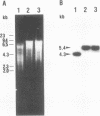
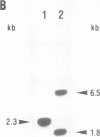
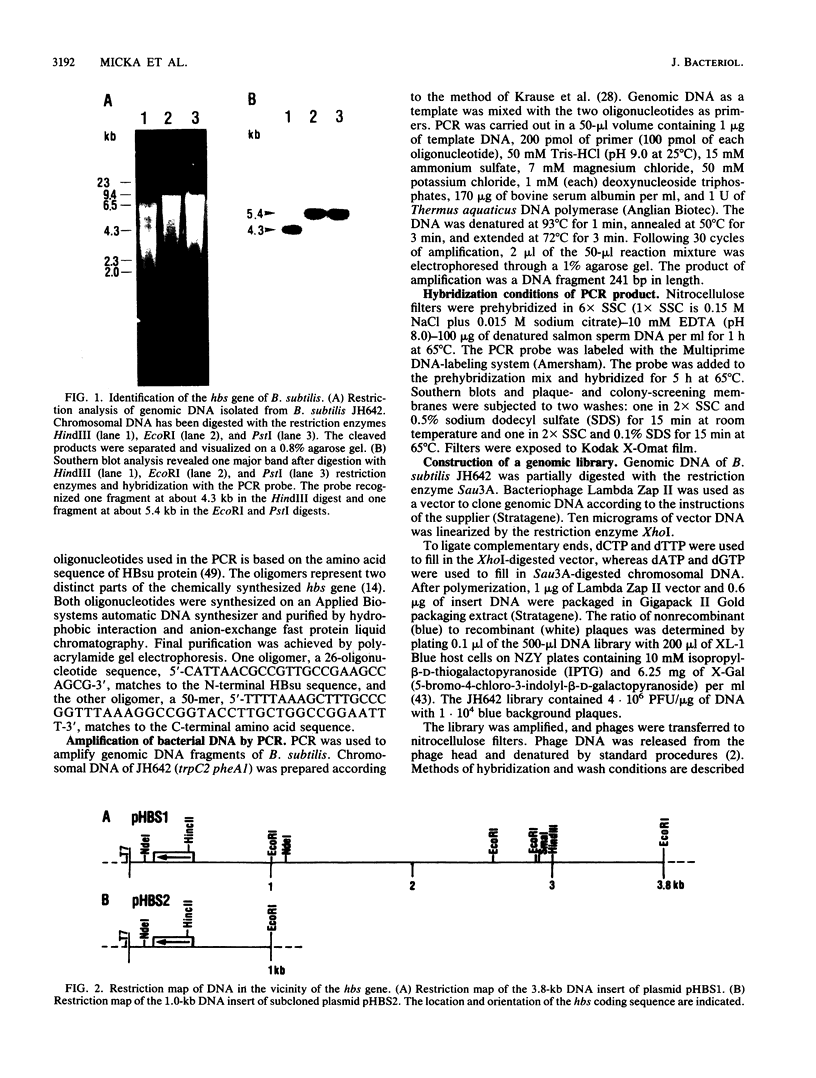
Abstract
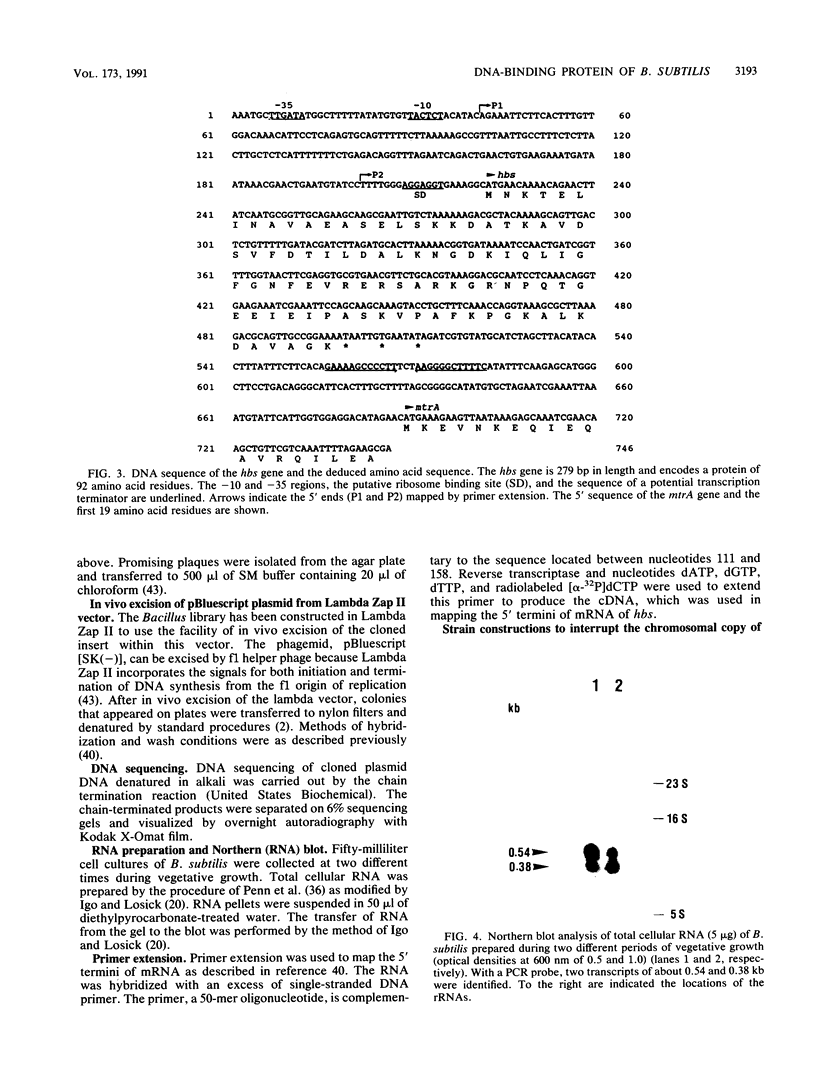
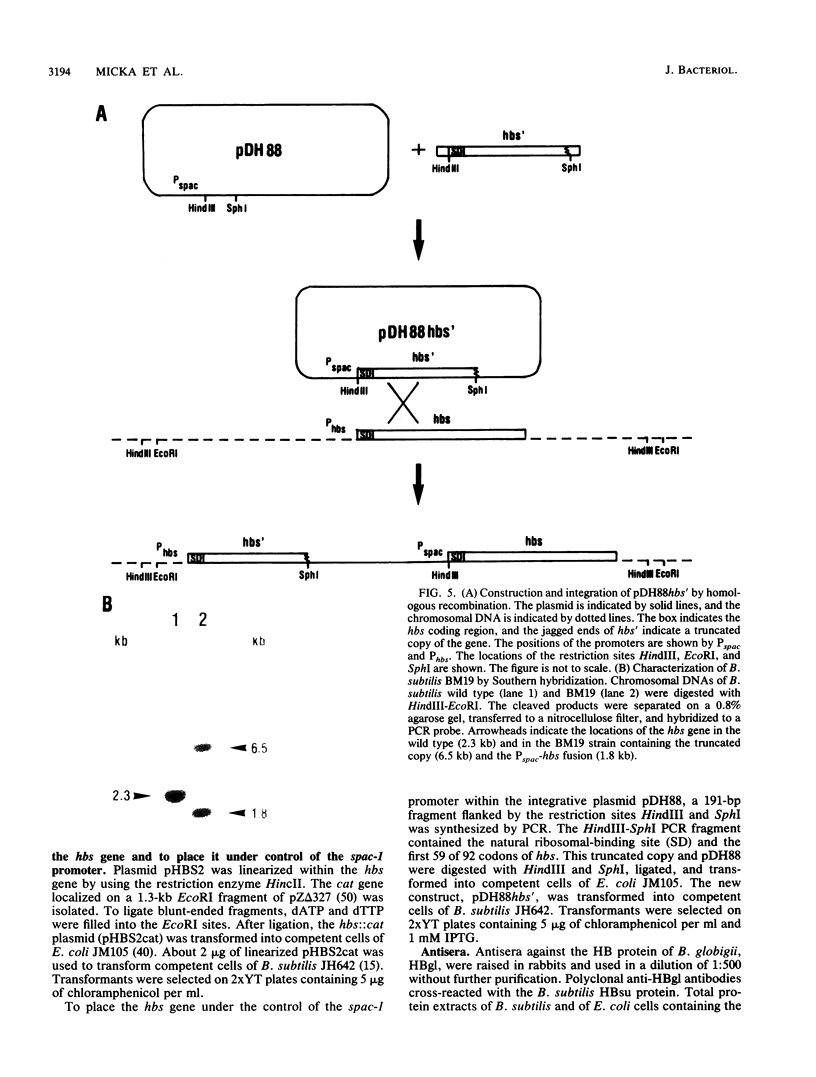
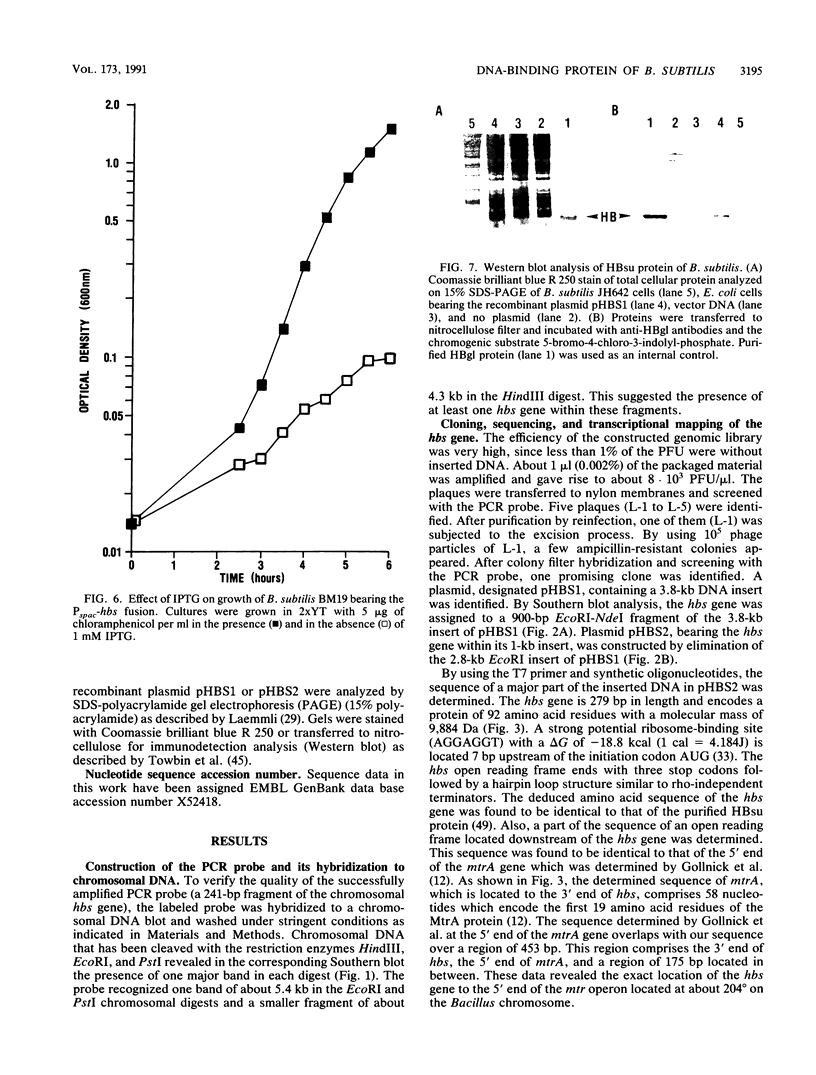
A homologous class of histonelike proteins which are believed to wrap the DNA and to condense the chromosome into highly folded nucleoid structures has been identified in different bacterial species. Bacillus subtilis encodes a homodimeric DNA-binding protein called HBsu. We have cloned the corresponding gene (hbs) on a 3.8-kb fragment. The gene was subcloned to a 1-kb fragment, sequenced, and characterized. It encodes a 92-amino-acid protein with a predicted molecular mass of 9,884 Da. Fortunately, analysis of the DNA sequence downstream of the 3' end of hbs revealed the location of the first 19 amino acid residues of MtrA. This finding located the hbs gene unequivocally to the 5' end of the mtr operon at about 204 degrees on the standard genetic map of B. subtilis. Northern (RNA) blot analysis and primer extension studies indicated the presence of two distinct hbs transcripts, which were found to be initiated at two different sites located about 160 bases apart. Several attempts to replace the hbs gene in the B. subtilis chromosome with a cat-interrupted copy (hbs::cat) through marker replacement recombination were unsuccessful. In order to study whether hbs is an essential gene, we have constructed a strain containing a truncated copy of the gene behind its own promoter and another intact copy under control of the isopropyl-beta-D-thiogalactopyranoside (IPTG)-inducible spac-1 promoter. In this strain (BM19), normal growth was found to depend on IPTG, whereas in the absence of IPTG, growth was severely affected. These results suggest an essential role for the hbs gene product for normal growth in B. subtilis.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck A., Dijk J., Reinhardt R. Ribosomal proteins and DNA-binding protein II from the extreme thermophile Bacillus caldolyticus. Biol Chem Hoppe Seyler. 1987 Feb;368(2):121–130. doi: 10.1515/bchm3.1987.368.1.121. [DOI] [PubMed] [Google Scholar]
- Broyles S. S., Pettijohn D. E. Interaction of the Escherichia coli HU protein with DNA. Evidence for formation of nucleosome-like structures with altered DNA helical pitch. J Mol Biol. 1986 Jan 5;187(1):47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- Dijk J., White S. W., Wilson K. S., Appelt K. On the DNA binding protein II from Bacillus stearothermophilus. I. Purification, studies in solution, and crystallization. J Biol Chem. 1983 Mar 25;258(6):4003–4006. [PubMed] [Google Scholar]
- Dixon N. E., Kornberg A. Protein HU in the enzymatic replication of the chromosomal origin of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(2):424–428. doi: 10.1073/pnas.81.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürrenberger M., Bjornsti M. A., Uetz T., Hobot J. A., Kellenberger E. Intracellular location of the histonelike protein HU in Escherichia coli. J Bacteriol. 1988 Oct;170(10):4757–4768. doi: 10.1128/jb.170.10.4757-4768.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Trach K., LeCoq D., Spence J., Ferrari E., Hoch J. A. Characterization of the spo0A locus and its deduced product. Proc Natl Acad Sci U S A. 1985 May;82(9):2647–2651. doi: 10.1073/pnas.82.9.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashner Y., Gralla J. D. DNA dynamic flexibility and protein recognition: differential stimulation by bacterial histone-like protein HU. Cell. 1988 Aug 26;54(5):713–721. doi: 10.1016/s0092-8674(88)80016-3. [DOI] [PubMed] [Google Scholar]
- Funnell B. E., Baker T. A., Kornberg A. In vitro assembly of a prepriming complex at the origin of the Escherichia coli chromosome. J Biol Chem. 1987 Jul 25;262(21):10327–10334. [PubMed] [Google Scholar]
- Geider K., Hoffmann-Berling H. Proteins controlling the helical structure of DNA. Annu Rev Biochem. 1981;50:233–260. doi: 10.1146/annurev.bi.50.070181.001313. [DOI] [PubMed] [Google Scholar]
- Gollnick P., Ishino S., Kuroda M. I., Henner D. J., Yanofsky C. The mtr locus is a two-gene operon required for transcription attenuation in the trp operon of Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8726–8730. doi: 10.1073/pnas.87.22.8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene J. R., Brennan S. M., Andrew D. J., Thompson C. C., Richards S. H., Heinrikson R. L., Geiduschek E. P. Sequence of the bacteriophage SP01 gene coding for transcription factor 1, a viral homologue of the bacterial type II DNA-binding proteins. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7031–7035. doi: 10.1073/pnas.81.22.7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Dubnau D. Construction and properties of chimeric plasmids in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1428–1432. doi: 10.1073/pnas.75.3.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henner D. J. Inducible expression of regulatory genes in Bacillus subtilis. Methods Enzymol. 1990;185:223–228. doi: 10.1016/0076-6879(90)85022-g. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Hillyard D. Primary structure and mapping of the hupA gene of Salmonella typhimurium. J Bacteriol. 1988 Dec;170(12):5751–5758. doi: 10.1128/jb.170.12.5751-5758.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillyard D. R., Edlund M., Hughes K. T., Marsh M., Higgins N. P. Subunit-specific phenotypes of Salmonella typhimurium HU mutants. J Bacteriol. 1990 Sep;172(9):5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., Faelen M., Girard D., Jaffé A., Toussaint A., Rouvière-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J Bacteriol. 1989 Jul;171(7):3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986 Oct 20;191(4):615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Imber R., Bächinger H., Bickle T. A. Purification and characterisation of a small DNA-binding protein, HB, from Bacillus globigii. Eur J Biochem. 1982 Mar 1;122(3):627–632. doi: 10.1111/j.1432-1033.1982.tb06485.x. [DOI] [PubMed] [Google Scholar]
- Imber R., Kimura M., Groch N., Heinemann U. DNA-binding properties and primary structure of HB protein from Bacillus globigii. Eur J Biochem. 1987 Jun 15;165(3):547–552. doi: 10.1111/j.1432-1033.1987.tb11474.x. [DOI] [PubMed] [Google Scholar]
- Johnson G. G., Geiduschek E. P. Specificity of the weak binding between the phage SPO1 transcription-inhibitory protein, TF1, and SPO1 DNA. Biochemistry. 1977 Apr 5;16(7):1473–1485. doi: 10.1021/bi00626a036. [DOI] [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Kano Y., Yoshino S., Wada M., Yokoyama K., Nobuhara M., Imamoto F. Molecular cloning and nucleotide sequence of the HU-1 gene of Escherichia coli. Mol Gen Genet. 1985;201(2):360–362. doi: 10.1007/BF00425687. [DOI] [PubMed] [Google Scholar]
- Kimura M., Wilson K. S. On the DNA binding protein II from Bacillus stearothermophilus. II. The amino acid sequence and its relation to those of homologous proteins from other prokaryotes. J Biol Chem. 1983 Mar 25;258(6):4007–4011. [PubMed] [Google Scholar]
- Krause M., Marahiel M. A., von Döhren H., Kleinkauf H. Molecular cloning of an ornithine-activating fragment of the gramicidin S synthetase 2 gene from Bacillus brevis and its expression in Escherichia coli. J Bacteriol. 1985 Jun;162(3):1120–1125. doi: 10.1128/jb.162.3.1120-1125.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laine B., Kmiecik D., Sautiere P., Biserte G., Cohen-Solal M. Complete amino-acid sequences of DNA-binding proteins HU-1 and HU-2 from Escherichia coli. Eur J Biochem. 1980 Feb;103(3):447–461. doi: 10.1111/j.1432-1033.1980.tb05968.x. [DOI] [PubMed] [Google Scholar]
- Marsh M., Hillyard D. R. Nucleotide sequence of the HU-1 gene of Salmonella typhimurium. Nucleic Acids Res. 1988 Jul 25;16(14B):7196–7196. doi: 10.1093/nar/16.14.7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende L., Timm B., Subramanian R. Primary structures of two homologous ribosome-associated DNA-binding proteins of Escherichia coli. FEBS Lett. 1978 Dec 15;96(2):395–398. doi: 10.1016/0014-5793(78)80446-3. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Nakayama T. Purification of B. subtilis DNA-binding protein related to E. coli HU. Biochem Biophys Res Commun. 1980 Nov 17;97(1):318–324. doi: 10.1016/s0006-291x(80)80170-7. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Wada M., Kano Y., Imamoto F., Okazaki T. DNA replication in Escherichia coli mutants that lack protein HU. J Bacteriol. 1989 Oct;171(10):5672–5679. doi: 10.1128/jb.171.10.5672-5679.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettijohn D. E. Structure and properties of the bacterial nucleoid. Cell. 1982 Oct;30(3):667–669. doi: 10.1016/0092-8674(82)90269-0. [DOI] [PubMed] [Google Scholar]
- Rouvière-Yaniv J., Yaniv M., Germond J. E. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell. 1979 Jun;17(2):265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- Salti V., Le Hégarat F., Hirschbein L. Isolation and characterization of small heat-stable acid-soluble DNA-binding proteins from Bacillus subtilis nucleoids. J Gen Microbiol. 1985 Mar;131(3):581–590. doi: 10.1099/00221287-131-3-581. [DOI] [PubMed] [Google Scholar]
- Schmid M. B. Structure and function of the bacterial chromosome. Trends Biochem Sci. 1988 Apr;13(4):131–135. doi: 10.1016/0968-0004(88)90069-2. [DOI] [PubMed] [Google Scholar]
- Sekimizu K., Bramhill D., Kornberg A. Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J Biol Chem. 1988 May 25;263(15):7124–7130. [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka I., Appelt K., Dijk J., White S. W., Wilson K. S. 3-A resolution structure of a protein with histone-like properties in prokaryotes. Nature. 1984 Aug 2;310(5976):376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Nedospasov S. A., Bakayev V. V., Bakayeva T. G., Georgiev G. P. Histone-like proteins in the purified Escherichia coli deoxyribonucleoprotein. Nucleic Acids Res. 1977 Aug;4(8):2725–2745. doi: 10.1093/nar/4.8.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Kano Y., Ogawa T., Okazaki T., Imamoto F. Construction and characterization of the deletion mutant of hupA and hupB genes in Escherichia coli. J Mol Biol. 1988 Dec 5;204(3):581–591. doi: 10.1016/0022-2836(88)90357-9. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Berger S. L. Screening colonies or plaques with radioactive nucleic acid probes. Methods Enzymol. 1987;152:415–423. doi: 10.1016/0076-6879(87)52048-1. [DOI] [PubMed] [Google Scholar]
- Watanabe F., Stankowski S., Schwarz G. Interaction of the HB protein of Bacillus globigii with nucleic acids. Analysis of the binding to DNA and polynucleotides. Eur J Biochem. 1984 Apr 2;140(1):215–219. doi: 10.1111/j.1432-1033.1984.tb08089.x. [DOI] [PubMed] [Google Scholar]
- Wilson K. S., Vorgias C. E., Tanaka I., White S. W., Kimura M. The thermostability of DNA-binding protein HU from bacilli. Protein Eng. 1990 Oct;4(1):11–22. doi: 10.1093/protein/4.1.11. [DOI] [PubMed] [Google Scholar]