distinct bodies that move in opposite directions toward the cell poles. During this time, the FtsZ ring forms at midcell and marks the site of the next division event (31, 35, 36). As the nucleoids become separated, localization of Spo0J at the end of each nucleoid toward the cell poles and away from the FtsZ ring becomes even more pronounced. (3) Before cell division, another round of DNA replication initiates in most cells and proceeds far enough such that the Spo0J binding region is replicated. This appears in our experiments as a nucleoid with two distinct sites of Spo0J, one toward the cell pole and the other closer to the FtsZ ring at midcell. This observation indicates that the newly replicated origin-proximal regions (including the Spo0J binding regions) are separated before the cell divides. (4) Finally, cell division occurs, and the FtsZ ring contracts and disassembles (31, 33). In a small percentage of the cells (Fig. 2A, class 1), division occurs before replication has reinitiated, or at least before a second Spo0J binding region is visible.
Figure 3.
Model for chromosome segregation and Spo0J localization during the cell cycle. (1) Immediately after cell division, there is no FtsZ ring and a partly replicated chromosome with two discrete sites of Spo0J staining oriented toward the cell poles. (2) As replication and cell growth continue, the FtsZ ring forms and the chromosomes continue to separate. Spo0J localization becomes markedly polar, with one site on each nucleoid oriented toward the cell pole and away from the FtsZ ring at midcell. (3) DNA replication reinitiates, most of the time on both chromosomes, and progresses far enough so that the Spo0J binding region is replicated. This is visualized as two discrete sites of Spo0J staining on each of the two nucleoids. One site on each nucleoid is toward a cell pole while the other site is closer to the FtsZ ring at midcell. (4) The FtsZ ring contracts and disassembles during septation and division, generating two cells with partly replicated chromosomes and two distinct sites of Spo0J staining.
Spo0J is an important component contributing to the fidelity of chromosome partitioning. The frequency of anucleate cells caused by a spo0J null mutation is ≈100-fold higher than that of wild-type cells (6). Spo0J (ParB) homologues have been found, based on chromosomal DNA sequence, in several other organisms, including P. putida (8), C. burnetii, and M. leprae. The Spo0J homologue of C. crescentus is involved in chromosome partitioning (42), and it seems likely that the ParB/Spo0J proteins from other organisms will be found to have similar roles in partitioning. These organisms do not undergo sporulation, indicating that Spo0J first evolved to help ensure the proper transmission of chromosomes (or plasmids) to dividing cells.
We suspect that during the course of evolution, B. subtilis adapted part of the chromosome segregation machinery (Spo0J) to create a checkpoint that couples the initiation of sporulation to cell cycle events. While Spo0J plays a functional role in chromosome segregation during vegetative growth and sporulation, it also plays a regulatory role during the initiation of sporulation. Spo0J acts to inhibit the function of the regulatory protein encoded by soj. Soj is a negative regulator of sporulation, functioning to inhibit or antagonize the function of the transcription factor Spo0A (6). The Spo0A transcription factor activates many of the early sporulation genes and is required for formation of the axial filament and to activate the switch from symmetric to asymmetric division (34, 40, 41). We suspect that at a particular step in the partitioning pathway, Spo0J acts to antagonize Soj, signaling that the chromosome segregation process is functioning normally.
The components of the bacterial chromosome segregation machinery have remained elusive. However, Spo0J of B. subtilis should now provide access to these components, including a centromere-like site, factors that assemble on this site, and the machinery that drives segregation of the chromosomes to dividing cells. It will be interesting to use Spo0J to identify these components and to determine how chromosomes are accurately segregated, how cell polarity is used to generate chromosomal orientation, how the various cell cycle processes are regulated and coordinated, and how they have been adapted to regulate developmental processes.
Acknowledgments
We thank C. Kaiser and P. Sorger and members of their labs for advice and for use of their fluorescence microscopes; C. Webb for advice and help with visualization of Spo0J-GFP; and J. Gober and J. Errington for communication of results prior to publication. We are grateful to members of our lab for useful discussions and comments on the manuscript, and to T. Baker, S. Bell, K. Ireton, R. Losick, and C. Webb for comments on the manuscript. D.L. was supported in part by a National Science Foundation predoctoral grant. P.L. was supported in part by Fellowship DRG1397 of the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation. This work was also supported in part by grants to A.D.G. from the Lucille P. Markey Charitable Trust and Public Health Services Grant GM41934 from the National Institutes of Health.
ABBREVIATIONS
- GFP
green fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Murray A, Hunt T. The Cell Cycle: An Introduction. New York: Freeman; 1993. [Google Scholar]
- 2.Hyman A A, Sorger P K. Annu Rev Cell Dev Biol. 1995;11:471–495. doi: 10.1146/annurev.cb.11.110195.002351. [DOI] [PubMed] [Google Scholar]
- 3.Hiraga S. Annu Rev Biochem. 1992;61:283–306. doi: 10.1146/annurev.bi.61.070192.001435. [DOI] [PubMed] [Google Scholar]
- 4.Rothfield L I. Cell. 1994;77:963–966. doi: 10.1016/0092-8674(94)90435-9. [DOI] [PubMed] [Google Scholar]
- 5.Wake R G, Errington J. Annu Rev Genet. 1995;29:41–67. doi: 10.1146/annurev.ge.29.120195.000353. [DOI] [PubMed] [Google Scholar]
- 6.Ireton K, Gunther IV N W, Grossman A D. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharpe M E, Errington J. Mol Microbiol. 1996;21:501–509. doi: 10.1111/j.1365-2958.1996.tb02559.x. [DOI] [PubMed] [Google Scholar]
- 8.Ogasawara N, Yoshikawa H. Mol Microbiol. 1992;6:629–634. doi: 10.1111/j.1365-2958.1992.tb01510.x. [DOI] [PubMed] [Google Scholar]
- 9.Ogura T, Hiraga S. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 10.Austin S, Abeles A. J Mol Biol. 1983;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- 11.Lane D, Rothenbuehler R, Merrillat A M, Aiken C. Mol Gen Genet. 1987;207:406–412. doi: 10.1007/BF00331608. [DOI] [PubMed] [Google Scholar]
- 12.Davis M A, Martin K A, Austin S J. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 13.Davey M J, Funnell B E. J Biol Chem. 1994;269:29908–29913. [PubMed] [Google Scholar]
- 14.Wu L J, Errington J. Science. 1994;264:572–575. doi: 10.1126/science.8160014. [DOI] [PubMed] [Google Scholar]
- 15.Wu L J, Lewis P J, Allmansberger R, Hauser P M, Errington J. Genes Dev. 1995;9:1316–1326. doi: 10.1101/gad.9.11.1316. [DOI] [PubMed] [Google Scholar]
- 16.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 17.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1990. [Google Scholar]
- 18.Youngman P, Poth H, Green B, York K, Olmedo G, Smith K. In: Methods for Genetic Manipulation, Cloning, and Functional Analysis of Sporulation Genes in Bacillus subtilis. Smith I, Slepecky R, Setlow P, editors. Washington, DC: Am. Soc. Microbiol.; 1989. pp. 65–87. [Google Scholar]
- 19.Kahana J A, Silver P A. In: Supplement 34: Use of the A. victoria Green Fluorescent Protein to Study Protein Dynamics in Vivo. Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. New York: Wiley; 1996. pp. 9.7.22–9.7.28. [Google Scholar]
- 20.Perego M, Spiegelman G B, Hoch J A. Mol Microbiol. 1988;2:689–699. doi: 10.1111/j.1365-2958.1988.tb00079.x. [DOI] [PubMed] [Google Scholar]
- 21.Youngman P, Perkins J B, Losick R. Plasmid. 1984;12:1–9. doi: 10.1016/0147-619x(84)90061-1. [DOI] [PubMed] [Google Scholar]
- 22.Sun D, Fajardo-Cavazos P, Sussman M D, Tovar-Rojo F, Cabrera-Martinez R M, Setlow P. J Bacteriol. 1991;173:7867–7874. doi: 10.1128/jb.173.24.7867-7874.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pringle J R, Adams E E M, Drubin D G, Haarer B K. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- 24.Jaacks K J, Healy J, Losick R, Grossman A D. J Bacteriol. 1989;171:4121–4129. doi: 10.1128/jb.171.8.4121-4129.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholson W L, Setlow P. In: Sporulation, Germination and Outgrowth. Harwood C R, Cutting S M, editors. Chichester, U.K.: Wiley; 1990. pp. 391–450. [Google Scholar]
- 26.Sterlini J M, Mandelstam J. Biochem J. 1969;113:29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harry E J, Pogliano K, Losick R. J Bacteriol. 1995;177:3386–3393. doi: 10.1128/jb.177.12.3386-3393.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pogliano K, Harry E, Losick R. Mol Microbiol. 1995;18:459–470. doi: 10.1111/j.1365-2958.1995.mmi_18030459.x. [DOI] [PubMed] [Google Scholar]
- 29.Spizizen J. Proc Natl Acad Sci USA. 1958;44:1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb C D, Decatur A, Teleman A, Losick R. J Bacteriol. 1995;177:5906–5911. doi: 10.1128/jb.177.20.5906-5911.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bi E, Lutkenhaus J. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 32.Addinall S G, Bi E, Lutkenhaus J. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, Lutkenhaus J. Mol Microbiol. 1993;9:435–442. doi: 10.1111/j.1365-2958.1993.tb01705.x. [DOI] [PubMed] [Google Scholar]
- 34.Levin P A, Losick R. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 35.Erickson H P. Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 36.Lutkenhaus J. Mol Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 37.Setlow P. Semin Dev Biol. 1991;2:55–62. [Google Scholar]
- 38.Setlow B, Magill N, Febbroriello P, Nakhimovsky L, Koppel D E, Setlow P. J Bacteriol. 1991;173:6270–6278. doi: 10.1128/jb.173.19.6270-6278.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb C D, Teleman A, Gordon S, Straight A, Belmont A, Lin D, Grossman A D, Wright A, Losick R. Cell. 1997;88:667–674. doi: 10.1016/s0092-8674(00)81909-1. [DOI] [PubMed] [Google Scholar]
- 40.Hoch J A. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 41.Grossman A D. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 42.Mohl D A, Gober J W. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]